Abstract
Mitochondrial dysfunction contributes to the pathophysiology of both acute and chronic neurodegenerative disorders. Quantification of mitochondrial bioenergetic properties generally requires the use of isolated brain mitochondria. However, the involvement of neuronal mitochondrial dysfunction in these disorders is limited by the lack of markers, and therefore isolation procedures, that distinguish neuronal compared with astrocyte mitochondria. To address this and other issues concerning neuronal mitochondria in the CNS, transgenic mice were generated that express a fluorescent protein targeted specifically to neurons. A neuron-specific promoter, CaMKIIα (calcium/calmodulin-dependent kinase IIα) driven tTA (tetracycline transactivator) mice were crossed with TRE (tetracycline responsive element) driven mitochondrial targeted enhanced yellow fluorescent protein (eYFP) mice. Expression of eYFP in the bigenic mouse brain was observed only in neuronal mitochondria of striatum, forebrain, and hippocampus and was enhanced by the removal of the tetracycline analog doxycycline (Dox) in the diet. The respiratory control ratio of synaptic and nonsynaptic mitochondria isolated from eYFP-expressing mice was the same as control mice, suggesting that neuronal mitochondria expressing eYFP maintain normal bioenergetic functions. More importantly, the development of Dox-inducible, neuron targeted mito/eYFP transgenic mice offer a unique in vivo model for delineating the participation of neuronal mitochondria in neuronal survival and death.
Keywords: cortex, hippocampus, neuron, mitochondria, transgenic, mice, eYFP, tetracycline, respiration, neurodegeneration, energy metabolism
Introduction
Mitochondria produce ATP via oxidative phosphorylation, regulate cellular Ca2+ homeostasis through its calcium uniporter and membrane potential, generate and detoxify reactive oxygen and nitrogen species, and regulate cell death pathways via release of apoptotic proteins (for review, see Nicholls and Budd, 2000). Alterations in these mitochondrial pathways have been implicated to play a causal role in aging and in age-associated neurodegenerative disorders (Novelli et al., 1988; Liu et al., 1996; Fiskum et al., 1999; Betarbet et al., 2000; Balaban et al., 2005; Beal, 2005). To study mitochondrial participation in neurodegeneration, mitochondria isolated from neurotoxin and genetic based mouse models are being used. Evidence suggests that isolated brain mitochondria show heterogeneous response. For example, when the swelling of brain mitochondria was examined by electron microscopy, a clear-cut heterogeneity in response to calcium was revealed (Brustovetsky and Dubinsky, 2000; Kristian et al., 2000, 2001, 2002). Although a small subpopulation of brain mitochondria showed typical characteristics of swelling (translucency, damaged cristae) and some mitochondria were without significant morphological alterations, the majority of mitochondria displayed shrunken matrices with expansion of the intermembrane space. This type of heterogenic response was preserved even in the presence of adenine nucleotides, a condition more closely resembling the intracellular environment. Mitochondria isolated from brain come from at least two major sources, neurons and glia. At present, there are no specific markers that can be used to identify or isolate either neuronal or glial mitochondria. To overcome this limitation, primary cultures of neurons and astrocytes are used (Almeida and Medina, 1998; Kristian et al., 2006). Using primary cultures, we and others have observed differences in mitochondrial calcium uptake capacity and differences in response to mitochondrial permeability transition between neuronal and astrocytic mitochondria (Bambrick et al., 2004, 2006; Brown et al., 2006). If neuronal and astrocytic mitochondria differ in their responses to calcium, then the heterogeneous response in isolated brain mitochondria could reflect differences in the cellular source of origin of mitochondria. Thus, there is a need to develop markers that can be used to identify and separate neuronal versus glial mitochondria.
Recent developments in transgenic technology provide an opportunity to target expression of marker fluorescent proteins to specific subcellular compartments and to specific cellular populations. Using the tetracycline system of regulated expression, we generated a transgenic mouse that expresses an enhanced yellow fluorescent protein (eYFP) specifically in neuronal mitochondria, and this expression is induced by the removal of doxycycline from the diet. Mitochondria isolated from the brains of the eYFP-expressing mice maintain normal mitochondrial functions. The development of the transgenic mice provides an in vivo model to study specifically the role of neuronal mitochondria in energy metabolism, in models of stroke and chronic neurodegenerative disorders.
Materials and Methods
Generation of pTet CaMKIIα-mito/eYFP transgenic mice.
Animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the University of Maryland Institutional Animal Care and Use Committee. Mitochondrial eYFP (mito/eYFP) encodes a fusion protein of mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase with eYFP. The fragment of mito/eYFP was generated by digesting the plasmid pEYFP-mito (Clontech, Mountain View, CA) with the restriction enzymes NheI and NotI. The mito/eYFP fragment was cloned between NheI and NotI sites of the tetracycline-responsive element (TRE) plasmid pTRE2-Hyg (Clontech). The insert devoid of vector sequences, containing the tet-responsive promoter upstream of the mito/eYFP coding sequence and β-globin poly(A) signal, was used for injection into fertilized C57BL/6 eggs to create pTRE-mito/eYFP transgenic founders. The mice developed in this study will be deposited in a resource open to the community.
Transgenic mice expressing the tetracycline-controlled transactivator protein (tTA) under regulatory control of the forebrain-specific calcium/calmodulin-dependent kinase IIα (CaMKIIα) promoter (pCaMKIIα-tTA) was purchased from The Jackson Laboratory (Bar Harbor, ME) (Mayford et al., 1996).
Bigenic mice that are positive for both pCaMKIIα-tTA and TRE-mito/eYFP (double-positive or bigenic) were obtained by crossing pTRE-mito/eYFP mice with pCaMKIIα-tTA mice.
Genotyping.
Animals positive for the transgene were identified by PCR using genomic DNA, isolated from the tails (Qiagen, Valencia, CA). The primers were designed to hybridize with the mito/eYFP coding region (forward primer, GCTGACCCTGAAGTTCATCTGC, and reverse primer, CATGATATAGACGTTGTGGCTGTTG) to amplify a 338 bp fragment. In the case of CaMKIIα, the primers (forward primer, GCTGTGGGGCATTTTACTTTAG, and reverse primer, CATGTCCAGATCGAAATCGTC) were designed to amplify a 450 bp fragment. Fifty nanograms of genomic DNA were used in the PCRs, with a program of one cycle of 94°C for 3 min, 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and one cycle of 72°C for 5 min. The PCR products were separated on 1% agarose gel, stained with ethidium bromide, and imaged.
Doxycycline-responsive regulation of eYFP expression.
The parents of the bigenic mice were fed a normal diet. Because pCaMKII-tTA is a tet-off system, the bigenic pups that inherits both pCaMKII-tTA and pTRE2-mito/eYFP genes expressed eYFP and exhibited fluorescence in the brain, which could be visualized through the skull at postnatal day 1 using a fluorescence visualization goggle (GFSP-5; Biological Laboratory Equipment, Budapest, Hungary). We examined doxycycline-responsive regulation of transgene expression in the bigenic mice by replacing doxycycline-free diet with doxycycline (200 mg/kg; BioServ, Frenchtown, NJ) containing diet.
Isolation of synaptic and nonsynaptic brain mitochondria.
Reagents were from Sigma (St. Louis, MO), unless otherwise indicated. Male (20 g) transgenic or wild-type C57BL/6 mice were used. Mice were decapitated, and brains were removed and homogenized in ice-cold isolation medium containing 225 mm mannitol, 75 mm sucrose, 5 mm HEPES, and 1 mm EGTA, pH 7.4, at 4°C. We used a slightly modified procedure by Dunkley et al. (1988) to separate synaptosomes and nonsynaptic mitochondria. After low-speed spin (1300 × g for 3 min) of the brain homogenate, the supernatant was centrifuged at 21,000 × g for 10 min. The pellet was suspended in 3% Percoll (Amersham Biosciences, Piscataway, NJ) and layered on top of preformed gradient of 24% (3.5 ml), 15% (2 ml), 10% (1.5 ml) of Percoll and centrifuged at 32,000 × g for 8 min. Nonsynaptic mitochondria sedimented at the bottom of the tube and the synaptosomes accumulated at the interface of 24 and 15% Percoll and also at the interface of 15 and 10% Percoll. The fraction of nonsynaptic mitochondria was diluted with isolation medium and centrifuged to sediment the purified nonsynaptic mitochondria. The synaptosomes were collected and the mitochondria were released from synaptosomes using a nitrogen cavitation technique (Brown et al., 2004; Kristian et al., 2006). The suspension was layered on to a preformed gradient of 40% (1.5 ml) and 24% (3.5 ml) Percoll. The mitochondria accumulate at the interface of 40 and 24% Percoll gradient. After the mitochondrial fraction was collected and diluted with isolation medium, it was centrifuged to pellet purified synaptic mitochondria. Both nonsynaptic and synaptic mitochondria were suspended in isolation medium containing 1 mg/ml bovine serum albumin (BSA) to remove free fatty acid from mitochondrial membranes. After centrifugation at 7500 × g, the mitochondrial pellet was suspended in isolation medium without EGTA.
Respiration measurements.
Mitochondrial respiratory functions were examined by measuring the oxygen consumption rates with a Clark-type oxygen electrode (Hansatech Instruments, Norfolk, UK). Mitochondria (0.25 mg of protein) were suspended in 0.5 ml of medium consisting of 125 mm KCl (ultrapure; EM Science, Fort Washington, PA), 2 mm K2HPO4, 1 mm MgCl2, 10 μm EGTA, 5 mm malate, 5 mm glutamate, and 20 mm HEPES-Tris, pH 7.0, at 37°C. State 3 respiration was initiated by the addition of 0.2 mm ADP. Approximately 2 min later, state 3 respiration was terminated and state 4o respiration (resting) was initiated with addition of 2.5 μg/ml oligomycin, an inhibitor of the mitochondrial ATP synthetase. The use of oligomycin eliminates the contribution of ATP cycling via hydrolysis by contaminating ATPases and resynthesis by the mitochondrial ATP synthetase to state 4 respiration. The oligomycin-induced state 4o rate of respiration is therefore a more specific indicator of mitochondrial proton cycling limited by passive proton leakiness of the inner membrane. The respiratory control ratio was defined as state 3 rate divided by state 4 rate. The rates of oxygen consumption are expressed as nanomoles of O2 per minute per milligram of mitochondrial proteins.
Protein concentrations were determined using a Lowry DC kit (Bio-Rad, Hercules, CA). Bovine serum albumin was used as standard.
Results were expressed as means ± SD. Statistical significance was determined using a paired Student t test.
Western blotting.
Western blot analysis against eYFP was performed using a peptide monoclonal antibody (JL-8; BD Biosciences, San Jose, CA). Aliquots of synaptic and nonsynaptic mitochondria were processed for Western blot as described previously (Kristian et al., 2006). The peptide monoclonal antibody of eYFP was used at 1:10,000 dilutions in 5% nonfat milk in PBST. Horse anti-mouse IgG coupled to horseradish peroxidase (1:10,000; Cell Signaling Technology, Danvers, MA) was used as secondary antibody. The immunoreactivity was visualized by chemiluminescence.
Immunohistochemistry.
Mice were perfusion-fixed with 4% paraformaldehyde in phosphate buffer, and brains were postfixed in 4% paraformaldehyde for 24 h, and transferred into 30% sucrose. For immunohistochemistry, coronal sections of 30 μm were cut and stored in cryoprotectant solution at −20°C. Sections were rinsed free of cryoprotectant with Tris-buffered saline, exposed to a 1% solution of sodium borohydride, and rinsed again. Tissue was then placed in preheated 0.01 m citrate buffer, pH 6.0, and heated further in a microwave to improve antigen accessibility. Sections were blocked (1:30 in dilution buffer of 1% NGS and 1% BSA in TBS plus 0.2% Triton X-100) for 1 h and then treated with anti-neuronal nuclei (NeuN) monoclonal antibody (Chemicon, Temecula, CA) at 1:10,000, anti-glial fibrillary acidic protein antibody (GFAP) antibody (DakoCytomation, Carpinteria, CA) at 1:10,000, anti-cytochrome oxidase subunit I (COX I) monoclonal antibody (Invitrogen, Carlsbad, CA) at 1:5000 in dilution buffer for 48 h at 4°C. Tissue was then exposed to a secondary antibody (goat anti-mouse conjugated to Alexa Fluor 594 or goat anti-rabbit conjugated to Alexa Fluor 555; Invitrogen) for 1 h at 1:600 in dilution buffer.
Results
Generation of neuron-specific mito/eYFP mice
To achieve eYFP expression within neuronal mitochondria, a neuron-specific promoter, CaMKIIα, driven tTA mice (pCaMKIIα-tTA) were crossed with TRE promoter regulated mito/eYFP transgenic mice (pTRE2-mito/eYFP). pCaMKIIα-tTA is a tet-off system; therefore, induction of expression of mito/eYFP in the bigenic mice occurs in the absence of doxycycline and is inhibited in presence of doxycycline. We observed that, by providing the mating pairs with normal diet, the pups that inherited both pCaMKIIα-tTA and pTRE2-mito/eYFP genes could be identified at postnatal day 1 by the expression of the fluorescent protein mito/eYFP, which could be visualized through the skull using fluorescence goggles. Tail DNA analysis confirmed the presence of both genes (Fig. 1 A). Positive bigenic mice and wild-type nontransgenic controls maintained on a normal diet were killed at 3 weeks of age, synaptic and nonsynaptic mitochondria were isolated, and the presence of eYFP was determined by Western blot analysis. The results (Fig. 1 B) confirmed the presence of eYFP within mitochondria of bigenic mice. Western blot analysis also showed an absence of eYFP in the individual pCaMKIIα-tTA and pTRE2- mito/eYFP transgenic mice (data not shown) and in wild type (Fig. 1 B).
Figure 1.
Genomic identification and mitochondrial expression of eYFP in the bigenic mice. A, Fifty nanograms of genomic DNA prepared from the tails were amplified by PCR. CaMKIIα-positive mice amplified a 450 bp PCR fragment (lane 1), mito/eYFP-positive mice amplified a 338 bp PCR fragment (lane 2), and the bigenic mice amplified both fragments (lane 3). M, Molecular weight marker. B, Western blot analysis of synaptic mitochondria (SM) and nonsynaptic mitochondria (NSM) isolated from bigenic and from control wild-type (WT) mice showed the presence of eYFP only in the bigenic mice. Porin was used as a mitochondrial marker protein.
No differences in appearance or in health were noted between the bigenic and nontransgenic C57BL/6 mice. The breeding pattern, the transmission of the transgene to progeny was normal, and eYFP expression was seen up to six generations.
Doxycycline regulates mito/eYFP expression
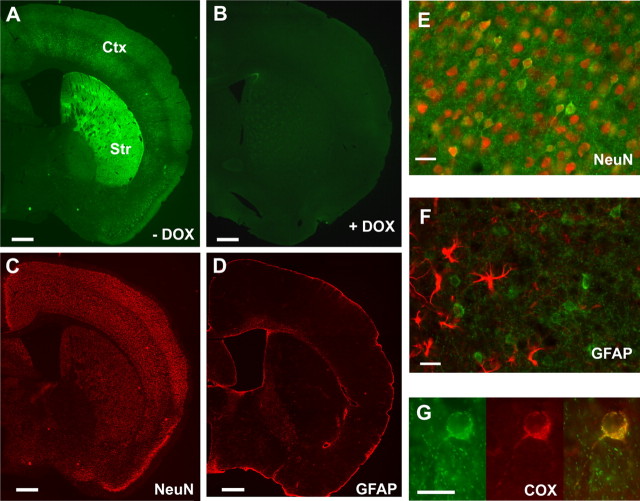
Doxycycline regulation of transgene expression was studied under the following two conditions: (1) regular doxycycline-free diet was replaced with doxycycline (200 mg/kg; BioServ) containing diet for 7–8 weeks (starting from postnatal day 1), expression of mito/eYFP was induced by feeding the animals with doxycycline-free diet for a period of 7 d, mice were either killed for isolation of mitochondria or their brains were perfused for microscopic examination; (2) regular doxycycline-free diet was replaced with doxycycline diet for 7–8 weeks (starting from postnatal day 1), expression of mito/eYFP was induced by feeding the animals with doxycycline-free diet for a period of 7 d, this was followed by doxycycline-containing diet for 3–4 weeks, mice were either killed for isolation of mitochondria or their brains were perfused for microscopic examination. The results on the expression of mito/eYFP and regulation by doxycycline are shown in Figure 2, A and B. The expression of mito/eYFP (Fig. 2 A) was highest in striatum, followed by the forebrain, neocortex, and hippocampus. The expression was absent in the cerebellum (data not shown) but was present in spinal cord motor neurons in the dorsal horn.
Figure 2.
Neuron-specific mitochondrial expression of eYFP and its regulation by doxycycline. A, B, Mito/eYFP expression was induced by feeding the bigenic mice with normal diet (A) or suppressed by feeding with doxycycline-containing diet (B). Coronal sections were visualized under fluorescence microscopy. C, D, Adjacent sections were counterstained with monoclonal anti-NeuN antibody (C) followed by secondary antibody (anti-mouse conjugated to Alexa Fluor 594) or with polyclonal anti-GFAP antibody (D) followed by secondary antibody (anti-rabbit conjugated to Alexa Fluor 555). Note colocalization of eYFP with NeuN staining. E, F, A higher magnification of eYFP fluorescence in cortical neurons shows colocalization with NeuN (E) but not with GFAP (F). G, Immunostaining of neurons from dentate gyrus with anti-cytochrome oxidase subunit I antibody (middle) showed eYFP expression (left) colocalized with the mitochondrial marker protein (right). Scale bars: A–D, 250 μm; E–G, 25 μm. DOX, doxycycline; Str, striatum; Ctx, cortex.
Mito/EYFP is expressed in neuronal mitochondria
To confirm eYFP expression specifically within neurons of the bigenic mice, brain sections were immunostained with anti-NeuN antibody (Fig. 2 C) or with anti- GFAP antibody (Fig. 2 D). The results showed clearly that eYFP expression is neuronal, corresponded with the neuronal marker, NeuN, and did not colocalize with GFAP staining. At higher magnification, the expression of eYFP was punctate, present in neuronal cell bodies and in neuropil, and corresponded with neurons (Fig. 2 E) and not with glial cells (Fig. 2 F). To ensure that eYFP is localized to mitochondria, brain sections that expressed eYFP (Fig. 2 G, left) were immunostained with anti-cytochrome oxidase subunit I antibody (Fig. 2 G, middle). EYFP expression colocalized with the mitochondrial marker (Fig. 2 G, right).
The results obtained in the hippocampal region of the brain are shown in Figure 3 A. At higher magnification in the CA1 region of the hippocampus (Fig. 3 B), expression of eYFP was clearly seen in neuronal cell bodies and in the processes. Immunostaining with NeuN antibody also showed that eYFP was not expressed in mitochondria of all neuronal cells (Fig. 3 C,D). Similar neuronal mitochondrial localization was observed in hippocampal CA2 and CA3 and dentate gyrus neurons. Again, mitochondrial eYFP fluorescence was not seen in all hippocampal neurons.
Figure 3.
Neuronal mitochondria-specific expression of mito/eYFP in the hippocampus. A, Coronal sections of bigenic mice were subjected to immunohistochemical assay using anti-NeuN antibody followed by secondary antibody (conjugated to Alexa Fluor 594). B, Imaging of hippocampal CA1 neurons (boxed region of A) at higher magnification shows presence of mito/eYFP in neuronal cell body and in processes. C, Combined imaging of mito/eYFP and neuronal staining shows neuronal mitochondria-specific presence of mito/eYFP. D, Distribution of neurons with anti-NeuN antibody alone is shown. Scale bars: A, 250 μm; B–D, 25 μm.
Mitochondrial function is normal in eYFP mitochondria
To study the role of neuronal mitochondria in physiology and pathology using these transgenic mice, it is important to determine whether the mitochondria that express eYFP functions normally. We isolated synaptic and nonsynaptic mitochondria and measured their rates of respiration using oxygen electrode. The results are shown in Figure 4. Both state 4 (oligomycin sensitive, basal) and state 3 (ADP-stimulated) rates were found to be unaffected by the expression of eYFP within the mitochondria. Synaptic mitochondria represent predominantly neuronal mitochondria. Similar rates of respiration in synaptic mitochondria of WT and transgenic mice suggests that eYFP expression does not interfere with normal mitochondrial bioenergetic functions.
Figure 4.
Mitochondrial respiration measurements. Mitochondrial state 3 (plus ADP) and state 4 (plus oligomycin) respirations were measured by the oxygen consumption rates with a Clark-type oxygen electrode in nonsynaptic (A) and synaptic (B) mitochondria isolated from bigenic and wild-type mice. No significant difference in the respiratory control ratio for nonsynaptic mitochondria [state 3/state 4 = 6.0 ± 1.2 in wild type (the 95% confidence intervals have minimum/maximum values of 5.1/6.9) and 6.5 ± 1.3 for eYFP (the 95% confidence intervals have minimum/maximum values of 5.1/7.3); n = 5; p = 0.797, two-tailed t test] and synaptic mitochondria [state 3/state 4 = 6.0 ± 0.7 in wild type (the 95% confidence intervals have minimum/maximum values of 5.4/7.0) and 6.4 ± 1.1 for eYFP (the 95% confidence intervals have minimum/maximum values of 5.6/7.6); n = 5; p = 0.429, two-tailed t test] was found between the bigenic and wild-type mice. Error bars indicate SD. WT, Wild type.
Discussion
The tetracycline (TET) system was chosen for the generation of the transgenic mouse because it allows combinatorial targeting of mitochondrial targeted yellow fluorescent protein (mito/eYFP) to be expressed in different cell populations using genetic crosses (Mayford et al., 1996; Kim et al., 2003). In addition, a temporal control overexpression could be achieved by feeding or withdrawing the tetracycline analog, doxycycline, from the diet. In this study, we crossed neuron-specific promoter driven tTA (tet-off) mice with TRE2-mito/eYFP mice to achieve expression of eYFP within neuronal mitochondria. We regulated the expression of eYFP with the doxycycline diet. Feeding the bigenic mice with normal diet induced the expression of eYFP (Fig. 2 A), whereas feeding the mice with doxycycline-containing diet abolished the expression of eYFP (Fig. 2 B). These results suggest that the tet-responsive regulation could be used to induce the expression of eYFP at different developmental stages.
Expression of eYFP was highest in striatum, followed by the forebrain, neocortex, and hippocampus, and was localized to neurons. The pattern of expression of eYFP corresponded with the expression of the endogenous CaMKIIα gene (Burgin et al., 1990). In the cortex and hippocampal subfields, expression of eYFP was not present in all neuronal populations. This suggests that the CaMKIIα promoter activity may vary among neuronal populations (Mayford et al., 1996; Hasan et al., 2004). Despite this restriction, sufficient number of neurons expressed mito/eYFP (20–50%, depending on the brain region), and thus the mito/eYFP transgenic provides a powerful tool to study neuronal mitochondrial function.
We used mitochondrial DNA-encoded cytochrome oxidase subunit I to determine mitochondrial localization of eYFP. Expression of eYFP colocalized with COX I in the majority but not all mitochondria. Expression of COX subunits varies among mitochondrial populations in brain tissue, reflecting differences in mitochondrial functional activity (Hevner and Wong-Riley, 1993). In contrast, mito/eYFP expression occurs in all mitochondria of CaMKIIα-responsive neurons regardless of their functional activity. Therefore, the immunoreactivity for cytochrome oxidase subunit I in some of the mitochondria can be below the threshold of detection. Because all the COX immunostaining colocalized with eYFP, we interpret these results to suggest that the expression of eYFP in the bigenic mice is specific to neuronal mitochondria.
The rates of respiration in synaptic and nonsynaptic mitochondrial fraction were unaffected by the expression of mito/eYFP, which suggests that eYFP expression does not interfere with normal mitochondrial bioenergetic functions. Additional evidence showed that expression of mito/eYFP does not alter mitochondrial functions and therefore can be used to monitor morphological, physiological, and pathological changes in mitochondria. Thus, for example, expression of mito/eYFP has been used to monitor the size and movement of mitochondria in primary neuronal cultures (Chang et al., 2006) in response to exposure to glutamate-induced excitotoxicity (Rintoul et al., 2003), mito/eYFP has been used to monitor mitochondrial changes in pH (Takahashi et al., 2001), and mito/eYFP or mito/GFP has been used as a marker to demonstrate mitochondrial fission and fusion in response to apoptotic stimuli (Karbowski et al., 2004). In all of these studies, primary cell cultures or cultures transfected with mito/eYFP were used.
In summary, the transgenic mice developed in this study provide opportunities to answer a number of existing questions. For example, (1) acute brain slices prepared from mito/eYFP-expressing mice could be combined with mitochondrial NADP(H) fluorescence or with dyes that measure mitochondrial membrane potential to determine neuronal mitochondrial changes in response to increased neuronal activity (Kasischke et al., 2004; Brennan et al., 2006) [or how does exposure to mitochondrial toxins such as MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) or rotenone affect neuronal mitochondria?]; (2) expression of mito/eYFP could be targeted to different cells, for example, to astrocytes, by crossing our mice with GFAP driven tet mice (GFAP-rtTA) (Kim et al., 2003) or to dopaminergic neurons using tyrosine hydroxylase (TH) promoter driven tet mice (TH-rtTA) (Gardaneh and O'Malley, 2004); (3) biochemical methods could be developed using mito/eYFP as a marker protein to isolate either neuronal or glial mitochondria (Kristian et al., 2006); and (4) the transgenic mice developed in this study could be crossed with other genetic models of chronic neurodegenerative diseases to evaluate the role of neuronal mitochondria in degeneration.
Footnotes
This work was supported by National Institutes of Health Grants R21NS050653 (T.K.), NS49425, and NS34152 (G.F.). We thank Dr. Zara Mehrabian for preparing primary neuronal cultures and Maryna Petrasheuskaya for maintaining and genotyping mice.
References
- Almeida A, Medina JM. A rapid method for the isolation of metabolically active mitochondria from rat neurons and astrocytes in primary culture. Brain Res Brain Res Protoc. 1998;2:209–214. doi: 10.1016/s1385-299x(97)00044-5. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Bambrick L, Kristian T, Fiskum G. Astrocyte mitochondrial mechanisms of ischemic brain injury and neuroprotection. Neurochem Res. 2004;29:601–608. doi: 10.1023/b:nere.0000014830.06376.e6. [DOI] [PubMed] [Google Scholar]
- Bambrick LL, Chandrasekaran K, Mehrabian Z, Wright C, Krueger BK, Fiskum G. Cyclosporin a increases mitochondrial calcium uptake capacity in cortical astrocytes but not cerebellar granule neurons. J Bioenerg Biomembr. 2006;38:43–47. doi: 10.1007/s10863-006-9004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Brennan AM, Connor JA, Shuttleworth CW. NAD(P)H fluorescence transients after synaptic activity in brain slices: predominant role of mitochondrial function. J Cereb Blood Flow Metab. 2006;26:1389–1406. doi: 10.1038/sj.jcbfm.9600292. [DOI] [PubMed] [Google Scholar]
- Brown MR, Sullivan PG, Dorenbos KA, Modafferi EA, Geddes JW, Steward O. Nitrogen disruption of synaptoneurosomes: an alternative method to isolate brain mitochondria. J Neurosci Methods. 2004;137:299–303. doi: 10.1016/j.jneumeth.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Brown MR, Sullivan PG, Geddes JW. Synaptic mitochondria are more susceptible to Ca2+overload than nonsynaptic mitochondria. J Biol Chem. 2006;281:11658–11668. doi: 10.1074/jbc.M510303200. [DOI] [PubMed] [Google Scholar]
- Brustovetsky N, Dubinsky JM. Limitations of cyclosporin A inhibition of the permeability transition in CNS mitochondria. J Neurosci. 2000;20:8229–8237. doi: 10.1523/JNEUROSCI.20-22-08229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DT, Honick AS, Reynolds IJ. Mitochondrial trafficking to synapses in cultured primary cortical neurons. J Neurosci. 2006;26:7035–7045. doi: 10.1523/JNEUROSCI.1012-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley PR, Heath JW, Harrison SM, Jarvie PE, Glenfield PJ, Rostas JA. A rapid Percoll gradient procedure for isolation of synaptosomes directly from an S1 fraction: homogeneity and morphology of subcellular fractions. Brain Res. 1988;441:59–71. doi: 10.1016/0006-8993(88)91383-2. [DOI] [PubMed] [Google Scholar]
- Fiskum G, Murphy AN, Beal MF. Mitochondria in neurodegeneration: acute ischemia and chronic neurodegenerative diseases. J Cereb Blood Flow Metab. 1999;19:351–369. doi: 10.1097/00004647-199904000-00001. [DOI] [PubMed] [Google Scholar]
- Gardaneh M, O'Malley KL. Rat tyrosine hydroxylase promoter directs tetracycline-inducible foreign gene expression in dopaminergic cell types. Brain Res Mol Brain Res. 2004;126:173–180. doi: 10.1016/j.molbrainres.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese G, Both M, Duebel J, Waters J, Bujard H, Griesbeck O, Tsien RY, Nagai T, Miyawaki A, Denk W. Functional fluorescent Ca2+ indicator proteins in transgenic mice under TET control. PLoS Biol. 2004;2:e163. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MT. Mitochondrial and nuclear gene expression for cytochrome oxidase subunits are disproportionately regulated by functional activity in neurons. J Neurosci. 1993;13:1805–1819. doi: 10.1523/JNEUROSCI.13-05-01805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305:99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- Kim BO, Liu Y, Ruan Y, Xu ZC, Schantz L, He JJ. Neuropathologies in transgenic mice expressing human immunodeficiency virus type 1 Tat protein under the regulation of the astrocyte-specific glial fibrillary acidic protein promoter and doxycycline. Am J Pathol. 2003;162:1693–1707. doi: 10.1016/S0002-9440(10)64304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian T, Gertsch J, Bates TE, Siesjo BK. Characteristics of the calcium-triggered mitochondrial permeability transition in nonsynaptic brain mitochondria: effect of cyclosporin A and ubiquinone O. J Neurochem. 2000;74:1999–2009. doi: 10.1046/j.1471-4159.2000.0741999.x. [DOI] [PubMed] [Google Scholar]
- Kristian T, Bernardi P, Siesjo BK. Acidosis promotes the permeability transition in energized mitochondria: implications for reperfusion injury. J Neurotrauma. 2001;18:1059–1074. doi: 10.1089/08977150152693755. [DOI] [PubMed] [Google Scholar]
- Kristian T, Weatherby TM, Bates TE, Fiskum G. Heterogeneity of the calcium-induced permeability transition in isolated non-synaptic brain mitochondria. J Neurochem. 2002;83:1297–1308. doi: 10.1046/j.1471-4159.2002.01238.x. [DOI] [PubMed] [Google Scholar]
- Kristian T, Hopkins IB, McKenna MC, Fiskum G. Isolation of mitochondria with high respiratory control from primary cultures of neurons and astrocytes using nitrogen cavitation. J Neurosci Methods. 2006;152:136–143. doi: 10.1016/j.jneumeth.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c . Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- Novelli A, Reilly JA, Lysko PG, Henneberry RC. Glutamate becomes neurotoxic via the N-methyl-d-aspartate receptor when intracellular energy levels are reduced. Brain Res. 1988;451:205–212. doi: 10.1016/0006-8993(88)90765-2. [DOI] [PubMed] [Google Scholar]
- Rintoul GL, Filiano AJ, Brocard JB, Kress GJ, Reynolds IJ. Glutamate decreases mitochondrial size and movement in primary forebrain neurons. J Neurosci. 2003;23:7881–7888. doi: 10.1523/JNEUROSCI.23-21-07881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Zhang Y, Centonze E, Herman B. Measurement of mitochondrial pH in situ. Biotechniques. 2001;30:804–808. doi: 10.2144/01304rv01. 810, 812. [DOI] [PubMed] [Google Scholar]