Abstract
Insulator elements affect gene expression by preventing the spread of heterochromatin and restricting transcriptional enhancers from activation of unrelated promoters. In vertebrates, insulator’s function requires association with the CCCTC-binding factor (CTCF), a protein that recognizes long and diverse nucleotide sequences. While insulators are critical in gene regulation, only a few have been reported. Here, we describe 13,804 CTCF binding sites in potential insulators of the human genome, discovered experimentally in primary human fibroblasts. Most of these sequences are located far from the transcriptional start sites, with their distribution strongly correlated with genes. The majority of them fit to a consensus motif highly conserved and suitable for predicting possible insulators driven by CTCF in other vertebrate genomes. In addition, CTCF localization is largely invariant across different cell types. Our results provide resource for investigating insulator function and possible other general and evolutionarily conserved activities of CTCF sites.
CTCF plays a critical role in transcriptional regulation in vertebrates (for reviews, see (Ohlsson et al., 2001) (Klenova et al., 2002) (Dunn and Davie, 2003)). It was first identified by its ability to bind to a number of dissimilar regulatory sequences in the promoter-proximal regions of the chicken, mouse, and human MYC oncogenes (Filippova et al., 1996; Lobanenkov et al., 1990). CTCF is a ubiquitously expressed nuclear protein with 11 zinc finger (ZF) DNA-binding domain (Filippova et al., 1996; Klenova et al., 1993). It is essential (Fedoriw et al., 2004) and highly conserved from Drosophila to mice and man (Moon et al., 2005). Point-mutations at the distinct DNA-recognition amino acid positions in ZF3 and ZF7 of CTCF have been identified in a variety of cancers selected for LOH at 16q22 where CTCF maps, suggesting its role as candidate tumor-suppressor gene (Filippova et al., 1998; Filippova et al., 2002).
Initial biochemical analyses revealed that CTCF contains two transcription repressor domains, and can act as a transcriptional repressor (Baniahmad et al., 1990; Burcin et al., 1997; Klenova et al., 1993; Lobanenkov et al., 1990). However, others have found that it could also function as a transcriptional activator in a different sequence context (Vostrov and Quitschke, 1997). Recent studies have identified CTCF to be the vertebrate insulator protein (Bell et al., 1999). So far, CTCF remains as the only major protein implicated in establishment of insulators in vertebrates (Felsenfeld et al., 2004), including those involved in regulation of gene imprinting and mono-allelic gene expression (Fedoriw et al., 2004) (Ling et al., 2006), as well as in X-chromosome inactivation and in the escape from X-linked inactivation (Filippova et al., 2005; Lee, 2003).
There has been a great interest in identifying where potential insulators are located in the eukaryotic genome, because knowledge of these elements can help understand how cis-regulatory elements coordinate expression of the target genes. Transcription of every eukaryotic gene begins with the assembly of an RNA polymerase preinitiation complex (PIC) at the promoter (Kadonaga, 2004), a process that is regulated by sequence specific transcription factors and cis-regulatory elements. Genetics studies in Drosophila first identified the importance of insulators in ensuring proper enhancer/promoter interactions (Udvardy et al., 1985). More recent studies have implicated insulators in the establishment of euchromatin/heterochromatin boundaries in vertebrates (Felsenfeld et al., 2004; Gerasimova and Corces, 2001; Jeong and Pfeifer, 2004). In addition, it has been demonstrated that an insulator in the IGF2/H19 locus is critical for the imprinting of the locus (Bell and Felsenfeld, 2000; Hark et al., 2000; Kanduri et al., 2000).
The mechanism of insulator function remains unclear. One model proposes that insulators, by formation of special chromatin structures, compete for enhancer-bound activators, preventing the activation of downstream promoters (Bulger and Groudine, 1999). Alternatively, insulators may facilitate the formation of loops, for example, via attachment of chromosomal regions to the nuclear membrane (Yusufzai et al., 2004), keeping the intermediate regions exposed for only local interactions between enhancers and promoters. Consistent with this model, it was recently shown that CTCF could mediate long-range chromosomal interactions in mammalian cells, providing a possible mechanism by which insulators establish regulatory domains (Kurukuti et al., 2006; Ling et al., 2006; Yusufzai et al., 2004). The extent at which each mechanism plays a role in shaping genome expression remains unresolved. Knowledge of insulators in the genome would provide a much-needed framework for understanding the genome organization and function.
The effort to computationally identify potential insulators in the human genome has been hampered by an incomplete understanding of the DNA recognition sequence of CTCF. Biochemical assays have indicated that the 11-zinc-finger protein can use different combinations of the zinc-finger domains to bind different DNA target sequences (Filippova et al., 1996; Ohlsson et al., 2001). Thus, the CTCF binding sites identified from in vitro protein/DNA interaction assays and a limited number of known insulators exhibit extensive sequence variation and not enough specificity for genome-wide prediction of CTCF binding (Ohlsson et al., 2001). Recently, an attempt has been made to systematically isolate insulators in the mouse genome through chromatin immunoprecipitation followed by cloning and sequencing (Mukhopadhyay et al., 2004). Unfortunately, due to a limited scale of the sequencing effort, only about 200 DNA-fragments with the enhancer-blocking activity, each driven by various CTCF binding sites, have been identified. However, no consensus of CTCF binding motif has been so far reported from this study.
As a first step towards understanding how insulators contribute to gene expression in human cells, we have located the sites of CTCF binding in the human genome using chromatin immunoprecipitation followed by detection with genome-tiling microarrays (Kim et al., 2005b; Kim and Ren, 2006). Our analyses have generated a high-resolution genomic map of CTCF binding, with on average 2.5 genes bounded by a pair of CTCF binding sites. We also identify a clear consensus of CTCF binding motif shared by a majority of the experimentally determined in vivo CTCF binding sites. We show that the sites of CTCF binding sequences in the human genome are highly conserved in other vertebrates, consistent with the widespread and fundamental role of CTCF in cellular function. In addition, we demonstrate that CTCF binding to DNA is largely invariant from cell to cell, with a subset interacting with the protein in a cell type dependent manner. Our results offer a general resource for understanding the role of CTCF in insulator function, gene regulation, and genome organization in human cells.
RESULTS
Genome-wide mapping of CTCF binding sites
Previously, we developed an improved genome-wide location analysis strategy to identify transcription factor binding sites throughput the genome in human cells (Kim et al., 2005b). This method, also known as ChIP-chip, involved the immunoprecipitation of transcription factor bound DNA from formaldehyde crosslinked cells, followed by detection with genome-tiling arrays. To identify CTCF binding sites in the human genome, we performed the same analysis with monoclonal antibodies against CTCF and chromatin extract from the primary human fibroblast, IMR90 cells. The CTCF-bound DNA was identified using a series of 38 arrays containing a total of 14.6 million 50-mer oligonucleotides, evenly positioned every 100 basepairs (bp) along the non-repeat sequence of the human genome. By applying a simple statistical filtering that requires the signals from four consecutive probes to be above a threshold (2.5 times the standard deviation of the average log ratios), we identified an initial list of 15, 221 genomic regions bound by CTCF (Figure 1a,b). To verify the binding of CTCF to these putative CTCF binding sequences, we designed a new oligonucelotide microarray representing these regions and the surrounding sequences at 100bp resolution. Using this array, we performed ChIP-chip analysis against CTCF with an independent chromatin sample of IMR90 cells and confirmed its binding to 13,804 regions.
Figure 1. Chromosomal distribution of CTCF binding sites.
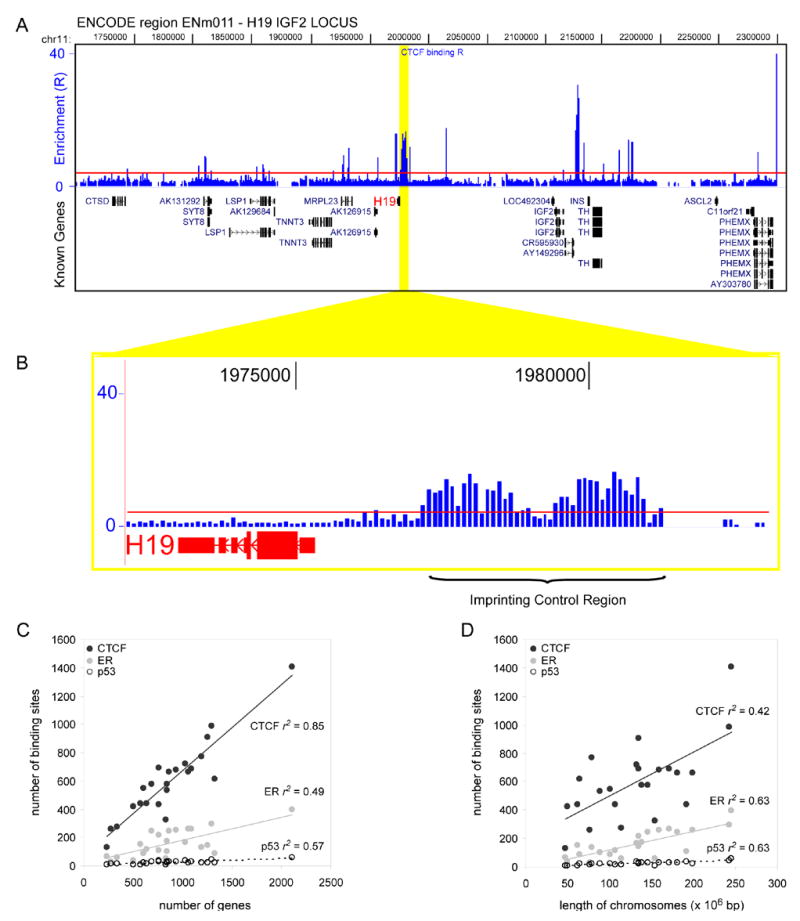
(a) ChIP-chip analysis results for IGF2/H19 locus are shown. (b) A view of the CTCF binding at the H19/IGF2 imprint control region. (c) Correlation analysis of number of CTCF, ER and p53 binding sites with gene number on each chromosome. (d) Correlation analysis of number of CTCF, ER and p53 binding sites with the length of each chromosome.
To assess the accuracy of these in vivo CTCF binding sites, we first randomly selected 84 (Supplemental Table 1) and performed conventional ChIP assays. This analysis validated the binding of CTCF to 80 (95%) tested sites (Supplemental Figure 2a), and suggested a high degree of specificity of our method.
Next, we examined CTCF binding on 60 previously characterized CTCF binding sites and insulators in the human genome, and found that 32 (~53%) were detected by our analysis (Supplemental Table 2). To determine whether the failure to detect CTCF binding at the remaining 28 sites was due to a moderate sensitivity of our method, we performed conventional ChIP assays and detected binding of CTCF to four of these sites (Supplemental Figure 2b, Supplemental Table 3). Since these known CTCF binding sites would be considered false negatives of our method, the sensitivity of our method was estimated to be about 88% (32 out of 36).
Third, we examined a multiple species sequence alignment score (PhastCon) for each CTCF binding site (Siepel et al., 2005) to determine their sequence conservation. A significant fraction (55%, P < 2.2×10−16) of the CTCF binding sites are conserved in vertebrates with a PhastCon score of 0.8 or higher (Supplemental Figure 2c), suggesting that most CTCF binding sites identified in our analysis are likely functional.
Distribution of CTCF binding sites in the genome
To characterize how the CTCF binding sites are distributed along the human genome, we compared their localization to a total of 20,181 well annotated human genes (Kent et al., 2002). We performed correlation analysis of CTCF binding sites with the number of genes or transcripts found on the chromosomes, or with the total nucleotide length of each chromosome (Figure 1c, d, Supplemental Figure 3a). As a control, we examined two enhancer binding proteins whose genomic binding sites were recently determined in human cells: estrogen receptor (ER) (Carroll et al., 2006) and p53 (Wei et al., 2006) (Supplemental Table 4). The results showed that CTCF binding correlates strongly with the number of genes on each chromosome (r2 = 0.85), and the degree of correlation is much higher than both ER and p53. In contrast, CTCF binding only weakly correlates with the chromosomal length (r2 = 0.42), and the degree of correlation is much less than that of the two transcription activator proteins (Carroll et al., 2006) (Figure 1c, d). Based on this analysis, we conclude that the distribution of CTCF binding sites along the genome is closely correlated with genes, and distinct from other known sequence specific transcription factors.
An independent analysis of CTCF localization along each chromosome also confirmed a strong correlation between CTCF binding and gene density. We segmented each chromosome with a sliding 2 Mbp window and calculated the correlation between numbers of CTCF binding sites and genes within each window. In general, the CTCF binding sites correlate strongly with genes, with a correlation coefficient of 0.786. In contrast, the average correlation coefficient between randomly generated genomic sites and genes is only 0.32 (Figure 2a). The degree of correlation between the CTCF binding sites and genes is similar to that between the TAF1 binding sites, mapped previously in the same cells, and genes (correlation coefficient of 0.792). This analysis indicates that CTCF binding is highly restricted to genes, displaying the same property as a general transcription factor. This property of CTCF distribution is consistent with its role at insulators, and suggests a widespread function of CTCF in the genome.
Figure 2. Distribution of CTCF binding sites relative to genes.
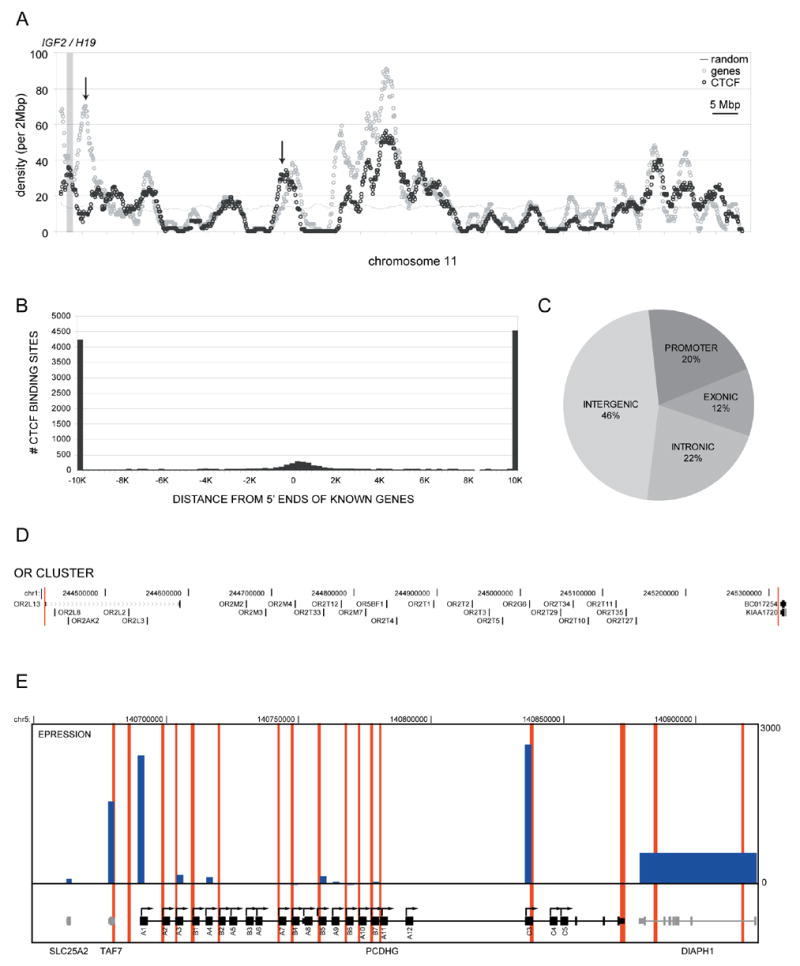
(a) A chromosomal view of the gene and CTCF binding site density of chromosome 11 is shown. Arrows indicate regions within the chromosome where overall correlation of CTCF binding sites and gene number deviate from the average. (b) A histogram summarizing the distribution of CTCF relative to the 5′ end of Known Genes. (c) A pie chart of CTCF binding sites mapping to exons, introns, promoters (within 2.5 Kb of the start sites), and intergenic regions of the genome. (d) Depletion of CTCF binding sites at clusters of related genes. A cluster of olfactory receptor (OR) genes is bounded by a pair of CTCF binding sites, indicated by a long red vertical lines. (e) An example of CTCF binding sites punctuating the alternate promoters in the protocadherin gamma locus. Red vertical lines indicate CTCF binding sites. The blue bars within the top panel show the relative expression of probes that map to the locus. The width of each bar represents the length of each gene.
While the distribution of CTCF binding sites resembles that of a general transcription factor such as TAF1, there are important differences between the two. The majority of TAF1 binding sites (89%) are within close proximity to the known 5′ ends of transcripts; in contrast, CTCF binding sites are generally very far from promoters, with an average distance of 48,000bp (Figure 2b). Nearly half (46%) of the CTCF binding sites are located in the intergenic (46%) regions, consistent with their potential role as insulators. Only about 20% CTCF sites are near transcription start sites. Unexpectedly, a significant number of CTCF binding sites fall within genes, with 22% in the introns and 12% in the exons (Figure 2c). There is no marked enrichment of CTCF binding sites near the polyadenylation sites (Supplemental Figure 3b). The binding of CTCF near promoters to a large extent is negatively correlated with gene activity, as most of these promoters (72%) are not occupied by the general transcription factor TAF1. This observation is consistent with the possibility that CTCF might function as a repressor at these promoters. The significance of CTCF binding within the introns and exons is not clear, but presumably it might be related to its insulator function to block enhancers and silencers present nearby these sequences. Combined together, these results demonstrate that CTCF binding sites are ubiquitous throughout the genome, and display unique distribution that is distinct from enhancers and promoters.
While CTCF binding sites are generally correlated with genes along the entire length of chromosomes, there are isolated regions that deviate from this trend (Figure 2a). Two notable types of loci can be defined: one type of loci is characterized by a relative depletion of CTCF binding sites and the other by an enrichment of CTCF binding sites. We can define CTCF depleted loci as those 2 Mbp windows that exhibit a lower than average density of CTCF binding sites (less than 2 per 2 Mbp, P < 0.05 for most chromosomes, Supplemental Table 5). Likewise, we can define CTCF enriched loci as those 2 Mbp windows that exhibit higher than average CTCF sites density (P < 0.001, Supplemental Table 6). We observe that the CTCF depleted domains tend to include clusters of related gene families and genes that are transcriptionally co-regulated, while CTCF enriched domains often have multiple alternative promoters (81% contain 2 or more alternative promoters). Both cases are consistent with the assumption of CTCF binding sites acting as insulators.
We have characterized these two types of regions further by considering only genes with multiple CTCF binding sites or clusters of genes with no CTCF binding sites. We have defined 13,766 genomic regions that are flanked by a pair of consecutive CTCF binding sites along the genome and named themCTCFpair defineddomains (CPD). About 43% (5969) of CPDs contain at least one gene locus in its entirety, while the remaining CPDs do not contain a complete gene. About 74% of all genes in the genome are surrounded in their entirety by the CTCF binding sites. The remaining genes are either telomeric to CTCF binding site (2.6% of genes) or contain internal CTCF binding sites (23% of genes). On average, about 2.5 genes are found in a CPD. The average size of a CPD is 212,090bp. A significant number of them (189 CPDs, P<0.001) contain 9 or more genes, with the largest one containing as many as 56 genes (P=3.42×10−56). Table 1a lists all CPDs with 15 or more genes, P=2.2×10−8. These CPDs often correspond to large clusters of related genes (Sproul et al., 2005), such as the Olfactory Receptor (OR) gene clusters (Figure 2d), ZNF gene clusters, KRTAP gene clusters (Supplemental Figure 4a), type I interferon (IFN) gene cluster, etc.
Table 1. Two distinct modes of CTCF binding site distribution.
(a) Gene clusters found within CPDs. (b) Genes with multiple CTCF binding sites.
| (a) Gene clusters found within CPDs | ||||
|---|---|---|---|---|
| coordinates | name | description | # genes | |
| chr11:48088265-56214717 | OR | olfactory receptors | 56 | |
| chrX:117723838-128460548 | unrelated | 41 | ||
| chr19:19616300-32957396 | ZNF14 | zinc finger protein | 32 | |
| chr17:36319559-36906400 | KRTAP | keratin associated proteins | 30 | |
| chr11:4616383-5358451 | OR | olfactory receptors | 27 | |
| chr1:244426810-245310724 | OR | olfactory receptors | 23 | |
| chr4:69540367-71551475 | UGT2B;CSN;HTN | UDP glycosyltransferase 2 family members; casein alpha, beta, kappa | 23 | |
| chr11:241380-652375 | IFITM | interferon induced transmembrane proteins | 22 | |
| chrX:139574141-148258030 | SPANX | sperm protein associated with the nucleus SPANX family proteins | 21 | |
| chr1:154908208-155781600 | CD1 | CD1 antigen; olfactory receptors | 20 | |
| chr16:1484561-1993646 | RP | ribosomal proteins | 20 | |
| chr9:122296512-122944407 | OR | olfactory receptors; zinc finger proteins | 20 | |
| chrX:153148757-153849359 | unrelated | 20 | ||
| chr1:149246363-149655393 | LCE | late cornified envelope proteins | 19 | |
| chr19:59681836-60291665 | LILRA, KIR3DL | leukocyte-associated immunoglobulin-like receptors | 19 | |
| chr5:140209093-140679714 | PCDHB | protocadherin beta | 19 | |
| chrX:150199685-151798057 | MAGEA | melanoma antigen family A proteins | 19 | |
| chr12:16404564-21817503 | SLCO | solute carrier organic anion transporter family proteins | 18 | |
| chr19:48981708-49695890 | ZNF | zinc finger proteins | 18 | |
| chr1:1097984-1346875 | TNFRSF | tumor necrosis factor receptors | 17 | |
| chr11:5662785-6228381 | OR | olfactory receptors | 17 | |
| chr11:59278847-60298781 | MS4A | membrane-spanning 4-domains | 17 | |
| chr12:10794287-11530870 | TAS2R | taste receptors | 17 | |
| chr19:62681155-63092088 | ZNF549 | zinc finger proteins | 17 | |
| chr2:27455959-27838284 | unrelated | 17 | ||
| chr21:44755457-45037442 | KRTAP | keratin associated proteins | 17 | |
| chr8:144686322-145048785 | unrelated | 17 | ||
| chrX:100477190-101961011 | ARMCX | armadillo repeat containing proteins | 17 | |
| chr1:165304985-167371954 | SEL | selectins | 16 | |
| chr14:37750277-44792052 | unrelated | 16 | ||
| chr16:54948373-55293378 | MT | metallothionein | 16 | |
| chr19:8625711-9402805 | OR | olfactory receptors | 16 | |
| chr6:26135302-26312482 | HIST | histones | 16 | |
| chr6:27868447-27970998 | HIST | histones | 16 | |
| chr10:73794396-74959208 | unrelated | 15 | ||
| chr14:19003935-19843534 | OR | olfactory receptors | 15 | |
| chr19:41311124-42099100 | ZNF | zinc finger proteins | 15 | |
| chr9:20931328-21385937 | IFN | interferons | 15 | |
| (b) Genes with multiple CTCF binding sites | ||||
| coordinates | gene | description | #CTCF | #starts |
| chr22:20777701-21573524 | IgL lambda | immunoglobulin lambda locus | 34 | 30 |
| chr16:68542310-73012791 | LOC348174 | secretory protein LOC348174 | 29 | 3 |
| chr22:18830549-20228404 | Q8IYP7 | similar to Peripheral-type benzodiazepine receptor-associated protein 1 | 27 | 2 |
| chr1:142300786-145375749 | Q8N4E8 | neuroblastoma breakpoint family, member 15 | 26 | 2 |
| chr7:71912639-74641641 | DKFZP434A0131 | DKFZp434A0131 protein isoform 1 | 25 | 6 |
| chr7:71884863-74669359 | LOC541473 | FKBP6-like | 25 | 2 |
| chr7:71882976-74679539 | Q8N4N6 | tripartite motif-containing 73 | 25 | 2 |
| chr16:14713046-18376428 | NPIP | nuclear pore complex interacting protein | 24 | 3 |
| chr16:14835163-18480935 | Q9H049 | NODAL modulator 2 | 24 | 3 |
| chr7:71912639-73751143 | DKFZP434A0131 | DKFZp434A0131 protein isoform 1 | 20 | 3 |
| chr17:31517173-33607593 | TBC1D3C | TBC1 domain family member 3C | 20 | 4 |
| chr10:46077353-49152919 | Q5RJ30 | FRMPD2 related 1 | 20 | 5 |
| chr15:82659067-83578998 | Q9BXM8 | similar to cis-Golgi matrix protein GM130 | 18 | 2 |
| chr16:14713046-16395314 | NPIP | nuclear pore complex interacting protein | 17 | 6 |
| chr17:31517173-33369298 | TBC1D3C | TBC1 domain family member 3C | 17 | 3 |
| chr1:151647667-151975780 | MUC1 | MUC1 mucin isoform 1 precursor | 16 | 6 |
| chr6:31529509-32034747 | P18615-2 | RD RNA binding protein | 16 | 2 |
| chr7:141638111-142017270 | TCRbeta | T cell receptor beta | 14 | 2 |
| chr11:130745778-131711925 | Q9P121-2 | neurotrimin | 14 | 3 |
| chr5:140690435-140872730 | PCDHGA1 | protocadherin gamma subfamily A | 13 | 46 |
| chr10:78299367-79067583 | KCNMA1 | large conductance calcium-activated potassium | 11 | 1 |
| chr14:21180948-22090938 | TCR alpha/delta | T cell receptor alpha locus | 11 | 14 |
| chr11:44537174-44929010 | TP53I11 | p53-induced protein | 11 | 1 |
| chr12:6304598-6648609 | O15420 | zinc finger protein 384 | 11 | 1 |
In contrast to depletion of CTCF binding sites within clusters of related genes, there is a significant concentration of CTCF binding sites at genes that display extensive alternative promoter usage. Forty nine genes contain significantly more CTCF binding sites (8 or more, P=0.0018, Table 1b) than expected by chance, including such genes as the protocadherin gamma (PCDHG), T cell receptor alpha/delta, beta, gamma loci (TCRα/δ TCRβ TCRγ), and immunoglobulin heavy chain locus (IgH), and light chain kappa and lambda loci (IgLκ IgLλ) (Supplemental Figure 4b). These genes all contain a large number of alternative promoters, most of which are separated from each other by CTCF binding sites (Figure 2e).
In conclusion, CTCF binding sites are distributed along the genome in a non-random fashion that is different from the general transcription factor and sequence specific activators previously characterized. In one aspect, the CTCF binding sites distribution is similar to a general transcription factor in that they both closely track the gene distribution on each chromosome. In comparison, the distribution of previously characterized sequence specific activators is less strongly correlated with the gene density but more significantly with chromosome length. However, unlike general transcription factors, which usually associate with the transcription start sites, the majority of CTCF sites are located remotely from the promoters. Such unique property of CTCF localization is consistent with its putative role as an insulator binding protein.
Most in vivo CTCF binding sites in putative insulators share a specific sequence motif
Previous studies have implicated divergent and variable modes of binding by CTCF and suggested that CTCF recognizes diverse sequences (Ohlsson et al., 2001). Identification of a large number of in vivo CTCF binding sites provides a unique opportunity to better define the in vivo recognition sequence for this DNA binding protein. Using the discriminating matrix enumerator (DME) algorithm (Smith et al., 2005b), we have identified a motif that best distinguishes the CTCF binding sites from their adjacent, control sequences (Figure 3a). This 20-basepair motif is similar to one particular form of CTCF binding consensus (Bell and Felsenfeld, 2000), but refines it significantly in six nucleotide positions (positions 7, 8, 9, 10, 13, and 17, Figure 3a). This motif is present in over 75% of the experimentally identified CTCF binding sites, but in less than 17% of the control, surrounding sequences. It is usually located in the middle of the experimentally identified CTCF binding fragments, as would be expected if they serve as the point of contact by the protein in vivo (Figure 3b).
Figure 3. CTCF binding sites are characterized by a 20-mer motif.
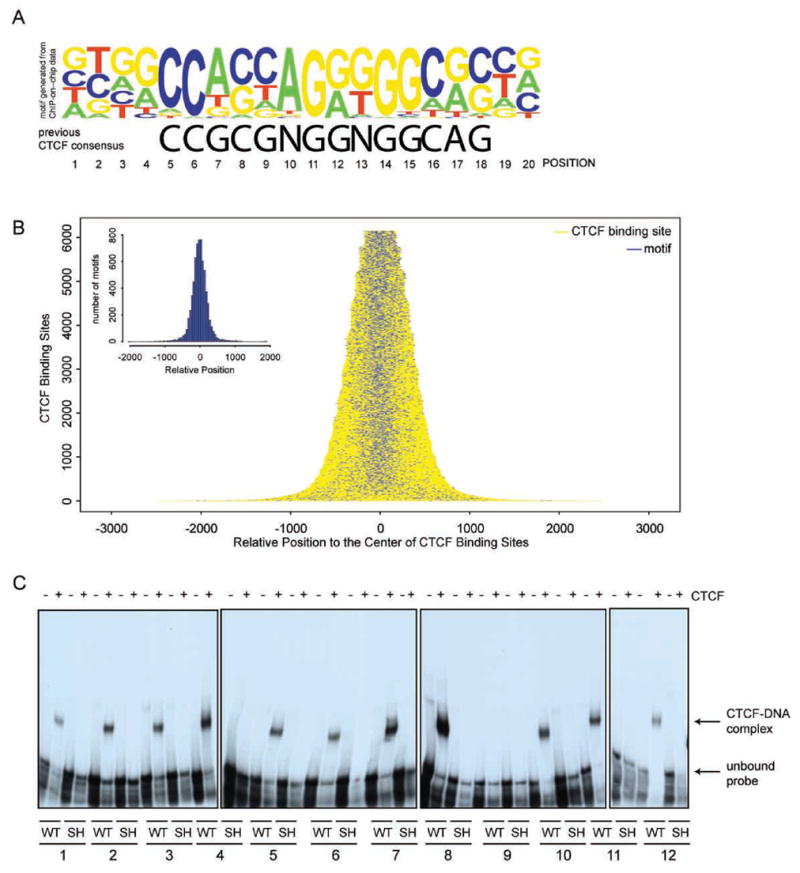
(a) DNA logo (Workman et al., 2005) representing the CTCF binding motif defined from ChIP-on-chip experiment and the previously reported consensus CTCF binding sites (Bell and Felsenfeld, 2000) are shown. Height of each letter represents the relative frequency of occurrence of the nucleotide at each position. (b) Distribution of high scoring motifs within the experimentally defined CTCF binding sites. Yellow horizontal lines represent each CTCF binding site, and short blue lines represent the position of a high scoring 20-mer motif found within the CTCF binding sites. (c) EMSA results for 12 CTCF (WT) and the corresponding shuffled (SH) probes (Supplemental Table 7) showing that 11 of 12 motifs found within the CTCF binding sites are specifically recognized by recombinant CTCF protein.
To test if this motif is indeed the CTCF recognition sequence, we performed electrophoretic mobility shift analysis (EMSA) with 12 randomly selected CTCF binding sites determined above. For each binding site, we designed an 80-mer EMSA probes with the recognizable 20-mer CTCF motif in the middle (Supplemental Table 7). We also designed a control probe by randomly shuffling the 20-mer CTCF motif within each test sequence. Eleven of the 12 probes were confirmed to interact specifically with a recombinant CTCF protein in this assay, while the shuffled probes did not (Figure 3c), indicating that CTCF indeed recognizes the newly identified motif. The one probe that failed to interact with CTCF protein may represent an inferior scoring motif that is more centrally located but may not correspond to the true in vivo CTCF binding site.
From these results, we conclude that under our experimental conditions CTCF binding in vivo appears to be mediated by a class of similar sequences that is well described by a consensus motif. However, a rather significant population of in vivo CTCF binding sites lacks this motif. Additional analysis has failed to identify any significantly overrepresented motifs within these regions. To test whether these sequence bind direct to CTCF in vitro, we have generated consecutive, overlapping DNA fragments to represent two randomly selected CTCF binding sites without the motif (Supplemental Table 8), and performed EMSA. Our results confirm that CTCF can indeed bind to both sequences in vitro (Supplemental Figure 5a, b). Therefore, a fraction of the in vivo CTCF binding sites might have a distinct binding mode and interact with this protein at different sequences. Additional experiments are required to resolve the binding sequence of CTCF at these sites.
The CTCF motif is highly conserved in vertebrates
The CTCF protein displays an unusually high conservation with over 95% amino acid sequence identity within its DNA binding domains among all vertebrate homologs. Moreover, the few amino acid substitutions within the CTCF DNA binding domain do not map to any residues predicted to make direct contacts with the DNA (Pabo et al., 2001). This high degree of sequence conservation supports an evolutionarily conserved function for CTCF, and predicts that the CTCF binding sites should also be conserved in other vertebrate genomes. Consistent with this prediction, the 20-mer motif sequence within each in vivo CTCF binding sites is highly conserved evolutionarily compared to randomly shuffled motifs (Supplemental Figure 6).
Furthermore, we have also searched the entire human genome for the occurrences of CTCF motif, extracted their aligned sequences in other vertebrate genomes where sequence information is available, and asked whether a high scoring CTCF motif is also present in the corresponding homologous sequences. To increase the specificity of computational prediction of CTCF binding sites, we have restricted the bases at position 6, 11, 14, and 16 to the nucleotide that is predominantly present within the experimentally defined CTCF binding sites (see Experimental Procedures for details). A total of 31,905 potential CTCF binding sites are identified in the human genome using this method. Of these sites, 19,271 can be aligned to the mouse genome, and 6,553 contained the CTCF consensus motif as defined above. In contrast, a similar search in the genome with a random matrix of the same length and base composition identifies an average of only 149 conserved occurrences, suggesting that the CTCF binding sequences are highly conserved (P=1.27×10−8, Figure 4a). In addition to the mouse genome, we have also examined the conservation of the predicted human CTCF binding sequences in other vertebrate genomes, finding 8,082 (P = 1.19×10−5), 8,154 (P = 3.84×10−6), 6,362 (P = 1.02×10−8), 263 (P = 5.09×10−5) and 204 (P = 5.48×10−5) to be significantly conserved in dog, cow, rat, chicken and zebrafish genomes, respectively (Figure 4a). In total, 12,799 (out of 31,905) computationally predicted CTCF binding sites in the human genome are conserved in at least one other vertebrate genome (excluding the chimp genome, Figure 4b). We define these highly conserved CTCF recognition sequences as potential CTCF binding sites.
Figure 4. CTCF recognition sites are highly conserved in other vertebrates.

(a) Distribution of CTCF binding motifs found in other vertebrate genomes is compared to the frequency of a randomly shuffled CTCF motif in each genome. (b) Venn diagram of computationally predicted CTCF binding sites in the human genome that are conserved in other vertebrates. The alignments on the right are examples of how each motif with different levels of conservation aligns to the corresponding sequences in other species. (c) EMSA results for 2 CTCF (WT) binding sites predicted in the chicken genomes and the corresponding shuffled (SH) probes (Supplemental Table 9).
The conserved CTCF recognition sequences in the human genome imply that the corresponding motifs in other species may also function as CTCF binding sites. To test this prediction, we have performed EMSA with two predicted CTCF binding sites in the chicken genome (Supplemental Table 9). The results confirm the binding of CTCF to both CTCF sites in vitro (Figure 4c).
Most CTCF binding sites are occupied in a different cell type
To evaluate the variability of CTCF binding in a different cell type, we have performed ChIP-chip analysis to identify CTCF binding sites in a hematopoietic progenitor cell line U937. We have focused our analysis on a set of 44 genomic regions representing 1% sampling of the human genome known as the ENCODE regions (Consortium, 2004; Kim et al., 2005a) (ENCODE arrays). These regions have been semi-randomly selected by the ENCODE consortium as a common platform for genomic research. We have used the previously described genome tiling arrays for this experiment (Kim et al., 2005a). These arrays contain PCR products as probes instead of the oligonucleotides. We have detected 232 sites in U937 cells at the confidence level of P < 0.000001 (Figure 5a, b), which overlap 151 of 225 (67%) CTCF sites detected within the same regions in IMR90 sites (Figure 5b). Less restricted criteria results in a larger degree of overlap (Supplemental Figure 7). This analysis shows that most of the CTCF binding sites detected in IMR90 cells are also occupied in another cell type, indicating that perhaps most CTCF binding sites in the genome are cell type invariant.
Figure 5. Comparison of CTCF binding in two cell types.
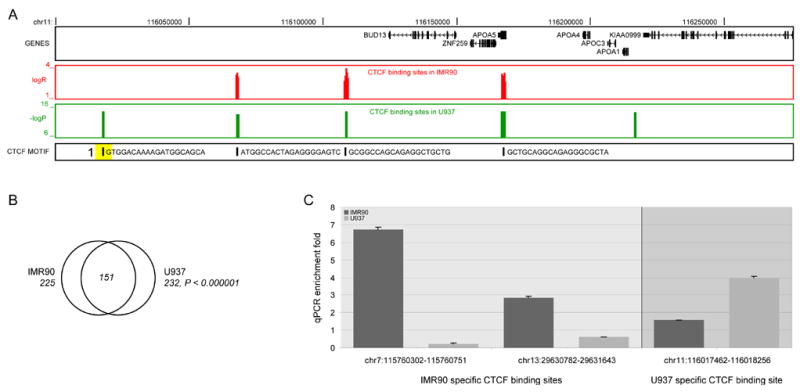
(a) Representative view of CTCF binding in IMR90 and U937 cells within the ENCODE regions. The first panel lists all known genes within the region. The second and third panels show the CTCF binding data within the region for the IMR90 and U937 cells, respectively. The fourth panel shows the predicted CTCF binding sites based on 20-mer motif. (b) A Venn diagram showing the overlap of CTCF binding in IMR90 and U937 cells at the confidence level, P < 0.000001. (c) Validation of three cell type specific sites by quantitative real-time PCR (Supplemental Table 10).
On the other hand, while the overlap between CTCF binding sites in U937 and IMR90 cells does increase with loosened criteria, it does not become 100%. A subset of the CTCF binding sites appears to interact with this protein in a cell type dependent manner. To confirm this, we have performed conventional ChIP assays to test the binding of CTCF to two IMR90-specific sites and one U937-specific site (Supplemental Table 10). The results indicate that the two IMR90 specific CTCF binding sites are indeed associated with the protein in IMR90 cells but not in U937 cells, while a U937 specific CTCF binding site interacts with this protein in an opposite way (Figure 5c). We conclude that a fraction of the CTCF binding sites in the genome may be subject to cell type dependent regulation, although the full extent of this population of CTCF sites remains to be determined.
Evolution of CTCF binding sites in the vertebrate genomes
Since we were able to computationally map CTCF binding sites in other vertebrate genomes, we were interested in knowing how these sites have evolved in different vertebrate species and whether the changes might reflect CTCF function. We have identified 14,352 nucleotide changes within the 12,799 evolutionarily conserved CTCF recognition sequences. Interestingly, the predominant base substitution occurs at the cytosine at position 16, which happens to be the dominant CG di-nucleotide within the consensus sequence (Figure 6). The cytosine to thymidine transition at this position accounts for nearly 17% of all nucleotide changes. One explanation for the unusually high rate of C-to-T substitution at this position is potential DNA methylation at the base (Jones and Baylin, 2002; Rideout et al., 1990), which is consistent with the regulation of CTCF binding by DNA methylation. This observation suggests an intriguing evolutionary model of deriving differential regulation of genes by simply altering CTCF binding in the genome, a process that can be facilitated by environmental and epigenetic factors.
Figure 6. CTCF binding site show a unique nucleotide change during evolution.
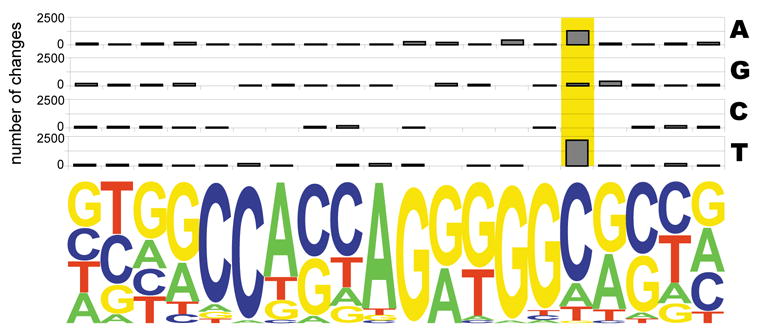
Nucleotide changes observed within the mapped CTCF motifs in all available vertebrate genomes. Distribution of base changes observed in the CTCF binding sites are plotted along the 20-mer motif.
DISCUSSION
In summary, we have generated a high-resolution map of CTCF binding sites in the human genome with unique distribution and sequence features. This map not only confirms most known insulators and CTCF binding sites, but also identifies over 13,000 novel CTCF binding sequences and potential insulators. Nearly 80% of the CTCF binding sites share a consensus motif that is highly conserved during evolution. We have found that CTCF binding sites are largely invariant between cell types. Our results represent a critical step toward comprehensive identification of CTCF-dependent insulators in the human genome.
Unique distribution of CTCF binding sites in the human genome
Unlike sequence specific transcription activators such as ER and p53, CTCF binding sites are ubiquitously and universally present throughout the genome, and its chromosomal distribution is strongly correlated with genes. In this aspect, CTCF resembles the behavior of general transcription factors. Yet, locations of CTCF binding sites are clearly different from that of general transcription factors. Except for a relatively small fraction (20%), the vast majority of CTCF binding occurs at sites remotely from the transcription start sites (Figure 2b). In contrast, nearly 90% of the TAF1 binding sites are located at promoters. This unique distribution of CTCF binding sites in the genome is consistent with the potential role of these sequences as insulators.
About a half of the CTCF binding sites are far away from genes. These distal sites likely define insulators and in many cases coincide with boundaries for gene clusters, such as olfactory receptor gene clusters. A number of genes in the mammalian genome are arranged into clusters, and existence of these clusters has implicated coordinated regulation of expression by shared long range elements such as locus control regions, as it is observed for the Hox and beta-globin gene clusters (Sproul et al., 2005). Recently, a study showed that the OR gene clusters located on separated chromosomes share a single enhancer that selectively interact with only one promoter, resulting in a highly exclusive activation of a single promoter out of about 1,500 others (Lomvardas et al., 2006).
Consistent with this gene segregation property of CTCF, the CTCF binding sites coincide with boundaries of genes that escaped X-inactivation (Filippova et al., 2005). X-inactivation has been shown to involve establishment of heterochromatin on one of the two X chromosomes of the female genome. A recent study shows that X-inactivation is not uniform along the inactive X chromosome (Carrel and Willard, 2005), and identified a number of gene clusters that can escape the chromosome-wide heterochromatin formation. If the CTCF binding sites indeed function as insulators, then one might expect them to segregate the gene clusters that escape inactivation on the X chromosome. Indeed, we have observed several domains on the X chromosome that are surrounded by CTCF binding sites (Supplemental Figure 8).
CTCF binding sites and selective usage of alternative promoters
While nearly half of the CTCF binding sites are found in sequences between genes, an equivalent number of CTCF sites are located within genes. It is not immediately obvious whether these sequences function as insulators. We note that many of them appear to segregate alternative promoters within a single gene and perhaps contribute to alternative promoter usage. Examples of this are provided by the protocadherin gamma locus (PCDHG, Figure 2e), T cell receptor alpha/delta, beta, gamma loci (TCRα/δ TCRβ TCRγ), immunoglobulin heavy chain (IgH), and light chain kappa and lambda loci (IgLκ IgLλ, Supplemental Figure 4b). In each case, CTCF binding segregates transcriptional start sites that display differential activities across tissues. About 52% of the human genes possess multiple promoters. While alternative promoter usage is very common (Carninci et al., 2005; Carninci et al., 2006; Kimura et al., 2006), the mechanisms are not clearly understood. It is generally assumed that different promoters employ distinct regulatory mechanisms to achieve tissue and temporal specific activities. The observation that CTCF binding sites punctuating alternative promoters may suggest involvement of insulator elements in the selection of promoters in distinct cell types.
A consensus motif can explain the majority of CTCF binding sites in possible insulators
One of the surprising findings of our study is that the vast majority of the experimentally identified CTCF binding sites are characterized by a specific 20-mer motif. We demonstrate that this motif is highly conserved in vertebrates and can be used to predict other potential CTCF binding sites in the genome. Furthermore, we show that the newly characterized CTCF consensus sequence specifically interacts with CTCF protein in vitro. Given the overwhelming diversity of sequences that CTCF may recognize in vitro, our finding of a single dominant CTCF binding consensus sequence within the in vivo CTCF binding sites is unexpected.
On the other hand, our results do not rule out the existence of additional CTCF binding motifs that may be recognized by the insulator binding protein along the genome. As a matter of fact, it is important to note 18% of the in vivo binding sites do not contain the newly characterized CTCF binding consensus sequence. When analyzed in vitro, some of these CTCF binding sites can indeed directly interact with CTCF, supporting the existence of different CTCF recognition sequences. Furthermore, quite a number of previously characterized CTCF binding sequences and insulators lack the newly identified motif. It is entirely possible that CTCF may bind to different classes of DNA sequences, either directly or in association with a partner. So far, our search has failed to yield another significant motif among this subset of in vivo CTCF binding sites.
In conclusion, we report here the first high-resolution map of CTCF binding in the human genome, which reveals several new aspects of CTCF function. Our results provide a much-needed resource for further investigation of CTCF’s role in insulator function, imprinting, and long-range chromosomal interactions.
Materials and Methods
A detailed description of the experimental methods and materials can be found at Cell onlinesupplemental materials. All raw and processed data are available at http://licr-renlab.ucsd.edu/download, the UCSC genome browser http://genome.ucsc.edu/, and Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/ (accession #GSE5559). Monoclonal CTCF antibodies used in this study have been characterized and described by E. Pugacheva and co-workers (Pugacheva et al., 2005) and are available from them upon request.
Chromatin immunoprecipitation and microarray experiments
IMR90 and U937 cells were grown and maintained according to the direction from American Type Culture Collection. Cells were harvested and crosslinked with 1% formaldehyde when they reached ~80% confluency on plates. Chromatin immunoprecipitation was performed as described (Kim et al., 2005b), with the use of 50ul of equimolar mixture of nine CTCF monoclonal antibodies, and three distinct array platforms - a whole human genome tiling arrays (Kim et al., 2005b), a condensed array which contained a total of 742,156 oligonucleotides, and PCR product arrays covering the ENCODE regions (Kim et al., 2005a). Microarray data analysis was carried out as described previously (Kim et al., 2005a; Kim et al., 2005b) (see onlinesupplemental materials).
Validation of ChIP-chip data
Quantitative real-time PCR was performed in duplicate with 0.5 ng of CTCF ChIP DNA and unenriched total genomic DNA, with iCycler™ and SYBR green iQ™ SYBR green supermix reagent (Bio-Rad Laboratories). Normalized Ct (ΔCt) values for each sample were calculated by subtracting the Ct value obtained for the unenriched DNA from the Ct value for the CTCF ChIP DNA (ΔCt = Ctctcf − Cttotal). The fold enrichment of the tested promoter sequence in ChIP DNA over the unenriched DNA was then estimated as described previously (Bernstein et al., 2005; Cawley et al., 2004). Primers used for this analysis are listed in the Supplemental Table 1.
Motif analysis
Motif discovery was performed as described in (Smith et al., 2005a; Smith et al., 2005b). All the CTCF binding sites were used as positive sequences and the flanking sequences as negative sequences. The overrepresented sequence motif found in the positive sequences compared to the negative was selected. Using this sequence motif, we generated an initial 20-bp position weight matrix (PWM). This 20-mer PWM was searched against the entire set of CTCF binding sites, and all the motifs found in the binding sites were used to generate the final PWM. The program Storm was then used to search the human genome (hg17) for presence of this motif. The high scoring motifs were selected for the presence key nucleotides C, G, G and C at positions 6, 11, 14 and 16. The resulting CTCF binding sites were then mapped to 14 vertebrate genomes using the available liftOver and genome alignment information available from UCSC genome browser. Each sequence was then scored using Storm and filtered for the critical nucleotides as per the human genome scan.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA were carried out as described (Pugacheva et al., 2005). Briefly, the DNA-binding domain of CTCF (11ZF) and Luciferase (Luc) were in vitro synthesized from pET-11ZF and T7 control plasmids, respectively (Awad et al., 1999; Filippova et al., 1996) by using TnT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI, Cat.# L1170). DNA fragments (Supplemental Table 2) were end-labeled at their 5′ ends using 32P-γ-ATP and T4 polynucleotide kinase. The labeled DNA were gel purified and combined with equal amounts of in vitro–synthesized protein, and incubated for 30 minutes at room temperature followed by electrophoresis on 5% non-denaturing polyacrylamide gels.
Analysis of statistical significance
Statistical significance of the computationally mapped CTCF sites was analyzed by comparing the number of mapped site to the distribution of number of sites mapped using random motif resulting from 1,000 iterations. The random PWM was derived from randomizing the position within the 20-mer CTCF motif. Statistical significance of observed gene clusters within CPDs and multiple CTCF binding sites within a gene was analyzed by calculating the expected probability of each number of observed genes per CPD or each number of CTCF binding sites per gene using Poisson distribution function. Statistical significance of observed evolutionary conservation of CTCF binding sites compared to random sites was analyzed by Mann-Whitney-Wilcoxon test.
Supplementary Material
Acknowledgments
We gratefully acknowledge computer resources made available to us by the Super Computer Center (NBCR award number P41 RR 08605 from NCRR, NIH). This research was supported in part by Ruth L. Kirschstein National Research Service Award F32CA108313 (THK), Ludwig Institute for Cancer Research (BR), U01HG003151 (BR), R33CA105829 (BR), R21CA116365-01 (RDG) and HG001696 (MQZ) from NIH, EIA-0324292 (MQZ) from NSF, and by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases (VVL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Awad TA, Bigler J, Ulmer JE, Hu YJ, Moore JM, Lutz M, Neiman PE, Collins SJ, Renkawitz R, Lobanenkov VV, Filippova GN. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Steiner C, Kohne AC, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505–514. doi: 10.1016/0092-8674(90)90532-j. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Consortium TEP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Dunn KL, Davie JR. The many roles of the transcriptional regulator CTCF. Biochem Cell Biol. 2003;81:161–167. doi: 10.1139/o03-052. [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Burgess-Beusse B, Farrell C, Gaszner M, Ghirlando R, Huang S, Jin C, Litt M, Magdinier F, Mutskov V, et al. Chromatin boundaries and chromatin domains. Cold Spring Harb Symp Quant Biol. 2004;69:245–250. doi: 10.1101/sqb.2004.69.245. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, Doggett NA, Lobanenkov VV. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- Filippova GN, Qi CF, Ulmer JE, Moore JM, Ward MD, Hu YJ, Loukinov DI, Pugacheva EM, Klenova EM, Grundy PE, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu Rev Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Jeong S, Pfeifer K. Shifting insulator boundaries. Nat Genet. 2004;36:1036–1037. doi: 10.1038/ng1004-1036. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Qu C, Van Calcar S, Trinklein ND, Cooper SJ, Luna RM, Glass CK, Rosenfeld MG, Myers RM, Ren B. Direct isolation and identification of promoters in the human genome. Genome Res. 2005a;15:830–839. doi: 10.1101/gr.3430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005b;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Ren B. Genome-Wide Analysis of Protein-DNA Interactions. Annu Rev Genomics Hum Genet. 2006 doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova EM, Morse HC, 3rd, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao ZH, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr Biol. 2003;13:R242–254. doi: 10.1016/s0960-9822(03)00162-3. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- Pugacheva EM, Tiwari VK, Abdullaev Z, Vostrov AA, Flanagan PT, Quitschke WW, Loukinov DI, Ohlsson R, Lobanenkov VV. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum Mol Genet. 2005;14:953–965. doi: 10.1093/hmg/ddi089. [DOI] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Das D, Zhang MQ. Mining ChIP-chip data for transcription factor and cofactor binding sites. Bioinformatics . 2005a;21(Suppl 1):i403–412. doi: 10.1093/bioinformatics/bti1043. [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Zhang MQ. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc Natl Acad Sci U S A. 2005b;102:1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Quitschke WW. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


