Abstract
Werner syndrome is a rare human disease characterized by the premature onset of aging-associated pathologies, cancer predisposition, and genomic instability. The Werner protein (WRN), which is defective in Werner syndrome (WS) patients, belongs to the RecQ family helicases and interacts with several DNA metabolic proteins, including DNA repair factors and telomere associated proteins. Nonhomologous end-joining (NHEJ) is an important pathway in the repair of DNA double strand breaks (DSBs), and the DNA-PK complex, composed of the heterodimer Ku 70/86 and the DNA-PK catalytic subunit (DNA-PKcs), together with the XRCC4-DNA ligase IV complex (X4L4), are major factors. One of the most prominent protein interactions of WRN is with Ku 70/86, and it is possible that WRN is involved in NHEJ via its associations with Ku 70/86 and DNA-PKcs. This study demonstrates that WRN physically interacts with the major NHEJ factor, X4L4, which stimulates WRN exonuclease but not its helicase activity. The human RecQ helicase, BLM, which possesses only helicase activity, does not bind to X4L4, and its helicase activity is not affected by X4L4. In a DNA end-joining assay, we find that a substrate, which is processed by WRN, is ligated by X4L4, thus further supporting the significance of their functional interaction.
DNA double strand breaks (DSBs1), if left unrepaired, are lethal to most dividing cells. DSBs are generated during normal DNA metabolism or by exposure to DNA damaging agents in the environment. In eukaryotic cells, DNA DSBs are repaired by nonhomologous end-joining (NHEJ) and homologous recombination (HR); however, NHEJ is thought to be the predominant pathway for DSB repair in human somatic cells (1). The DNA-PK complex, which comprises DNA-PKcs and the Ku 70/86 heterodimer, and the X4L4 complex are essential factors for NHEJ and V(D)J recombination (2-4). Recently, Cernunnos/XLF was identified as an XRCC4 interactor and was shown to bind to the X4L4 complex (5, 6). Cells deficient in any of these proteins are deficient in DNA DSB repair and V(D)J recombination (7-11). NHEJ is initiated by tight binding of Ku 70/86 to DNA ends. DNA-PKcs is then recruited to the Ku/DNA complex, the DNA-PKcs kinase is activated, and DNA-PKcs autophosphorylation results in DNA end access. X4L4 is also recruited to the DNA ends and performs ligation. DSBs are mainly produced by ionizing radiation or reactive oxygen species, leaving complicated DNA ends. Some of these are not readily ligatable, and processing is needed before they can be ligated by X4L4. The NHEJ factors that participate in this processing include Artemis nuclease and DNA polymerase μ (pol μ), DNA polymerase λ (pol λ), and TdT, members of the X family of DNA polymerases (12).
The WRN protein, defective in the autosomal recessive disorder Werner syndrome possesses a 3′-5′ exonuclease activity in addition to DNA-dependent ATPase, 3′—5′ DNA helicase, and strand annealing activities. WRN is the only human RecQ family protein that has both helicase and exonuclease activities (13, 14). WS is characterized by the premature onset of aging-related pathologies after adolescence. Individuals with WS frequently experience cancer predisposition at an earlier age than normal individuals. In addition, the cellular phenotype of WS is characterized by genomic instability, variegated translocation mosaicism, and an elevated mutation rate (15).
A role for WRN in NHEJ is supported by the following observations: (1) WRN accumulates at the sites of DSBs (16); (2) Ku binds to WRN with high affinity and stimulates WRN exonuclease activity (17); (3) WRN is a substrate for DNA-PK, and phosphorylation by DNA-PK affects the exonuclease and helicase activities of WRN (18); (4) WRN stimulates flap cleavage by FEN-1, a possible NHEJ end-processing factor (19); (5) WRN-deficient cells are weakly sensitive to γ-irradiation, but less sensitive than cells deficient in known NHEJ factors (20). Therefore, WRN is probably not an essential component of general genome NHEJ, but more likely acts as an accessory protein during NHEJ-mediated DNA repair or may act in NHEJ under certain circumstances.
The BLM protein is also a member of the RecQ family helicases. A defect in the BLM protein results in Bloom syndrome (BS), which is characterized by cancer predisposition and genome instability. Chicken cells lacking BLM show no increased sensitivity to a topoisomerase II inhibitor, which is known to induce DSBs (21). However, genetic interactions between BLM and NHEJ were reported in Drosophila, suggesting an undefined potential functional interplay between BLM and the NHEJ pathway in mammalian cells (22). So et al. suggested the presence of an alternative, DNA ligase IV-independent NHEJ pathway, which was affected by the loss of BLM (23). Several groups have performed in vivo or in vitro end-joining assays in BS cells (24-26). However, the results are contradictory, and a role of BLM in NHEJ pathway in mammalian cells still remains in question.
On the basis of the above observations, we speculated that WRN enzymatic functions, especially its exonuclease activity, may serve as an end-processing nuclease prior to ligation by X4L4. Herein, we have explored the interaction of WRN and BLM with X4L4 and find that WRN but not BLM interacts with the X4L4 complex. WRN binds to XRCC4 itself, a subunit of X4L4. We have also tested the effect of the X4L4 interaction on WRN enzymatic activities and found that the X4L4 complex, but not XRCC4 itself, stimulates WRN exonuclease activity. The complex stimulated neither WRN nor BLM helicase activities. Taken together, we suggest that the interaction between X4L4 and WRN enables WRN exonuclease to serve as a DNA end-processing factor during NHEJ. Consistent with this notion, a DNA end-joining assay shows that X4L4 ligates the DNA ends in the presence of Ku, a stimulator of the ligase activity, upon the removal of a few nucleotides from the noncomplimentary DNA ends by WRN.
EXPERIMENTAL PROCEDURES
Proteins
Recombinant His-tagged wild-type WRN (27), untagged human XRCC4/His-tagged human ligase IV complex (28), and human Ku 70/86 (28) were purified using a baculovirus expression system as described previously. Recombinant N-terminus His-tagged human XRCC4 was expressed in Sf9 insect cells using a baculovirus expression system. Cells were grown to 1.5 × 106 cells/mL and infected with XRCC4 baculovirus, harvested by centrifugation after 96 h, and resuspended in lysis buffer [50 mM Tris-HCl (pH 8.0), 0.5 mM β-mercaptoethanol, and 1% Nonidet P-40] supplemented with protease inhibitors (Roche Molecular Biochemicals). The lysate was cleared by centrifugation and incubated with TALON metal affinity resin (Clontech) for 2 h at 4 °C. The resin was washed with lysis buffer, then washed 3 times with wash buffer [20 mM Tris-HCl (pH 8.0), 0.5 mM β-mercaptoethanol, and 12.5 mM imidazole) containing 500 mM NaCl, and finally washed twice with wash buffer containing 100 mM NaCl. His-tagged XRCC4 was eluted in E buffer [20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5 mM β-mercaptoethanol, and 100 mM imidazole]. Purified protein was dialyzed and concentrated in buffer [20 mM Tris-HCl (pH 8.0) and 50 mM NaCl] on 0.5 mL Vivaspin columns (10 kDa cutoff size). Aliquots of the purified complexes were snap-frozen in liquid nitrogen and stored at −80 °C in the presence of 10% glycerol. The purity of the preparation was verified by SDS–polyacrylamide gel stained with Colloidal Coomassie blue G-250 (Sigma). The protein concentration of XRCC4 was determined by measuring UV absorption at 280 nm using an extinction coefficient of 29,870 M−1 cm−1 estimated from the amino acid sequence (ProtParam). The result was confirmed by comparing Coomassie Blue staining intensity for experimental samples and known BSA standards.
DNA Substrate Preparation
The oligonucleotides 37A (5′-TTT TTT TTT TTT TTT GAG TGT GGT GTA CAT GCA CTA C-3′), 37B (3′-TTT TTT TTT TTT TTT CTC ACA CCA CAT GTA CGT GAT G-5′), 49A (5′-TTT TTT TTT TTT TTT TTA GGG TTA GGG TTA GGG TTA GGG CAT GCA CTA C-3′), 49B (3′-TTT TTT TTT TTT TTT AAT CCC AAT CCC AAT CCC AAT CCC GTA CGT GAT G-5′), 49C (5′-TTT TTT TTT TTT TTT GGT GAT GGT GTA TTG AGT GGG ATG CAT GCA CTA C-3′), 49D (3′-TTT TTT TTT TTT TTT CCA CTA CCA CAT AAC TCA CCC TAC GTA CGT GAT G-5′), 64A (5′-GTA CCA GCT GGG AAT TCC ATA TGA GCG CTG CAG ATG CAC TTG CTC GAT AGA TCT AAC ATG AGC G-3′), 64B (3′-GTC GAC CCT TAA GGT ATA CTC GCG ACG TCT ACG TGA ACG AGC TAT CTA GAT TGT ACT CGC CAT G-5′), 66A (5′-GTA CCA GCT GGG AAT TCC ATA TGA GCG CTG CAG ATG CAC TTG CTC GAT AGA TCT AAC ATG AGC GGT-3′), and 66B (3′-TGG TCG ACC CTT AAG GTA TAC TCG CGA CGT CTA CGT GAA CGA GCT ATC TAG ATT GTA CTC GCC ATG-5′) were from Midland Certified Reagent Co. or Integrated DNA Technologies. The oligonucleotides (37A, 49A, 49C, 64A, and 66A) were 5′ end-labeled using [γ-32P] ATP (Amersham Biosciences) and T4 polynucleotide kinase (New England Biolabs), and the labeled oligonucleotides were annealed to their unlabeled complementary strands (37B, 49B, 49D, 64B, and 66B) at a 1:1 molar ratio, respectively.
Immunoprecipitation (IP)
HeLa nuclear extracts were either purchased from 4C Biotech (Seneffe, Belgium) or prepared as described previously (29). 250 μg of extracts were incubated at 4 °C for 3 h with mouse anti-XRCC4 antibody (genetex, GTX70293), rabbit anti-XRCC4 antibody (Serotec, AHP 387), rabbit polyclonal anti-WRN antibody (Abcam, ab200), mouse control IgG, or rabbit control IgG in IP buffer [100 mM KCl, 30 mM NaCl, 0.05% triton X-100, 2% glycerol, 20 mM Hepes (pH 7.5), 0.1 mM EDTA, 50 μg/mL BSA, 0.5 mM PMSF, and 2 mM DTT], followed by 1 h of incubation with protein A-agarose beads (Santa Cruz Biotechnology, sc-2001). The beads were washed 4 times with IP buffer. The bound fraction was then eluted with SDS-containing buffer and analyzed by SDS–PAGE. Western blot was performed using rabbit anti-WRN antibody, rabbit anti-BLM antibody (Abcam, ab476), rabbit anti-DNA ligase IV antibody (Serotec, AHP554), or mouse XRCC4 antibody.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA was conducted as described previously with modifications (30). Wells in 96 wells plates were coated overnight at 4 °C with WRN or BLM protein in Figure 1D, with X4L4 or XRCC4 protein in Figure 1E, or with BSA as a background control. Wells were washed and incubated with blocking buffer (1× PBS, 0.5% Tween 20, and 3% BSA) at 37 °C for 1 h. After blocking, X4L4 (Figure 1D) or WRN (Figure 1E) was added at the indicated concentration and incubated at 37 °C for 1 h 30 min. Following washing, mouse anti-XRCC4 antibody (Genetex) or rabbit anti-WRN antibody (H-300) (Santa Cruz Biotechnology) was added and incubated at 37 °C for 1 h. Wells were washed, and secondary antibody (goat antimouse or -rabbit IgG-horseradish peroxidase; Santa Cruz Biotechnology) was added and incubated at 37 °C for 1 h. Wells were washed again, and secondary antibody was detected with o-phenylenediamine dihydrochloride (Sigma). Reactions were terminated with 3 M H2SO4. Absorbance was read at 490 nm, and values were corrected for background signal as needed. The dissociation constant (Kd) was calculated on the basis of the fraction of immobilized protein. Data were analyzed using a Hill plot, as described previously (30).
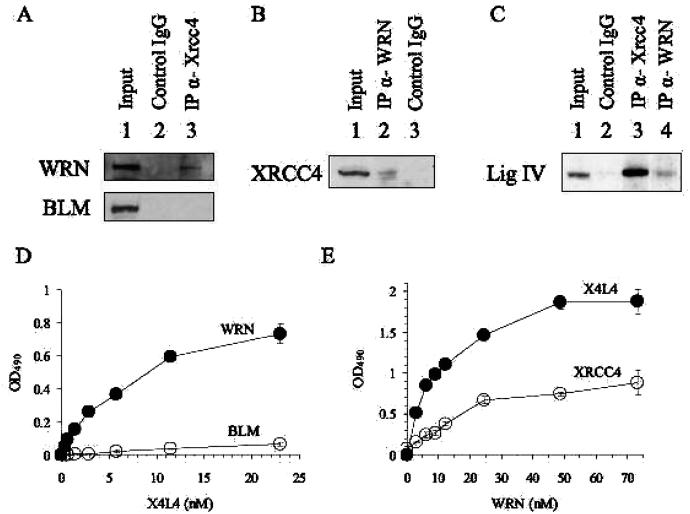
Figure 1.
WRN interacts with X4L4. (A, B, and C) Co-IP assay. Protein complexes were immunoprecipitated from nuclear extracts using control mouse IgG (lane 2 in A) or mouse antibody against XRCC4 (lane 3 in A), using control rabbit IgG (lane 3 in B) or antibody against WRN (lane 2 in B), or using control rabbit IgG (lane 2 in C), rabbit antibody against XRCC4 (lane 3 in C), or rabbit antibody against WRN (lane 4 in C). Proteins immunoprecipitated from HeLa cell nuclear extracts were analyzed by Western blot analysis with rabbit antibody against WRN (upper panel in A) and BLM (lower panel in A), with mouse antibody against XRCC4 (B), or with rabbit antibody against DNA ligase IV (C). 10% of IP input is shown in lane 1 (A, B, and C). (D) ELISA. Wells were coated with 9 nM WRN (●) or BLM (○) and incubated with increasing concentrations of X4L4 (0.36, 0.72, 1.4, 2.9, 5.8, 12, and 23 nM). Bound X4L4 was detected using mouse anti-XRCC4 antibody. (E) ELISA. Wells were coated with 14.4 nM X4L4 (●) or dimeric XRCC4 (○). Increasing amounts of WRN (3.0, 6.1, 9.2, 12, 24, 49, and 73 nM) was added to the wells. Bound WRN was detected using rabbit polyclonal anti-WRN antibody. Absorbance values were corrected for BSA background and plotted against X4L4 (D) or WRN (E). Values and error bars are the mean of at least two independent experiments.
Helicase Assay
Reactions were performed as described previously (31). Briefly, the indicated amount of WRN or BLM and X4L4 were preincubated on ice and incubated with 10 fmol [32P] labeled DNA substrate 37A/37B in 20 μL of standard reaction buffer. Reaction mixtures were electro-phoresed in nondenaturing 12% polyacrylamide gels, and the results were analyzed using a PhosphorImager and ImageQuant software (Molecular Dynamics).
Exonuclease Assay
Reactions were performed as described previously with modifications (31). Ten microliter reactions contained 10 fmol [32P] labeled 49A/49B (Figure 3A) or 49C/49D (Figure 3B) in a buffer containing 40 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 5 mM DTT, 100 μg/mL BSA, and 2 mM ATP. Protein concentrations are indicated in the figure legends. Reaction mixtures were incubated for 15 min at 37 °C and terminated by the addition of an equal volume of formamide-containing stop solution. Heat-denatured products were electrophoresed in a 14% denaturing polyacrylamide gel and were visualized and quantified using a PhosphorImager and ImageQuant software (Molecular Dynamics).
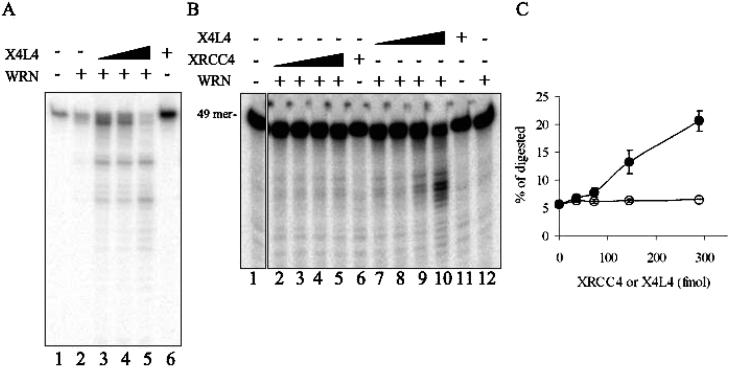
Figure 3.
X4L4 stimulates WRN exonuclease activity. Exonuclease assays. (A) 100 fmol of WRN (lanes 2–5) and increasing amounts of X4L4 (25 fmol in lane 3, 50 fmol in lane 4, and 100 fmol in lanes 5 and 6) were preincubated on ice for 10 min, followed by the addition of 10 fmol of DNA substrate (49A/49B). Reactions were analyzed by denaturing PAGE. (B) Increasing amounts of XRCC4 or X4L4 (36, 72, 144, and 288 fmol) and WRN (36 fmol) were preincubated on ice for 10 min, followed by the addition of 10 fmol of the radiolabeled substrate (49C/49D) in lanes 2–5 and 7–10. XRCC4 (288 fmol) alone, lane 6; X4L4 (288 fmol) alone, lane 11; WRN (36 fmol) alone, lane 12. (C) The amount of the labeled probe excised by WRN was calculated as a fraction of the total DNA for each lane in B.
DNA End-Joining Assay
X4L4, Ku, and WRN were preincubated on ice in a reaction (25 mM Tris-HCl (pH 8.0), 100 mM NaCl, 50 μg/mL BSA, 1 mM DTT, 5 mM MgCl2, 100 uM ATP, and 10% polyethylene glycol). Ten nanomolar [32P] labeled DNA substrate 66A/66B or 64A/64B was added to the reaction and incubated at 37 °C for 15 min. Reactions were terminated by adding two volumes of formamide loading dye. Reaction products were analyzed using 8% denaturing polyacrylamide gel electrophoresis. Products were visualized using PhosphoImager and ImageQuant software (Molecular Dynamics).
RESULTS
WRN Interacts with X4L4 in Vivo and in Vitro
If WRN cooperates with X4L4 to promote the NHEJ-mediated repair of DNA DSBs, it is possible that WRN and X4L4 interact physically in vivo. This was tested by analyzing proteins that Co-IP from HeLa nuclear extracts with the antibody to XRCC4 (Figure 1A, lane 3). The results demonstrated complex formation between WRN and XRCC4. The interaction among BLM, another member of the RecQ helicase family, and XRCC4 was also tested, and no interaction was detected. The reverse Co-IP using an antibody against WRN confirmed the interaction (Figure 1B). Moreover, we also tested the interaction between WRN and DNA ligase IV (Figure 1C). The results demonstrated a tight complex between ligase IV and XRCC4, as reported previously (32, 33). We also detected a less stable association between ligase IV and WRN. This is the first observation of an association between WRN and XRCC4, or ligase IV. Next we performed Co-IP between WRN and XRCC4 from cells irradiated by X-ray or treated with hydroxyurea (Figure S1, Supporting Information). This showed a clearly detectable interaction, which was not affected after IR or hydroxyurea treatment, suggesting that the interaction is not in response to DSBs or replication fork arrest.
The Co-IP between WRN and XRCC4 or ligase IV detected in Figure 1A, B, and C did not necessarily imply direct binding between WRN and XRCC4 or ligase IV. It is possible that one or more other proteins such as Ku 70/86, which bind to both WRN and ligase IV, mediate the interaction between WRN and ligase IV or XRCC4. We therefore performed ELISA using purified recombinant proteins and demonstrated that the X4L4 complex directly bound to WRN (Figure 1D), suggesting that the complex formation between WRN and ligase IV or XRCC4 demonstrated in Figure 1A, B, and C is not mediated by Ku 70/86. ELISA experiments were also performed using purified BLM, another member of the RecQ helicase family (Figure 1D). These experiments indicated that BLM and X4L4 did not interact physically, at least under conditions that support the binding of X4L4 to WRN.
In order to address whether ligase IV or XRCC4 binds WRN, WRN protein was also incubated with XRCC4. The X4L4 complex contains two subunits of XRCC4 and one subunit of ligase IV; therefore, the concentration of XRCC4 is displayed as a dimer in Figure 1E. This experiment showed that WRN bound directly to XRCC4 (Figure 1E); however, it had higher affinity for X4L4 than for XRCC4. The WRNX4L4 and WRN-XRCC4 interactions were not disrupted by ethidium bromide (data not shown), suggesting that the complex is stable in the absence of DNA. The Kd values for the interaction of WRN with the X4L4 complex and XRCC4 were 7.8 nM and 12.8 nM, respectively. (DNA ligase IV is unstable in the absence of XRCC4, precluding direct assays for an interaction between WRN and the soluble monomeric form of DNA ligase IV.) We conclude that WRN binds more strongly to X4L4 than to XRCC4, suggesting that ligase IV stabilizes the binding between WRN and XRCC4 as it forms the complex with XRCC4.
X4L4 Did Not Affect WRN Helicase Activity
Experiments were then performed to test whether WRN and X4L4 interact functionally. The effects of X4L4 on WRN helicase activity were tested using a DNA substrate containing a 22 bp duplex region and 15 base arms, a preferred substrate for WRN (Figure 2) (31). BLM helicase, which does not bind to X4L4, was also tested. Some models suggest that WRN helicase participates in NHEJ to unwind the strands after Ku binding to search for microhomology (34). WRN itself unwound the substrate as reported previously (Figure 2, lane 2) (35). WRN-catalyzed unwinding was not affected by X4L4 (Figure 2, lanes 3–5). BLM helicase activity was also not affected by X4L4 (Figure 2, lanes 9–11).
Figure 2.
X4L4 does not stimulate WRN helicase. Helicase assays. WRN (lanes 2–5) or BLM (lanes 8–11) and increasing amount of X4L4 (lanes 3–6 and 9–12) were preincubated on ice. DNA substrate (37A/37B) was added to the proteins and incubated. Products were analyzed by nondenaturing PAGE.
X4L4 Stimulates WRN Exonuclease Activity
We next tested the effect of X4L4 on WRN exonuclease activity using a DNA substrate containing a 34 bp duplex region and 15 base arms (referred to as a Y-substrate), a preferred substrate for WRN helicase and exonuclease (31). Slightly shorter DNA products were detected with WRN alone, showing exonuclease activity of WRN on the substrate (Figure 3A, lane 2). This exonuclease activity of WRN was stimulated by the addition of X4L4 in a concentration-dependent manner (Figure 3A, lanes 3–5).
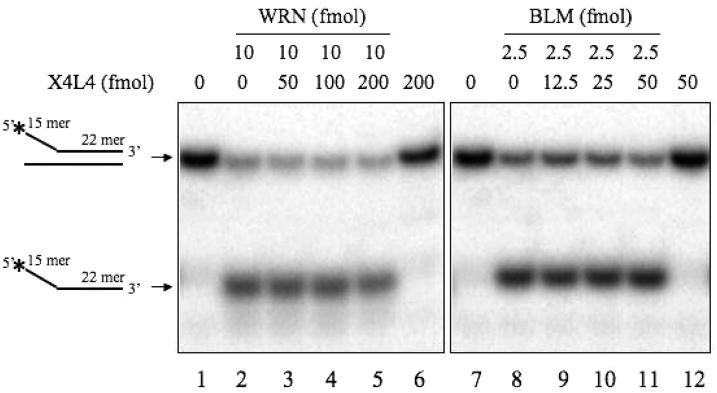
To address whether XRCC4 alone can stimulate WRN exonuclease activity, this activity was also measured on the Y-substrate in the presence of XRCC4 and X4L4 (Figure 3B and C). X4L4 significantly stimulated WRN exonuclease at a 4:1 stoichiometry, and at an 8:1 ratio, it stimulated WRN exonuclease 4-fold (Figure 3B, lanes 9 and 10, and Figure 3C). In contrast, XRCC4 did not stimulate WRN exonuclease activity. Although XRCC4 itself directly binds to WRN (Figure 1E), this interaction alone does not stimulate WRN exonuclease. The association with the X4L4 complex is required for the stimulation of WRN exonuclease. To address the substrate specificity for the stimulation of WRN by X4L4, we utilized a wide variety of double stranded DNA substrates, such as 3′-recessed ends, blunt ends (21, 34, 80, or 150 bp in length), 3′ overhang ends, or NHEJ intermediates (Figure S2, Supporting Information and data not shown). However, there was no stimulation of WRN exonuclease by X4L4 on those substrates. Together, these data suggest that the X4L4 complex possibly induces a conformational change of WRN, which promotes its exonuclease activity on Y-substrates.
Ku also stimulates WRN exonuclease activity on Y-substrates. To investigate the effect of Ku on X4L4 stimulation of WRN exonuclease, we examined WRN exonuclease activity in the presence of X4L4 and Ku (Figure S3, Supporting Information). The pattern of the digested products by WRN in the presence of X4L4 was different from that of WRN with Ku alone, and the effect of Ku on WRN exonuclease activity was dominant when both Ku and X4L4 were present in the WRN exonuclease assay. We also tested the effect of another NHEJ factor, DNA-PKcs (Figure S3, Supporting Information). DNA-PKcs inhibited WRN exonuclease activity on Y-substrates, and the X4L4 stimulation of WRN exonuclease was diminished in the presence of DNA-PKcs. These results suggested that each of these NHEJ factors affect WRN exonuclease activity and that the effect of X4L4 was less prominent than that of Ku and DNA-PKcs.
Specificity of X4L4 Stimulation on WRN Exonuclease Activity
To determine the specificity of the functional interaction between X4L4 and WRN exonuclease, we performed exonuclease assays with some other 3′ to 5′ exonucleases, DNA polymerase I large subunit (Klenow), and E.coli exonuclease III using the same DNA substrate that we had used for the WRN exonuclease assay (Figure S4, Supporting Information). Increasing amounts of Klenow and exonuclease III were used in the exonuclease assays (Figure S4A, Supporting Information), and they were active on the substrate. The effect of X4L4 was tested using the intermediate amount of Klenow and exonuclease III to be able to detect potential stimulation or inhibition by X4L4 if present. Neither Klenow nor exonuclease III were stimulated by X4L4 (Figure S4B and C, Supporting Information). These data indicate that the ability of X4L4 to stimulate 3′ to 5′ exonuclease activity may not be a general phenomenon but may be limited to and specific for the WRN 3′ to 5′ exonuclease.
WRN Produces a Substrate for X4L4 from DNA with Noncohesive Ends
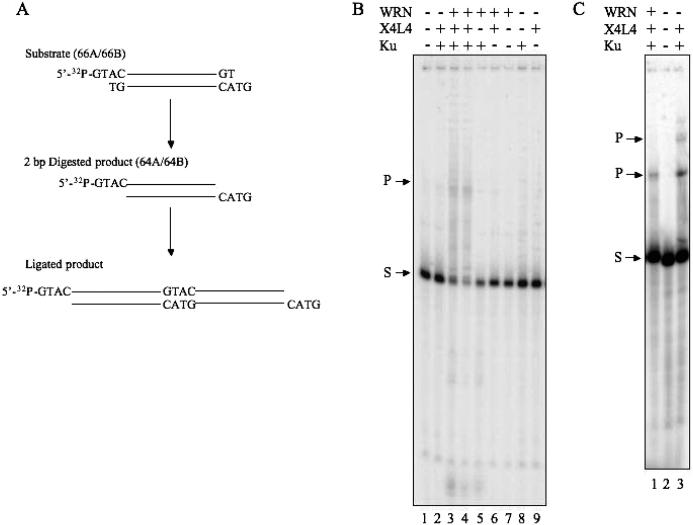
DNA end-processing factors facilitate the generation of ligatable substrates containing single-stranded tails for X4L4 during NHEJ. To test whether WRN contributes as an end-processing factor to NHEJ, we used DNA with noncohesive ends containing a complementary sequence (Figure 4). If WRN can excise the two nucleotides at the end of the substrate and produce ligatable DNA ends, X4L4 can then ligate the substrate as represented in the model in Figure 4A. We preincubated the substrate with Ku 70/86 prior to X4L4, followed by the addition of WRN to start the reaction. The presence of Ku 70/86 was not sufficient to provoke X4L4 ligation on this substrate, although X4L4 plus Ku could ligate several fully incompatible DNA ends that do not share even 1 bp of terminal microhomology (Figure 4B, lane 2) (36). There was no activity of WRN exonuclease on the substrate, either alone or in the presence of X4L4 (Figure 4B, lanes 6 and 7), although we have shown that X4L4 stimulates WRN exonuclease activity on another type of substrate (Figure 3). Ku stimulates WRN exonuclease activity on a wide variety of DNA substrates and enables WRN to act on substrates on which it has no exonuclease activity by itself (Figure 4, lanes 5 and 8) (37). In the presence of both WRN and Ku 70/86, X4L4-catalyzed ligation was observed (Figure 4B, lanes 3 and 4). To examine whether WRN produced substrates that have complementary sequence at the ends as depicted in the middle of Figure 4A, we synthesized DNA substrates which have those ends, and the substrates were incubated with Ku and X4L4 (Figure 4C, lane 3). The products from 66A/66B by WRN, Ku, and X4L4 were migrated similar to the one from 64A/64B by Ku and X4L4 (Figure 4C, lanes 1 and 3). These results suggested that WRN removed two nucleotides from DNA ends to reveal cDNA ends by its exonuclease activity. This is the first article demonstrating that WRN can function as an NHEJ end-processing factor.
Figure 4.
WRN generates ligatable DNA ends. (A) Schematic representation of the reaction is indicated. (B) Ku (100 fmol in lanes 2–5 and 8), X4L4 (400 fmol in lanes 2–4, 6 and 9), and WRN (200 fmol in lane 3, and 400 fmol in lanes 4–7) were preincubated. The DNA substrate (66A/66B) was added to the reaction and incubated. Reaction products were analyzed by denaturing PAGE. (C) Ku (200 fmol in lanes 1 and 3), X4L4 (400 fmol in lanes 1 and 3), and WRN (200 fmol in lane 1) were preincubated. The DNA substrates (66A/66B in lanes 1 and 2, and 64A/64B in lane 3) were added to the reaction and incubated. S indicates substrates, and P indicates ligated products in B and C.
DISCUSSION
This is the first demonstration of a physical and functional interaction between WRN and a key protein complex in NHEJ, X4L4. This complex stimulates the exonuclease activity of WRN on Y-substrates that mimic replication structures (Figure 3). WRN does not affect the ligase activity of X4L4 on substrates with 3′ and 5′ overhangs, cohesive ends, or blunt ends (data not shown). However, the physical and functional interaction between WRN and X4L4 implies that WRN may function to process certain types of DNA ends for NHEJ when breaks occur in a specific context. Indeed, we were able to show ligated products from substrates with incompatible DNA ends using X4L4 and Ku in the presence of WRN.
To search for this cooperative function between WRN, Ku, and X4L4 in vivo, we used DNA end-joining assays with extracts from WRN siRNA knockdown cells (Figure S5, Supporting Information). We detected no deficiency in end-joining activity in WRN knockdown cells compared to control cells using plasmid DNAs with compatible 5′ overhang ends. We have considered several possibilities to explain these observations. (i) It is possible that WRN may compete with other nucleases. Perhaps, in the absence of WRN, other nucleases can access DNA ends and complement WRN function. (ii) There appears to be backup pathways of NHEJ, dependent on DNA ligase III and independent of ligase IV (38). It is possible that such a backup pathway can overcome the lack of WRN. (iii) It is possible that the NHEJ reaction involving WRN takes place under specific cellular circumstances. Such events may not be detected by a simple plasmid end-joining assay. Perry et al. reported elevated microhomology-mediated repair in Werner cells by transfecting linear blunt end plasmid DNA-containing terminal direct repeats of 6 base pairs (39).
We have performed WRN exonuclease assays in the presence of X4L4 on a wide variety of substrates. Results indicate that purified X4L4 stimulates WRN exonuclease specifically on Y-substrates but not on substrates such as recessed or blunt-end DNA molecules, or NHEJ intermediates (Figure 3 and Figure S2, Supporting Information). This specificity for Y-substrates, which resemble DNA replication intermediates, suggests that WRN may participate in the repair of DSBs proximal to replication forks. Taken together, WRN may participate in the repair of DSBs in S phase. In addition, WRN interacts with a number of proteins involved in DNA replication, including replication protein A (RPA), FEN1, proliferating cell nuclear antigen (PCNA), and DNA polymerase δ (19, 40-43). Recently, it has been reported that the recruitment of X4L4 by DNA-PK to DSBs prevents the formation of long single stranded DNA ends at DSBs during the S phase. WRN might occupy the end of DSBs through its interaction with X4L4 and protect them from being digested by other robust nucleases.
WRN and BLM share several protein partners such as RPA and PCNA, which are related to DNA replication. However, the results presented here show that X4L4 binds to WRN but not to BLM (Figure 1A). It suggests that the function of WRN in NHEJ may be specific to WRN exonuclease. The observation that BLM does not bind to X4L4 is consistent with the data that BLM acts on the repair of DSBs through the DNA ligase IV-independent end-joining pathway, suggesting that BLM affects end-joining activity by participating in an alternative end-joining pathway and not in the classical end-joining pathway through the interaction with X4L4 (23).
Previous studies demonstrated a physical association of pol μ with X4L4 (44). It was also shown that the X family of DNA polymerases including pol μ are involved in NHEJ (45). It was also reported that pol μ and X4L4 form a complex with DNA in the presence of Ku. However, in the absence of Ku, pol μ and X4L4 together do not form a complex with DNA (44). We found that the interaction between WRN and ligase IV was much weaker than that of XRCC4 and ligase IV, and that WRN and X4L4 do not form a complex with DNA (Figure 1C and data not shown). These results suggest that even if complex formation with X4L4 is weak, WRN may function together with X4L4. The interaction between NHEJ polymerases and X4L4 suggests that this complex formation can couple the catalytic activities of these factors and may guard against unproductive polymerization during NHEJ (44, 46). The same may be the situation for the association of WRN with X4L4. The WRN-X4L4 complex may allow ligation by X4L4 to occur immediately after WRN digests minimum DNA ends required to form a ligation substrate. X4L4 may be a key protein for controlling both end-processing and polymerization during end-joining. It is possible that X4L4 senses whether the DNA ends need to be polymerized or digested and selects WRN or a DNA polymerase as a partner. WRN may edit DNA if there is misincorporation of nucleotides by NHEJ polymerases. It is of interest to know whether an X4L4-WRN-DNA polymerase complex exists. The NHEJ polymerases belong to the X-family of DNA polymerases. We have previously shown that WRN can assist one of the X-family of DNA polymerases, DNA polymerase β (pol β), as a proof reader for mismatches (47). When a primer terminus contains a mismatched nucleotide, pol β is unable to extend the primer. The addition of WRN to this reaction can enable pol β to synthesize DNA.
In conclusion, we demonstrate a new protein interaction for WRN with X4L4. This implies that WRN interacts with three of the core NHEJ factors in functional interactions and suggests that WRN plays a role in NHEJ. We could not detect this function in vivo using cell extracts, and also, WS cells are only partially sensitive to IR. We speculate that the role of WRN in NHEJ is particularly important under specific cellular conditions, which are yet to be defined.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Dr. Ian D. Hickson for purified BLM. We thank Drs. Jason W. Aulds and Wen-Hsing Cheng for their critical reading of this manuscript.
Footnotes
This work was supported in part by the Intramural Research Program of the NIH, National Institute on Aging, as well as NIH grant CA84442 (to D.A.R). The work in A.V.'s laboratory is supported by a grant from the Human Frontier Science Program, by an FIRB grant of MIUR (Ministero dell'Istruzione dell'Universita' e della Ricerca), and by grant no. 02.00648.ST97 from Consiglio Nazionale delle Ricerche, Rome. R.K. was supported by a Research Fellowship from Toyobo Biotechnology Foundation (Japan).
Abbreviations: WRN, Werner protein; WS, Werner syndrome; NHEJ, nonhomologous end-joining; DSBs, double strand breaks; DNA-PKcs, DNA-PK catalytic subunit; X4L4, XRCC4-ligase IV; HR, homologous recombination; pol μ, DNA polymerase μ; pol λ, DNA polymerase λ; BS, Bloom syndrome; IP, immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; RQC, RecQ conserved; pol β, DNA polymerase β; RPA, replication protein A; PCNA, proliferating cell nuclear antigen.
SUPPORTING INFORMATION AVAILABLE
Materials and methods for supplementary data, references, interaction between WRN and XRCC4, X4L4 and WRN exonuclease activity, Ku and WRN exonuclease activity, X4L4 and Klenow and exonuclease III, WRN knockdown cells and end-joining of DNA, and oligonucleotide sequences. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair. 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair. 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat. Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 5.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Buck D, Moshous D, de CR, Ma Y, le DF, Cavazzana-Calvo M, Fischer A, Casanova JL, Lieber MR, de Villartay JP. Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur. J. Immunol. 2006;36:224–235. doi: 10.1002/eji.200535401. [DOI] [PubMed] [Google Scholar]
- 7.Kurimasa A, Ouyang H, Dong LJ, Wang S, Li X, Cordon-Cardo C, Chen DJ, Li GC. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang H, Nussenzweig A, Kurimasa A, Soares VC, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M, Iliakis G, Chen DJ, Li GC. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J. Exp. Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nussenzweig A, Chen C, da CS,V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 11.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 13.Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′—>5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin GM. Genetics and aging; the Werner syndrome as a segmental progeroid syndrome. Adv. Exp. Med. Biol. 1985;190:161–170. doi: 10.1007/978-1-4684-7853-2_5. [DOI] [PubMed] [Google Scholar]
- 16.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 17.Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 18.Karmakar P, Piotrowski J, Brosh RM, Jr., Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J. Biol. Chem. 2002;277:18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- 19.Brosh RM, von KC, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Seki M, Narita Y, Sonoda E, Takeda S, Yamada K, Masuko T, Katada T, Enomoto T. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000;19:3428–3435. doi: 10.1093/emboj/19.13.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusano K, Johnson-Schlitz DM, Engels WR. Sterility of Drosophila with mutations in the Bloom syndrome gene-complementation by Ku70. Science. 2001;291:2600–2602. doi: 10.1126/science.291.5513.2600. [DOI] [PubMed] [Google Scholar]
- 23.So S, Adachi N, Lieber MR, Koyama H. Genetic interactions between BLM and DNA ligase IV in human cells. J. Biol. Chem. 2004;279:55433–55442. doi: 10.1074/jbc.M409827200. [DOI] [PubMed] [Google Scholar]
- 24.Rünger TM, Kraemer KH. Joining of linear plasmid DNA is reduced and error-prone in Bloom's syndrome cells. EMBO J. 1989;8:1419–1425. doi: 10.1002/j.1460-2075.1989.tb03523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langland G, Elliott J, Li Y, Creaney J, Dixon K, Groden J. The BLM helicase is necessary for normal DNA double-strand break repair. Cancer Res. 2002;62:2766–2770. [PubMed] [Google Scholar]
- 26.Onclercq-Delic R, Calsou P, Delteil C, Salles B, Papadopoulo D, mor-Gueret M. Possible anti-recombinogenic role of Bloom's syndrome helicase in double-strand break processing. Nucleic Acids Res. 2003;31:6272–6282. doi: 10.1093/nar/gkg834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orren DK, Brosh RM, Jr., Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brosh RM, Jr., Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 31.Opresko PL, Laine JP, Brosh RM, Jr., Seidman MM, Bohr VA. Coordinate action of the helicase and 3′ to 5′ exonuclease of Werner syndrome protein. J. Biol. Chem. 2001;276:44677–44687. doi: 10.1074/jbc.M107548200. [DOI] [PubMed] [Google Scholar]
- 32.Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat. Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 33.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 34.Opresko PL, Cheng WH, von KC, Harrigan JA, Bohr VA. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- 35.Brosh RM, Jr., Majumdar A, Desai S, Hickson ID, Bohr VA, Seidman MM. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J. Biol. Chem. 2001;276:3024–3030. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- 36.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Comai L. Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein-Ku70/80 complex. J. Biol. Chem. 2001;276:9896–9902. doi: 10.1074/jbc.M008575200. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Wind-hofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 39.Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat. Struct. Mol. Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 40.Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brosh RM, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 42.Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 43.Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner Syndrome protein and DNA polymerase δ. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase μ (pol μ) with Ku and ligase IV: role for pol μ in end-joining double-strand break repair. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 46.Derose EF, Clarkson MW, Gilmore SA, Galban CJ, Tripathy A, Havener JM, Mueller GA, Ramsden DA, London RE, Lee AL. Solution structure of polymerase μ's BRCT domain reveals an element essential for its role in nonhomologous end joining. Biochemistry. 2007;46:12100–12110. doi: 10.1021/bi7007728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrigan JA, Wilson DM, III, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase β. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.