Abstract
Expression QTL mapping by integrating genome-wide gene expression and genotype data is a promising approach for identifying functional genetic variation, but is hampered by the large number of multiple comparisons inherent to such studies. A novel approach for addressing multiple testing problems in genome-wide family-based association studies is screening candidate markers using heritability or conditional power. We apply these methods for the setting in which microarray gene expression data are used as phenotypes, screening for SNPs near the expressed genes. We perform association analyses for phenotypes using a univariate approach. Simulations were also performed on trios with large numbers of causal SNPs to determine the optimal number of markers to use in a screen. We demonstrate that our family-based screening approach performs well in the analysis of integrative genomic datasets, and that screening using either heritability or conditional power produce similar, though not identical, results.
Keywords: gene expression, association study, SNP, power, heritability, screening, multiple testing
The advent of high-throughput genotyping platforms has revolutionized disease gene mapping by making genome-wide association (GWA) studies both physically and technically feasible. The typical GWA study entails genotyping several hundreds of thousands of genetic variants in large, well-characterized cohorts and subsequently testing these variants for evidence of association with clinical disease or health-related quantitative phenotypes. Initial studies have resulted in the successful mapping of a number of disease susceptibility traits, including body mass index [1] (but see [2; 3; 4; 5] for comments), and susceptibility loci for age-related macular degeneration [6], Crohn’s disease [7] and type 2 diabetes [8]. Despite these early successes and the ever-increasing number of GWA studies being performed, a consensus regarding the optimal statistical approach for analyzing these large datasets remains elusive. Of particular interest is the ability to detect significant association when the functional variants confer only modest individual genetic effect sizes, such as are expected in complex diseases where many genes each contribute only modestly to the complex clinical phenotype.
Furthermore, while some adjustment for the large number of comparisons performed is required in order mitigate against overwhelming type I error, most multiple comparison adjustment methods (such as Bonferroni or False Discovery Rate [9]) further compromise this limited statistical power. One promising approach to address the multiple comparisons dilemma is the family-based screening algorithm described by van Steen et al. (2005), a two-stage procedure whereby all testable SNP are first ranked according to their expected influence on phenotype (measured either as SNP-specific heritability or conditional power - step 1) and then only those markers with the highest expected effect are formally tested for genetic association (step 2). As such, only those markers with the highest pre-test probability of demonstrating association are actually tested, thereby dramatically reducing the number of comparisons performed (Bonferroni adjustment only needs to be done for this smaller number of comparisons). The two stages are statistically independent and are robust to the effects of population stratification [10] and the approach was used to identify a genetic determinant of obesity, a finding which has subsequently been replicated in no fewer than six populations (Herbert et al. 2006; Lyon et al. 2007).
While this family-based screening approach largely addresses the multiple comparison issues of GWA studies, it, like all other multiple comparison corrections, does not address the prevailing issue, namely that most genetic variants influencing common, complex clinical phenotypes likely confer relatively small genetic effects, making their identification via direct association mapping very difficult, even with extremely large sample sizes. For complex diseases, phenotypes are typically indirectly related to underlying genetics. This is so not only because such complex traits largely manifest through the combined interaction of several (if not many) genes with multiple environmental factors, but also because the majority of common genetic variation influencing complex phenotype likely exert their function by subtly altering gene expression (regulatory variation) rather than radically altering the structural integrity of genes (coding variation) [12; 13].
This latter realization has prompted the development of new experimental models for the rapid identification of regulatory variation. One such design that is gaining increasing attention as a practical and feasible approach is that of expression quantitative trait locus (eQTL) mapping. First proposed by Jansen and Nap [14], eQTL mapping aims to identify regulatory polymorphism using an integrative genomics approach whereby both genome-wide gene expression data and genome-wide genotype data are measured in a population, and thousands of GWA studies are performed in which each measured gene expression as a separate quantitative trait. Using this approach, SNPs can then be tested as potential disease-susceptibility variants. Using SNPs is appealing because the direct genetic effects of such variants on gene expression is likely considerably stronger (i.e. explain a larger proportion of the total genetic variance) than the variants’ ultimate effects on the complex clinical trait (which result from interactions of multiple genes and multiple environmental factors); thus the power to map such regulatory variation is greater than that required to map clinical phenotypes. Moreover, if eQTL mapping is limited to genes whose expression has been implicated in the pathobiology of the disease in question, those variants that associated with both gene expression and clinical phenotype are more likely to be truly functional (or in linkage disequilibrium with functional variants) than SNPs who show some association with clinical phenotype but have no demonstrated regulatory impact.
Proof of concept eQTL studies in both animal models [for example, 15] and humans [16] have demonstrated the potential power of this approach. However, because these studies involve performing repeated GWA studies for thousands of expression traits, the number of multiple comparisons quickly balloons. We suggest that in family-based eQTL association studies, the family-based screening approach may be a powerful approach for analyzing this data. Herein we explore this potential application using both simulated and real data.
The Screening Algorithm
The screening algorithm is based on the method described by van Steen et al. 2005. The algorithm is based on only performing FBATs on only a subset of the markers in the original study so that Bonferroni adjustment only needs to be done for a small number of multiple comparisons. The subset of markers can be chosen based on some criterion such as heritability or conditional power (see methods).
For this paper, association tests are only performed on the k marker-phenotype combinations that satisfy some criterion independent of the FBAT p-values, such as heritability or conditional power [17]. Note that in this setting, the same SNP may be analyzed more than once (if it is close to more than one gene), and likewise the same expressed gene may be used more than once if it is close to several SNPs.
We consider using heritability and conditional power as criteria. These are computed using parental markers and expected offspring phenotype markers given the parental markers (see [18]). Because conditional power and heritability are independent of observed offspring genotypes conditional on the parents, the conditional power and heritability are statistically independent of the FBAT p-value. Therefore the number of tests that need to be accounted for when doing multiple comparisons is the number of marker-phenotype combinations in the screen. Because there are many fewer marker-phenotype combinations surviving the screen than the original number of combinations, it is possible to use a conservative multiple testing correction such as Bonferroni and still obtain genome-wide significance.
The screening algorithm can be outlined as follows:
Choose k, the number of marker-phenotype combinations to be in the screen. E.g. k = 10 or k = 100.
Compute heritability or conditional power estimates for all SNP-gene expression combinations.
Rank (or sort in decreasing order) SNP-gene expression combinations according to either heritability or conditional power.
Compute FBAT p-values for the top k combinations, where k was chosen in step 1.
SNP-gene expression combinations are genome-wide significant if p ≤ α/k, where α is the chosen significance level.
Results
Simulation
We simulated trios to show the power of the screening methodology in a genome-wide setting and to find optimal numbers of marker-phenotype combinations to retain in a screen when there a large number of causal loci, as would be expected when looking for cis-regulatory SNPs. For each individual, we generated 1000 independent SNPs with 10 gene expression values each, resulting in 10,000 marker-phenotype combinations. Of these, 100 of the SNPs were chosen to be causal for one expression level, resulting in 100 out of the 10,000 marker-phenotype associations being true associations. This was done for 200 trios. The genetic effect size a is a function of the heritability and minor allele frequency. Expression levels were drawn from a N(aX, 1.0) distribution, where a is the effect size, and X is the number of minor alleles at the causal SNP. Here a = 0 for phenotypes with no cis-acting SNP. This procedure was repeated 1000 times. The number of SNPs detected based on screening the k SNP-phenotype combinations with the highest conditional power was then recorded for k = 1, …, 10000.
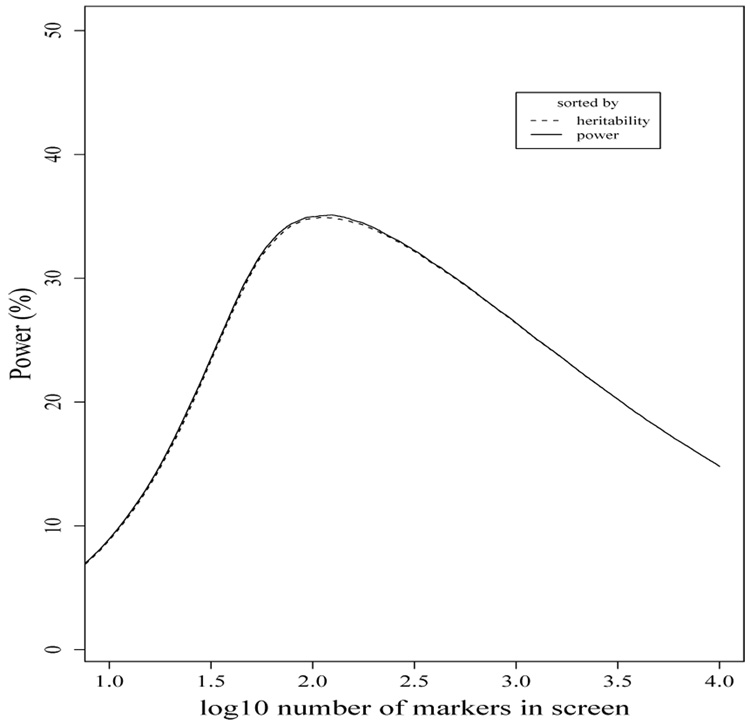
We used the screening method of van Steen, et al. (2005) to rank marker-phenotype combinations, and pick the top k combinations, where k = 1, …, 10000, to show the power of the screening method for every choice of k. The case of k = 10, 000 corresponds to the classical Bonferroni correction. Figure 1 shows the power of the screening method as a function of k using both heritability and power as screening criteria. The results are very similar, with conditional power being slightly more powerful for each k. In spite of the large number of potential tests, we found the optimal choice of k to be much smaller than the number of tests. Using heritability as a criterion, the maximum number of true significant associations found was with k = 108 and k = 110, which tied for an estimated power of 34.9%. The power was over 34% for k = 76 to k = 194, and over 30% for any k between 49 and 500. Thus power decays only gradually from the optimum choice for the number of markers in the screen. Using power as a criterion gave very similar results, with an optimal k = 124, which had an average power of 35.1%. In this case, power was over 34% for k = 73 to k = 204, and power was over 30% for any k between 48 and 505. Although sorting by power had slightly higher estimated power for most (5609 out of 10,000) choices of k, neither criterion was uniformly more powerful.
Figure 1.
Average number (out of 100) of causal SNPs detected as a function of the log10 number of the top k marker in the PBAT screen, k = 1, …, 10, 000, based on 1000 simulations and using either heritability or power as the screening criterion.
We note that optimal choices of k are slightly higher than, but still fairly close to the true number of causal marker-phenotype combinations in the study. The Bonferroni correction method, however, detected an average of 14.8% of the causal SNPs, or approximately 42% of the maximum power of the screening method. This is also lower power than any choice of k > 17 for either screening criterion.
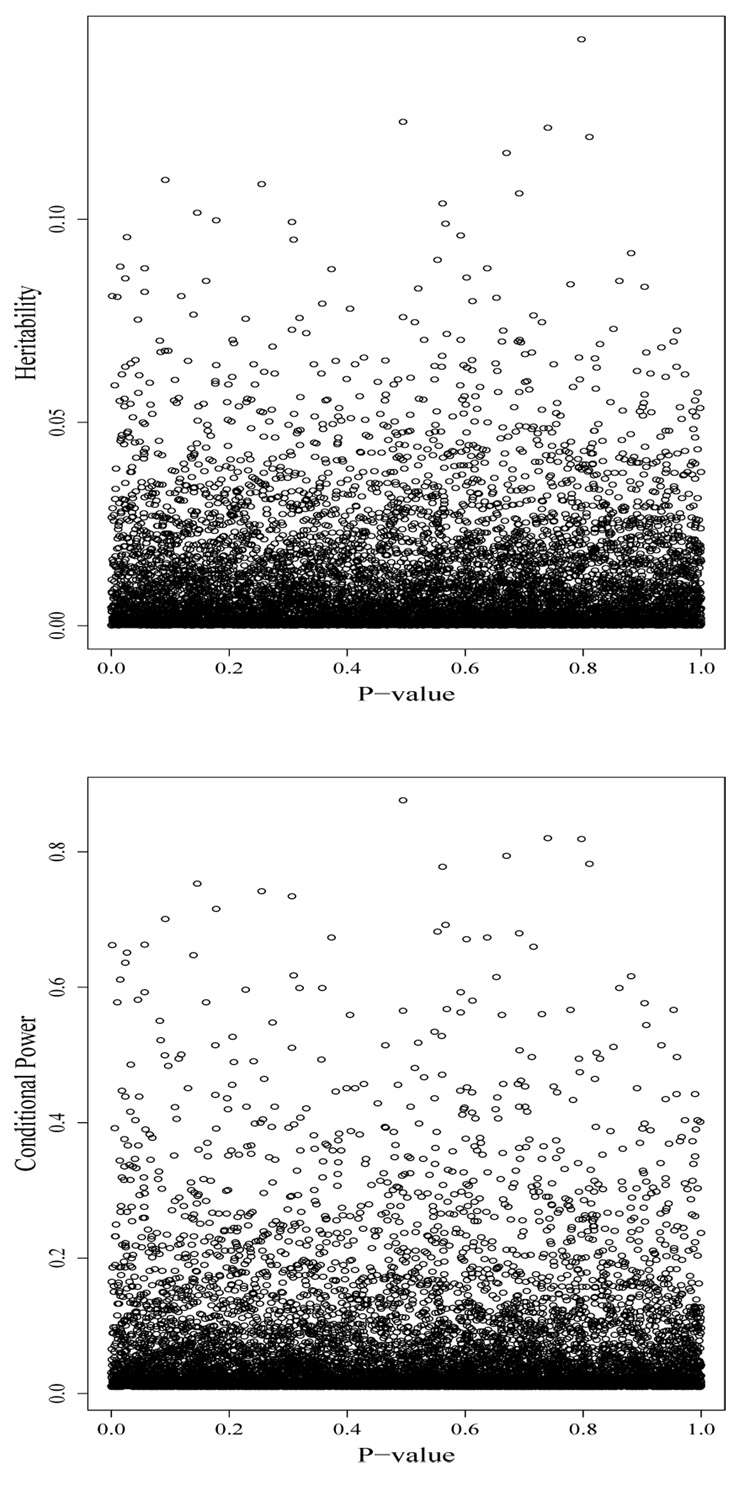
The simulations also illustrate the independence (under the null hypothesis of no association) of the FBAT p-value with heritability and conditional power. To show this, a single run of the simulation is shown with heritability and conditional power plotted against the p-value for the 9,900 marker-phenotype combinations in which the SNP was noncausal, resulting in correlations of −0.005 and 0.002 respectively (Fig. 2).
Figure 2.
Heritability (top) and conditional power (bottom) plotted against p-values for one of the 1000 simulated data sets using noncausal SNPs only.
Data Analysis
We report the candidate marker-phenotype associations that had nominal p-values below 0.05 and that survived screening the top k marker-phenotype combinations based on both the absolute value of heritability (k = 110) and conditional power (k = 124). Choices of k in each analysis are based on the results of the simulation study.
Screening using either heritability or conditional power resulted in 12 marker-phenotype associations with p < .05. There were seven associations that overlapped between the two criteria. For both methods, only markers for which there were at least five informative families were included in the screen; however, this did not affect the screening algorithm when conditional power was used since only those markers with relatively large numbers of informative families were highly powered; however, it did affect the ranks when sorting by heritability.
Based on the simulation study, we used α = 0.05/110 = 0.000455 as our significance level when sorting by heritability and α = 0.05/124 = 0.000403 when sorting by conditional power. There were three genome-wide significant associations found using either criteria. Two of the genome-wide significant associations were for two SNPs (rs178814 and rs178815) located 1226 bp apart that were associated with the same gene (Contig47134_RC) (p = 1.39 × 10−5 and p = 1.36 × 10−5, respectively). These associations were both detected using either heritability or conditional power as the screening method. Using heritability, a significant association between rs2172962 and Contig45657 was found (p = 4.46 × 10−4. Screening by conditional power did not detect this combination, but did result in one significant association not detected using heritability: rs925197 and Contig20565_RC (p = 8.28 × 10−5). Thus each screening criteria was able to find a genome-wide significant association not detected by the other criterion. There was substantial overlap in the SNP-phenotype combinations detected under the two types of screening, with seven associations (including the two that were genome-wide significant) detected under both screening criteria (Table 1 and Table 2). To compare gene expression for genes that survived the screen versus those that did not, we averaged expression levels across all parents (to avoid the nonindependence of children) for each gene. Using the top 118 uniquely occurring genes when sorting by power (six of the 124 top genes occurred twice in the screen), these genes had similar means, medians, ranges, and variances in terms of expression (p > 0.05 in each case). Although these measures did not show dramatic differences, those genes that survived the screen tended to have more SNPs within 1.0 Mb (p = 4.9 × 10−9), with an average of 3.67 SNPs, compared to an average of 40227/14219 = 2.83 SNPs per gene for the entire dataset (Table 3).
Table 1.
Marker-phenotype combinations within the top 100 heritability values where the association had FBAT p-values < 0.05. Rank refers to the heritability within the top 110. Distance refers to the minimum distance of the SNP to either end of the gene.
| Marker | Minor allele frequency | Gene | Chrom | Distance | Informative families | FBAT p-value | Conditional power | Heritability | Rank | in Table 2? |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2172962 | 0.367 | Contig45657 | 1 | 40022 | 7 | 0.000446* | 0.238 | 0.535 | 8 | NO |
| rs178815 | 0.482 | Contig47134_RC | 17 | 212582 | 9 | 0.0000136* | 0.743 | 0.514 | 10 | YES |
| rs178814 | 0.482 | Contig47134_RC | 17 | 213808 | 9 | 0.0000139* | 0.732 | 0.510 | 11 | YES |
| rs1411875 | 0.396 | NM_001081 | 10 | 338648 | 8 | 0.003438 | 0.712 | 0.473 | 23 | YES |
| rs734910 | 0.481 | Contig28617_RC | 19 | 602979 | 9 | 0.019150 | 0.041 | 0.457 | 33 | NO |
| rs940287 | 0.294 | NM_007203 | 9 | 145148 | 10 | 0.021949 | 0.644 | 0.437 | 44 | YES |
| rs2143544 | 0.430 | Contig31391_RC | 20 | 164659 | 9 | 0.025907 | 0.291 | 0.429 | 49 | NO |
| rs1015416 | 0.202 | NM_005024 | 18 | 13362 | 11 | 0.002059 | 0.958 | 0.428 | 50 | YES |
| rs2008734 | 0.489 | NM_016055 | 11 | 132030 | 10 | 0.036777 | 0.618 | 0.403 | 73 | YES |
| rs740951 | 0.295 | Contig35752 | 7 | 268660 | 6 | 0.002635 | 0.491 | 0.395 | 79 | NO |
| rs740672 | 0.253 | AB033102 | 4 | 67855 | 11 | 0.007647 | 0.723 | 0.388 | 84 | YES |
| rs584109 | 0.428 | NM_015368 | 11 | 97619 | 11 | 0.001409 | 0.536 | 0.384 | 92 | NO |
Table 2.
Marker-phenotype combinations within the top 100 conditional power values where the association had FBAT p-values < 0.05. Rank refers to conditional power within the top 124.
| Marker | Minor allele frequency | Gene | Chrom | Distance | Informative families | FBAT p-value | Conditional power | Heritability | Rank | in Table 1? |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1015416 | 0.202 | NM_005024 | 18 | 13362 | 11 | 0.002059 | 0.958 | 0.428 | 2 | YES |
| rs1997034 | 0.201 | Contig27827_RC | 1 | 600645 | 12 | 0.030215 | 0.786 | 0.306 | 38 | NO |
| rs178815 | 0.482 | Contig47134_RC | 17 | 212582 | 9 | 0.0000136* | 0.743 | 0.514 | 43 | YES |
| rs178814 | 0.482 | Contig47134_RC | 17 | 213808 | 9 | 0.0000139* | 0.732 | 0.510 | 46 | YES |
| rs740672 | 0.253 | AB033102 | 4 | 67855 | 11 | 0.007647 | 0.723 | 0.388 | 49 | YES |
| rs2009989 | 0.157 | NM_005024 | 18 | 30768 | 10 | 0.003973 | 0.720 | 0.257 | 51 | NO |
| rs586853 | 0.223 | Contig16931 | 5 | 394577 | 12 | 0.005587 | 0.712 | 0.290 | 54 | NO |
| rs1411875 | 0.396 | NM_001081 | 10 | 338648 | 8 | 0.003438 | 0.712 | 0.473 | 55 | YES |
| rs925197 | 0.216 | Contig20565_RC | 5 | 17139 | 8 | 0.0000828* | 0.710 | 0.268 | 57 | NO |
| rs940287 | 0.294 | NM_007203 | 9 | 145148 | 10 | 0.021849 | 0.642 | 0.309 | 84 | YES |
| rs2008734 | 0.489 | NM_016055 | 11 | 132030 | 10 | 0.036777 | 0.618 | 0.403 | 104 | YES |
| rs1968867 | 0.135 | AL109691 | 15 | 744682 | 10 | 0.006775 | 0.607 | 0.205 | 111 | NO |
Table 3.
Number of Marker-phenotype combinations for the CEPH dataset within a given distance from the gene. The same SNP may occur in more than one combination, and similarly, the same expressed gene may occur in more than one combination.
| Distance | Marker-phenotype combinations | Number of SNPs | Number of Expressed Genes | Genes per SNP |
|---|---|---|---|---|
| 50 kb | 2241 | 1090 | 1495 | 2.06 |
| 100 kb | 4181 | 1462 | 2388 | 2.86 |
| 500 kb | 19913 | 2122 | 8757 | 9.38 |
| 1 Mb | 40227 | 2279 | 14219 | 17.65 |
| 2 Mb | 79009 | 2314 | 19352 | 34.14 |
Discussion
Heritability versus conditional power
An open question in using screening methodology is whether it is better to screen based on heritability or power. In particular, heritability would seem to be a promising criterion because the phenotypes can be much more highly heritable than is typical in genetic association studies. Also, the CEPH dataset used has a small sample number of families and therefore low power. Not surprisingly, sorting by power tended to favor marker-phenotype combinations that had higher numbers of informative families than did sorting by heritability (Table 1 and Table 2). Based on this study, however, we find the performance of the two methods to be very similar in the number of SNPs detected, both in the simulation study and using real data. Since more SNPs can be detected using both approaches than using one alone, we cannot recommend using one criterion exclusively. A possibility to explore would be combining criteria. For example, a method for combining objective criteria for ranking genes in expression studies was developed by [19].
Computational issues
The amount of computation time is linear in the number of marker-phenotype combinations. We note that this problem is highly parallel, with each of the 40,227 associations being computed independently. For this data set, the total computation time was under 24h running PBAT [20] on 20 processors in parallel on a linux cluster. Thus, if parallel processing is available, an entire genome-wide dataset with this type of data can be analyzed quickly by using the strategy of analyzing SNP-phenotype associations where SNPs are within some window of the expressed genes. The method could also be used for extended pedigrees by taking advantage of parallel computing, although computation time can increase significantly compared to nuclear families for some combinations of missing parental and grandparental markers.
Conclusions
In the analysis presented, we limited the number of associations by only considering SNPs on the same chromosome as the gene transcripts which were within 1.0 Mb of the gene. Although this method is useful for looking for possible cis-acting SNPs, the method could be extended to look for trans-acting SNPs by considering markers further from the gene transcripts or even on different chromosomes. This would have the cost of increasing the number of conditional power and heritability estimates to be computed, but this could be accomplished with parallel computing. A strategy for reducing the number of associations to be computed is to use principle components (FBAT-PC; [21]), in which multiple phenotypes are analyzed for each SNP by finding the linear combination of phenotypes which maximizes heritability (the first principle component). This linear combination is then analyzed as a single phenotype, thus greatly reducing the total number of phenotypes (and therefore SNP-phenotype combinations) to be analyzed. Although generally the large number of gene expression values increases the severity of the multiple testing problem in association studies, the methods of this paper demonstrate the feasibility of incorporating integrative genomics data in GWA studies in a family-based setting.
Methods
We used data from 15 CEPH families (IDs 1334, 1340, 1345, 1346, 1349, 1350, 1358, 1362, 1375, 1377, 1408, 1418, 1421, 1424, 1477) also used in Monks et al. (2004). To use only nuclear families we did not include grandparents in the analysis. This resulted in 142 individuals, 112 of whom were offspring. Gene expression data were available for 24 of the 30 parents and 91 of the offspring. Data were available for 23,880 expressed genes and 2322 genotyped SNPs on the 22 autosomal chromosomes. We performed association analysis on the 40,227 SNP-gene expression combinations where the SNP was within 1.0Mb of one end of the gene. This resulted in 2279 SNPs and 14219 expressed genes, with an average of 17.65 genes per SNP (Table 3).
To screen for cis-acting SNPs, we considered all combinations of markers and phenotypes where the SNP was within 1.0 Mb of either end of the expressed gene, resulting in 40,227 marker-phenotype combinations. See Table 3 for the number of combinations as a function of distance from genes. Because of computational difficulties when there were missing markers in both parents and grandparents, we limited our analysis to nuclear families obtained by only considering parents and offspring; i.e., by excluding the grandparental generation. Univariate family-based association analyses were then performed on the marker-phenotype combinations. An additive genetic model was used in all analyses. The dataset was then sorted using two different criteria: heritability and conditional power.
Heritability is defined as the proportion of phenotypic variability that can be explained by genotypic variability [23]. We use SNP-specific heritability. The conditional power is computed from the conditional mean model using the methods in [24]. In the model, the genotype of the offspring is a random variable conditioned on the genotypes of the parents, and the phenotype Yij (e.g., the gene expression level) depends on the expected conditional offspring genotypes:
| (1) |
Here observed genotypes of the jth offspring in the ith family, Xij , have been replaced by their expected values given the parental values. Under an additive inheritance model, Xij is the number of minor alleles for the individual at a locus. If parental values are missing, sufficient statistics for the parental markers can be used instead [25]. The term Zij allows for covariate adjustment.
In each analysis, the top 100 combinations among the 40,227 were retained for statistical testing, i.e. computing FBAT p-values when there is a continuous phenotype, the gene expression value. The FBAT test statistic is
| (2) |
where Yij represents the phenotype (e.g. gene expression) of the jth offspring in family i. Because an additive genetic model is being used, Xij denotes the number of minor alleles for the jth offspring in the ith family. The parameter µ is an offset parameter which is estimated from the data [18]. The FBAT statistic has an approximate standard normal distribution under the null hypothesis of no linkage and no association. For an overview of generalizations of the FBAT statistic, see Laird and Lange (2006).
Acknowledgments
JHD and CL were supported by National Institutes of Health Grant MH59532. BAR was supported by K08 HL074193 and R01 HL086601A from the National Institutes of Health / National Heart Lung and Blood Institute. We thank the anonymous reviewers for comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu XF, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 2.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, Charles MA, Balkau B, Froguel P. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315:187. doi: 10.1126/science.1129402. [DOI] [PubMed] [Google Scholar]
- 3.Loos RJF, Barroso I, O’Rahilly S, Wareham NJ. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315:187. doi: 10.1126/science.1130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosskopf D, Bornhorst A, Rimmbach C, Schwahn C, Kayser A, Krüger A, Tessmann G, Geissler I, Kroemer HK, Völzke H. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315:187. doi: 10.1126/science.1130571. [DOI] [PubMed] [Google Scholar]
- 5.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu XF, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. Comment on “A common genetic variant is associated with adult and childhood obesity. Science. 2007;315:187. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 6.Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, Olson L, Kenealy SJ, Hauser M, Gilbert JR, Pericak-Vanc MA. Functional candidate genes in age-related macular degeneration: Significant association with VEGF, VLDLR, and LRP6. Investigative Ophthalmology & Visual Science. 2006;47:329–335. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 7.Daly MJ, Pearce AV, Fisher LFSA, Latiano A, Prescott NJ, Forbes A, Mansfield J, Sanderson J, Langelier D, Cohen A, Bitton A, Wild G, CM CML, Annese V, Mathew CG, Rioux JD. Association of DLG5 R30Q variant with inflammatory bowel disease. European J. of Human Genetics. 2005;13:835–839. doi: 10.1038/sj.ejhg.5201403. [DOI] [PubMed] [Google Scholar]
- 8.Kovac IP, Havlik RJ, Foley D, Peila R, Hernandez D, Vrieze FW-D, Singleton A, Egan J, Taub D, Rodriguez B, Masaki K, Curb JD, Fujimoto WY, Wilson AF. Linkage and association analyses of type 2 diabetes/impaired glucose metabolism and adiponectin serum levels in Japanese Americans from Hawaii. Diabetes. 2007;56:537–540. doi: 10.2337/db06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Steen K, McQueen MB, Herbert A, Raby B, Lyon H, DeMeo DL, Murphy A, Su J, Datta S, Rosenow C, Christman M, Silverman EK, Laird NM, Weiss ST, Lange C. Genomic screening and replication using the same data set in family-based association testing. Nature Genetics. 2005;37:683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 11.Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, Zhu X, Thorleifsson G, Gunnarsdottir S, Walters GB, Thorsteinsdottir U, Kong A, Gulcher JR, Nguyen TT, Scherag A, Pfeufer A, Meitinger T, Brvnner G, Rief W, Soto-Quiros ME, Avila L, Klanderman B, Raby BA, Silverman EK, Weiss S, Laird N, Ding X, Groop LC, Tuomi T, Isomaa B, Bengtsson K, Butler JL, Cooper R, Fox CS, O’Donnell CJ, Vollmert C, Celedon JC, Wichmann HE, Hebebrand J, Stefansson K, Lange C, Hirschhorn JN. The association of a SNP upstream of INSIG2 with Body Mass Index is reproduced in several but not all cohorts. PLoS Genet. doi: 10.1371/journal.pgen.0030061. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fay JC, McCullough HL, Sniegowski PD, Eisen MB. Population genetic variation in gene expression is associated with phenotypic variation in saccharomyces cerevisiae. Genome Biol. 2004;5:R26. doi: 10.1186/gb-2004-5-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight JC. Regulatory polymorphisms underlying complex disease traits. J. Mol. Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 15.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 16.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:269–270. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 17.Laird NM, Lange C. Family-based designs in the age of large-scale geneassociation studies. Nat. Rev. Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 18.Lange C, Laird NM. On a general class of conditional tests for family-based association studies in genetics: The asymptotic distribution, the conditional power, and optimality considerations. Genet. Epi. 2002;23:165–180. doi: 10.1002/gepi.209. [DOI] [PubMed] [Google Scholar]
- 19.Hero AO, Fleury G. Pareto-optimal methods for gene ranking. J. VLSI Sig. Proc. 2004;38:259–275. [Google Scholar]
- 20.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: Tools for family-based association studies. Am. J. Hum. Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange C, van Steen K, Andrew T, DeMeo DL, Raby B, Murphy A, Silverman EK, MacGregor A, Weiss ST, Laird NM. A family-based association test for repeatedly measured quantitative traits adjusting for unknown environmental and/or polygenic effects. Stat. Appl. Genet. Mol. Biol. 2004;3:17. doi: 10.2202/1544-6115.1067. [DOI] [PubMed] [Google Scholar]
- 22.Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE. Genetic inheritance of gene expression in human cell lines. Am J. Hum. Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. London: Longman; 1997. [Google Scholar]
- 24.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. Using the non-informative families in family-based association tests: a powerful new testing strategy. Am. J. Hum. Genet. 2003;73:801–811. doi: 10.1086/378591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Human Heredity. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]