Abstract
Blood borne macrophage ingress into brain in HIV-1 associated neurocognitive disorders governs the tempo of disease. We used superparamagnetic iron-oxide particles loaded into bone marrow-derived macrophages (BMM) injected intravenously into HIV-1 encephalitis mice to quantitatively assess BMM entry into diseased brain regions. Magnetic resonance imaging tests were validated by histological coregistration and enhanced image processing techniques. The demonstration of robust BMM migration into areas of focal encephalitis provide ‘proof of concept’ for the use of MRI to monitor macrophage migration into brain.
Keywords: image coregistration, blood-brain barrier, monocyte-derived macrophages, HIV-1 encephalitis
1. Introduction
Migration of blood-borne macrophages across the blood brain barrier (BBB) into diseased brain tissue governs the onset and progression of HIV-1 associated neurocognitive disorders (HAND) (Antinori et al. 2007; Ciborowski and Gendelman 2006; Glass et al. 1995). This cell migratory process perpetuates the viral reservoir and provides a continuous source of cellular and viral toxins that influence disease progression (Kadiu et al. 2005; Persidsky and Gendelman, 2003; Reynolds et al. 2007). Interestingly, the ability of macrophages to migrate to the sites of infection makes them an attractive drug delivery system for disease and specifically for antiretroviral and neuroprotective therapies (Dou et al 2006; Kinglsey et al. 2006).
Amongst the techniques used to monitor macrophage migration from blood to tissues, including the central nervous system (CNS), superparamagnetic iron oxide (SPIO) enhanced magnetic resonance imaging (MRI) stands as the most sensitive and well established method (Bendszus and Stoll 2003; Berger et al. 2006; Engberink et al. 2007; Gorantla et al. 2006; Kleinschnitz et al. 2003; Linker et al. 2006; Oweida et al. 2004; Oweida et al. 2007; Rausch et al. 2004; Smirnov et al. 2006). Indeed, macrophage MRI tracking using SPIO labeling was in past works, in our laboratories and others to track cell migration for multiple sclerosis, ischemic stroke lesions, tumors, and HIV-1 encephalitis (HIVE) (Anderson et al. 2004; Boska et al. 2004; Kabanov and Gendelman 2007; Kleinschnitz, et al. 2005; Smirnov et al. 2006; Zelivyanskaya et al. 2003). However, the ability to harness this approach with precision to quantitatively monitor cell migration remains elusive.
To monitor macrophage brain ingress as a function of disease we injected SPIO labeled BMM intravenously in HIVE mice. Accumulation of SPIO particles in brain tissue was readily observed in areas of active neuroinflammation resulting in increased magnetic T2/T2* relaxivity of 1H nuclei and signal loss. The distribution of MRI detected BMM was registered to histology demonstrating macrophage ingress into areas of focal HIVE. Registration of MRI to histology was performed using intermediate digital brain images obtained during cryosectioning (blockface images) as the spatial reference. Robust methods to extract brain images from whole mouse head MRI were developed. Moreover, a semi-automated feature extraction method was applied to modified thin plate splines for warping MRI and processed histology to blockface images. These data demonstrate, for the first time, an ability to quantitatively monitor the migration of BMM from blood to brain. This observation along with the specificity of cell brain entry into areas of focal viral encephalitis demonstrates the accuracy of macrophage quantitation in diseased brain subregions by histology and MRI registration.
2. Materials and Methods
2.1. Murine HIVE
Four-week-old male C.B.-17 severe combined immunodeficient (SCID) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center. Animals were allowed to acclimate to the animal center for one week before the experiment. Monocytes were isolated by centrifugal elutriation methods isolated from peripheral blood mononuclear cells that were acquired from leukopaks from HIV-1,2 and hepatitis B seronegative donors. Monocytes were cultivated in suspension cultures within Teflon jars for seven days in Dulbecco’s Modified Eagles Media supplemented with 10% human sera and 1,000 Units/ml of recombinant macrophage colony stimulating factor (MCSF, a generous gift from Wyeth Inc., Cambridge, MA). After differentiation into monocyte-derived macrophages (MDM), the cells were infected with the macrophage tropic ADA strain of HIV-1 at a multiplicity of 0.1 infectious viral particles/cell (Gendelman et al. 1988). One day after infection, HIV-1ADA-infected MDM (5×105 cells in 5 μl) were injected intracranially into the caudate and putamen (n=5) by stereotactic procedures (Persidsky et al. 1996). Replicate animals were sham operated (n=5).
2.2. BMM Preparation and Injections
BMM were obtained from mouse femurs and cultured in Teflon flasks (Dou et al. 2006; van der Meer et al. 1978). BMM were labeled with SPIO (Feridex, Berlex Inc., Wayne, NJ) by incubation with 2.5 mg/ml of Feridex/107 BMM/ml in complete media for 1 hour at 37°C (Zelivyanskaya et al. 2003). Cells were washed twice with DMEM and each recipient mouse received 1 × 107 SPIO-labeled BMM in 100 μl in media intravenously through the tail vein.
2.3. MRI Acquisition
For HIVE and sham operated control mice three dimensional (3D) high resolution MRI scans were performed for each animal immediately prior to and five days after saline injections or adoptive transfer of SPIO labeled BMM. Mice were anesthetized by inhalation of 1.5% isoflurane in a 70% nitrous oxide/30% oxygen mixture prior to MRI data acquisition. Anesthetized animals were placed in a custom made stereotactic holder with a breathing monitor and placed into the MRI system for imaging. Anesthesia was maintained during the acquisition at .5% to 1.5% isoflurane by maintaining the breathing rate at 40-80 breaths per minute. Breathing rate was monitored using an MRI compatible physiological monitoring system (Model 1025, SA Instruments, Stony Brook, NY). The presence of SPIO-labeled BMM in tissues was evaluated by 3D high resolution (150 μm isotropic) spoiled gradient recalled echo (T2*-weighted) MRI using a Bruker (Karlshure, Germany) Pharmascan 7T MRI system operating Paravision 3.0.2. High resolution 3D gradient recalled echo MRI scans of mouse head were acquired using a 25 mm birdcage volume coil with acquisition parameters of TE = 5 ms, TR = 50 ms, 30% echo, flip angle = 35 degrees, Averages = 2, field of view = 20 × 20 × 20 mm with a resolution of 128 × 128 × 128 (voxel size = 150 × 150 × 150 μm), total acquisition time = 30 min. Mice were euthanized after the final MRI. Brains were extracted, fixed and embedded for histological evaluation. Blockface images were used as an intermediate modality to register in-vivo MRI to stained histological sections. Coregistered MRI and histology were used to assess quantitatively BMM migration to the brain.
2.4. Blockface imaging and 3D reconstruction
Blockface images were acquired using a digital camera (Canon EOS Digital Rebel 300D with a Canon Ultrasonic EFS 60mm f/2.8 Macro USM lens) mounted to the front of the cryostat with a custom mount and triggered by a remote switch. Digital images were acquired every 50 micrometers through the entire brain volume. Slices were numbered to allow registration within the volume after histological processing and staining. Individual blockface slices were aligned to reconstruct the 3D volume using the block outlines to account for jitter in the position of the cryostat head. The brain volume was then automatically segmented using seed based region growing algorithm in the Analyze software package (AnalyzeDirect, Lexena, KS). In this method a seed is selected in the blockface volume and the upper and lower bounds of the intensity are assigned. The region to be segmented grew from the seed to include the pixels connected to the region and within the intensity limits. The intensity limits of the image were defined to prevent the region from growing into areas outside of the brain.
2.5. Histology and Immunohistochemistry
Slices from the brain were collected every 25 μm and their positions numbered. Brain sections obtained through the injection line were stained with Prussian blue, counterstained with hematoxylin and mounted for cell volume analyses and MRI coregistration. Immunocytochemical assays were performed as previously published (Dou et al. 2005). Human HIV-1 infected MDM were detected with Vimentin and astrocytes by glial fibrillary acidic protein (GFAP, Dako). Neuronal Nuclei (NeuN) assessed neuronal numbers and integrity (Chemicon International Inc., Temecula, CA). Twenty five micron sections were cut from formalin-fixed paraffin-embedded blocks through the entire thickness of the HIV-1 infected human MDM injected brain tissue. The imaging parameters were set using the caged control on any given slide.
2.6. Coregistration of MRI with histology
Coregistration of MRI and histology uses the blockface images as the spatial reference. The process is illustrated in Figure 1. The mouse brain images were first extracted from the whole head MRI. The brain MRI was then registered to the blockface volume using the automatic image registration package (AnalyzeDirect, Lexena, KS). The brain MRI was then resliced to create coronal slices that are registered to the blockface images. Both histological slices and resliced MRI were then warped to the blockface images. The details of the brain extraction and warping techniques (boxes in Figure 1) are described in the following paragraphs.
Figure 1.
Flowchart of the procedure of the coregistration between MRI and histological slices. Texts without box represent data; boxed texts represent data processing procedures.
2.6.1. Brain extraction
The brain extraction technique was developed based on a level set method (Li et al. 2005). Level set methods represent a 3D surface C as the cross section of a four dimensional (4D) function (m= φ(x,y,z))taken at m=0. This can be formulated by: C = {(x,y,z) | Φ(x,y,z) = 0}. The 4D function Φ is referred to as the level set function; the surface C is referred to as the zero level surface (Osher and Sethian 1988). Level set methods start with an initial zero level surface and update the surface iteratively towards the object boundaries. The movement of the zero level surface is represented by C(t) = {(x,y,z) | Φ(t,x,y,z) = 0} resulting from the evolution of the level set function Φ can be written in the following general form (Osher and Sethian 1988):
| (1) |
which is the level set equation (Osher and Sethian 1988). The function F is the speed function. For this study, the function F depends on the image gradient and the level set function Φ. Equation (1) can be written as:
| (2) |
where Δ is the Laplacian operator, δ is the Dirac function, and g is the gradient map. The first term at the right hand side corresponds to the internal force, penalizing the deviation of the level set function from the previous iteration. The second and third terms correspond to the external force that drives the zero level surface towards the locations with high gradient (edges). μ, λ and v are the weighting factors of the forces. By choosing the appropriate values for the weighting factors, the internal and external forces together drive the surface towards and converge at the brain boundary.
Constraints were introduced to the level set method by localizing the weighting factors in the head MRI. We designated this method as the “pinned” level set method since the constraints were designated at particular points (pins) extracted from brain contours drawn on one sagittal and two axial slices. Contrary to the edges which attract the surface, the constraints trap the surface and block its propagation. To incorporate the constraints in the level set method, the weighting factors were localized (assigned to each voxel). Instead of being constants, they are varied at the pins by increasing μ (deviation penalty), decreasing λ, and changing the sign of v (both are for external force). In this study, μ = 0.05, λ = 0.0, and v = 1.5 at the pins, and μ = 0.04, λ = 5.0, v = -3 at all other points in the image.
The initial zero level surface was constructed from the pins using a 2-D stacking procedure. In each coronal slice, the pins were connected by line segments to form a closed polygon. The polygons were stacked to form the closed initial surface. The brain extraction procedure illustrated in Table 1 was performed using a computer program coded in Matlab (MathWorks, Natick, MA).
Table 1.
The procedure to extract the brain parenchyma from a whole mouse head MRI using a computer program coded in Matlab
| 1. | Draw brains contours on different sagittal and axial slices. |
| 2. | Extract pins from these contours. |
| 3. | Set the weighting factors of internal penalty and external image gradient forces with appropriate values. |
| 4. | Set the weighting factors of internal penalty and external image gradient forces at the pins. |
| 5. | Construct the initial surface from the pins. |
| 6. | Update level set function and zero level surface iteratively. |
2.6.2. Coregistration of MRI/histological sections to blockface
The MRI was registered to the blockface volume using affine transformation (AnalyzeDirect, Lexena, KS). Normalized mutual information was used as the cost function for the registration. The registered MRI volume was then resliced in the coronal plane to register with the blockface images.
Both MRI and histological slices were warped to blockface images for comparison. The warping procedure developed in this work consisted of matching similar anatomical contours from the source images (MRI and histological slices) to target images (blockface) using thin plate splines. The thin plate splines are a control point (or landmark) based technique, in which the control points are matched between the source to the target image and other parts of the source image are interpolated using the thin plate splines. Excellent reviews of the details of thin plate splines are available in the literature (Bookstein 1989). The manual selection of control points for the traditional thin plate splines is time-consuming and often lacks of accuracy. A semi-automatic technique was developed for this study to allow effective and accurate control point selection. Control points were defined by delineating the contours of the brain boundary and/or corpus callosum and/or ventricles (if they appear on each slice) on the source and target images. The contours on the source image were then sampled by a user-defined accuracy (usually 10-20 pixels) to extract control points. The corresponding target control points were found by minimizing a cost function consisting of a match matrix (Lee et al. 2003). The elements of the match matrix M are assigned as:
| (3) |
where S = {Sj}pj=1 and T = {Tk}pk=1 are the source and target control points, respectively. d(Sj, Tk) is the distance between S and T. The symbols kjS and kkT represent curvature of contours fS and fT at control point position j and k, respectively, and are defined as: Equation (3) incorporates both Euclidean distance and local curvature at control points, by which two geometrically different points would not match only because they are spatially close. The weightings of distance and curvature in (3) are determined by coefficients α and β, respectively. A constrained optimization using the Broyden-Fletcher-Goldfarb-Shanno (BFGS) (Broyden 1970; Fletcher 1970; Goldfarb 1970; Shanno 1970) Quasi-Newton method with a cubic line search procedure was employed to find the optimal positions of the target control points by minimizing the match matrix. The target control points were constrained on the anatomical contours during the optimization. Based on the control point correspondences that were found, the source image was warped with minimum bending energy to the target image. The control point optimization and warping using the thin plate splines were implemented using Matlab. The accuracy of the registration was confirmed using the target registration error (TRE) method (Fitzpatrick et al. 1998; Fitzpatrick and West 2001):
| (4) |
where s and t are corresponding points on the source and target images, respectively, and f is the warping function. Points s and t were placed on anatomical features and not used to effect the warping. Three pairs of points (n=3) were selected in this study for each warping evaluation.
2.7. Detection of BMM by Pre-Post-Injection MRI Signal Subtraction
The intracranial injection of HIV-1ADA-infected MDM induces a focal HIVE at the injection site (Persidsky et al. 1996). The injuries can cause signal loss and generate similar intensities to those of SPIO in T2*-weighted MRI. To differentiate SPIO-labeled BMM from the injuries, the warped post-injection MRI (acquired five days after BMM injection) was subtracted from the warped pre-injection MRI (obtained before BMM injection) from the same animal, generating positive values only in the region of the SPIO labeled BMM. Histogram normalization of intensities between the two data sets was performed before the subtraction. Results of subtraction were windowed and leveled to eliminate noise and the resulting positive signals were superimposed on the pre-injection MRI for comparison to histology. A statistical analysis was performed to calculate the correlation between the quantities of BMM detected by MRI and stained in histology. The spatial correlation of the BMM distribution between MRI and histology was analyzed by calculating the matching ratios of the BMM clusters in coregistered 3D MRI and histological volumes in each animal.
3. Results
3.1. Pathobiology of Murine HIVE
Migration of BMM across the BBB is a defining component of the pathogenesis of HIV-1 infection. Thus, to examine the process and extent of cell migration, we developed in-vivo cell tracking methods validated by histology and applied these methods to evaluate cell migration in the murine model of HIVE. Microscopic imaging demonstrated human cells, astrogliosis, microgliosis, and neuronal impairment (Figure 2). Robust expression of GFAP was demonstrated in and around the areas of focal encephalitis as was reduced NeuN immunostaining. These observations provide a putative mechanism for macrophage blood-brain barrier migration and an opportunity and foundation to explore the functional dynamics of cell migration to assess the potential of cell-packaged drug delivery in disease-affected brain regions.
Figure 2.
Histological and immunocytochemical evaluation of BMM macrophage migration in focal murine HIVE. SCID mice were stereotactically injected with human HIV-1 infected MDM. After four days, murine BMM containing SPIO were administered intravenously through the tail vein and ingress of BMM from blood to brain in areas of the brain injection sites were monitored by Prussian blue staining (BMM, Blue) and immunocytochemistry for density of human Vimentin+ MDM (A and B, brown), signs of GFAP+ astrogliosis (C and D, brown), and NeuN+ neuronal integrity (E and F, brown). Sham operated animals were used as controls and showed no evident BMM ingress or signs of astrogliosis or neuronal loss (A, C, and E). In sharp contrast HIVE mice (B, D, and F) showed ingress of BMM specifically into sites of pathology as evidenced by astrocyte activation (D), and diminished viable neurons (F).
3.2. MRI Tracking of SPIO-Labeled BMM
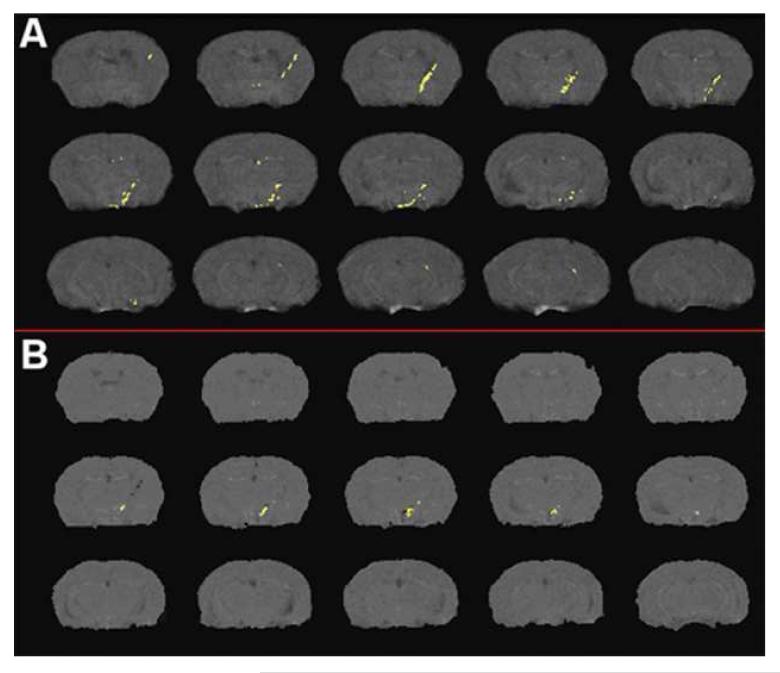
The SPIO-labeled BMM detected by the MRI is shown in Figure 3A. Most BMM cells are clustered at the injection site of HIV-1ADA-infected MDM, in the region of focal encephalitis. Some BMM cells appeared in the ventricles indicating a possible mode of entry through structures with no blood-brain barrier adjacent to the ventricular system such as the subfornical organ. False positive results were observed in control animals (Figure 3B), likely due to healing of the injection site between initial imaging session and the second imaging session five days later. The regions with false positive results were few and small compared to the true positive results in experimental animals. The false positives were primarily located around the tip of the injection line.
Figure 3.
Overlay of subtraction results used for cell detection from MRI in (A): mouse I.V. injected with SPIO labeled cells and (B) negative control mouse (no cell injection). Effects of healing of the injection site between initial imaging session and the second session five days later are apparent in (B).
3.3. Coregistration of Histology and MRI Using Blockface Imaging
The 3D head MRI volume is shown in Figure 4A using the volume rendering in an image visualization software Amira (Visage Imaging, Chelmsford, MA). It can be seen that brain parenchyma and peripheral tissues have similar intensities. Figure 4B shows the brain contours drawn on sagittal and axial slices. The pins extracted from the contours were shown in Figure 4C. Figure 4D demonstrated the initial surface constructed from the pins. After 60 iterations, the final surface reached the brain boundary and was shown in Figure 4E. The segmented brain volume is shown in Figure 4F. The sagittal slice on which a contour was drawn is shown in Figure 4G to provide clear visualization of the contour. The pins, the initial and final surface contours on a central coronal slice were shown in Figure 4H-J, respectively. With the incorporation of the constraints, the surface correctly identifies the brain boundary (Figure 4E) and the level set method accurately segmented the brain tissue (Figure 4F).
Figure 4.
Results of brain extraction using the pinned level set method. (A) A mouse head MRI. (B) Brain contours were drawn sagittal and axial slices. (C) Pins were extracted from the contours. (D) The initial zero level surface was constructed using the pins as vertices. (E) The final surface after 60 iterations. (F) The segmented brain using the pinned level set method. (G) The contour drawn on the sagittal slice. (H) Pins extracted from the contours on a coronal slice. (I) The initial surface contour on the coronal slice. (J) The final surface contour on the coronal slice.
Figure 5A shows the subimaged MRI brain volume which has been registered to the blockface volume (Figure 5B). The affine transformation correctly compensated the global brain deformation caused by perfusion and extraction from the skull. The coregistration between MRI and the blockface volume allowed processed histology slices (Figure 5C) to be registered with the transformed MRI using affine transformation of the processed histological slices to the corresponding slices of the blockface volume.
Figure 5.
Visual representation of brain volume processing. A mouse brain volume (A) was registered to the blockface volume (B) in which slices about the injection line (C) were coregistered and warped to the blockface volume for histological analysis.
The warping procedure to register MRI and histological slices to blockface images is shown in Figure 6. The control points on the source images were extracted from the anatomical contours at the accuracy (10-20 pixels between control points) set by the user (Figure 6A and D). The target control points were extracted from the anatomical contours and optimized using the match matrices in Equation (3) (Figure 6B and E). The final warping results using the control points are shown in Figure 6C and 6F. The accuracy of the warping was satisfactory as determined by the target registration error method (TRE < 1 in Equation 5).
Figure 6.
Image warping using thin plate splines. Left panel: A histological slice was warped to a blockface image. Right panel: A MRI slice was warped to the same blockface image. The contours of the brain boundary and corpus callosum on the histological slice were traced manually and sampled at the accuracy of 10 pixels to generate control points (A). The corresponding contours on the blockface were traced and sampled to generate the homologues of the control points (B). Similar to (A) and (B), the control points on MRI (C) and blockface(D) were generated with the sampling accuracy of 20 pixels. The sampling accuracy was chosen depending on the image resolution. (E, F): The warped histological slice (E) and MRI (F) using thin-plate splines.
3.4. Comparison of BMM on MRI to histology
Two analyses were performed to compare the BMM volume and distribution between MRI and histology. The correlation of the volumes of tissue containing BMM between MRI and histology was calculated, and the spatial distribution of BMM around the injection line was compared. The total volumes of cells by MRI were compared to the volumes detected by histology to correlate the BMM quantity between MRI and histology. Statistical analysis showed a high correlation between the volume occupied by BMM detected in MRI versus histology (R = 0.95, Figure 7).
Figure 7.
Correlation of BMM containing tissue volumes between MRI and histology. Volumes are expressed as a percentage of total brain volume. Points are from the five animals injected with SPIO labeled BMM, and the solid line is the linear regression with the regression results displayed at the upper left of the figure.
The distribution of BMM in brains detected by MRI and histology were compared by visual inspection and quantitative biostatistical analyses. The MRI and histological slices were warped to the blockface images and reconstructed into 3D volumes. The BMM clusters in the MRI and histological volumes were labeled with different pseudo-colors. The overlapped volumes of the two BMM clusters were also labeled for visualization. The matching ratios of the two BMM clusters were calculated using C∩ /Chist, C∩/MRI and Chist/CMRI to investigate the accuracy of the MRI detection of BMM, where CMRI and Chist are the BMM containing voxels in the MRI and histological volumes, respectively, and C∩ is the overlapping pixels of the two BMM clusters. The matching ratios of the two BMM clusters are shown in Table 2. It can be seen in Table 2 that MRI overestimated the volume of BMM containing tissue by an average of 178%.
Table 2.
Matching ratios of the BMM clusters detected by MRI and stained in histology
| histology Animal ID | C∩ / Chist | C∩ / CMRI | Chist / CMRI |
|---|---|---|---|
| M1 | 0.69 | 0.12 | 0.17 |
| M2 | 0.29 | 0.091 | 0.32 |
| M3 | 0.74 | 0.35 | 0.47 |
| M4 | 0.75 | 0.34 | 0.48 |
| M5 | 0.72 | 0.24 | 0.33 |
| mean±s.d. | 0.64±0.20 | 0.23±0.12 | 0.36±0.15 |
C∩ : overlapped volume of the two BMM clusters Chist: BMM cluster a histological volume CMRI: BMM cluster in a MRI volume
The overlap of the stained BMM cluster in histology in one animal to the 3D MRI volume is shown in Figure 8. Figure 8B shows the BMM clusters detected by MRI (labeled in red) and stained in histology (labeled in blue). The overlap volume of the two BMM clusters was labeled in yellow and shown in Figure 8C. In this figure, it is clear that the histology BMM cluster is largely contained within the MRI-detected BMM cluster.
Figure 8.
Three dimensional overlay of MRI and histology detected cell locations. (A) MRI detected BMM locations (red) overlapped with histology detected BMM locations (blue). (B) MRI (red) and histology (blue) detected BMM clusters. (C) Overlap volume of the two BMM clusters (yellow).
4. Discussion
We demonstrated robust migration of blood borne macrophages into HIVE affected brain subregions using registration of MRI and histology techniques. Such observations is a first, to our knowledge, using these combined tests to track disease onset and progression relevant to the neuropathogenesis of HIV-1 infection. Of interest, such approaches can also be modified to track other cell types as well as cell-based drug delivery systems into the brain, making these observations broadly applicable for neuroscience research. Nonetheless, we acknowledge and explain the potential limitations of the data set. Migration of BMM was seen in the area of the needle track, raising the concern that simple BBB disruption elicited the selective migratory response which was observed. We posit that this was likely not the case. Previous studies from our laboratories demonstrated that the BBB is healed within 2-3 days of any induced trauma (Persidsky et al. 1996). Moreover, parallel studies performed in our laboratories of cell migration in sham-operative and lipopolysaccharide-induced neuroinflammation confirm a specific BMM migratory response to inflammation (H. Gendelman, unpublished observations). Indeed, no BMM ingress into brain was seen following trauma but correlated with demonstrable neuroinflammatory responses including elevated macrophage chemotactic protein-1 levels. These findings, taken together, support that the inflammatory cascade induced by virus-infected macrophages was responsible for the migratory events across the BBB into the area of focal encephalitis.
Implementation and the development of state-of-the-art image processing methods allow whole brain detection of migrating cells, validated by accurate histological coregistration. Development of blockface imaging, pinned level set method for image segmentation and modified image warping methods provide the groundwork to validate MRI results using standard histopathological assessments.
A number of automated or semi-automated brain extraction techniques have been developed for human head MRI (Boesen et al. 2004; Chiverton et al. 2007; Hartley et al. 2006; Lee et al. 2003b; Rex et al. 2004; Segonne et al. 2004; Smith 2002; Zhuang et al. 2006). However, these techniques are prone to failure when used for mouse brain extraction due to the small or non-existent gap between the brain and the non-brain tissue. We presented a technique based on level set methods with the incorporation of higher level constraints obtained from prior knowledge and understanding of the mouse brain anatomy. Level set methods were first introduced two decades ago (Osher and Sethian 1988), and their application has received great attention in image segmentation in recent years. As describe above, similar intensity of brain parenchyma and peripheral tissues and narrow or disappearing brain-skull gap (Figure 9A and 9B) causes the traditional level set methods to miss the brain-skull border (Figure 9C). A method to prevent the surface from moving beyond the brain-skull border is incorporating high level constraints into the level set methods, which, even with their increasing applications in medical image segmentation, has rarely been studied (Mitchell et al. 2002). Generally, several properties are desirable for the constraints including 1) the ability to effectively represent the prior knowledge of the object to be segmented; 2) ease of definition by the user, 3) seamless incorporation into level set methods. The strategy used in this study assures that the constraints meet these desired properties. With minimum knowledge of the mouse brain anatomy, and by quickly browsing through the head MRI, a researcher can locate the areas with weak edges, and thus define brain contours to constrain these locations. The contours were defined on orthogonal (sagittal and axial) 2-D slices to avoid the complex 3-D visualization and manipulation. Their seamless incorporation was achieved in this study by localizing the weighting factors of the internal and external forces. Additionally, the pins were used to automatically define the initial surface by using them as the vertices of the surface, eliminating the need for user intervention (Figure 9D). This also assured the initial surface to be close to the brain boundary, which reduced the time required for the surface to converge to the brain boundary. The level set method with incorporation of high level constraints developed in this study can also be used for the segmentation of other organs and tissues.
Figure 9.
The utility of the pinned level set in an area of mouse brain with no significant signal gradient. (A) Disappearing of the gap between the brain and skull in a posterior slice, (B) Image gradient, (C) A traditional (unpinned) level set method (the red closed contour) missed the brain-skull border (pointed by the arrow) on the posterior slice, (D) The initial zero level surface contour (the red closed contour) as defined from the pins (green) on a posterior coronal slice, (E) the final surface correctly segments the brain, (F) the mouse brain volume extracted from the whole head MRI using the pinned level method.
MRI needs to be coregistered to histology to evaluate the accuracy of the MRI to monitor cell trafficking to brain. Histology is difficult to register with MRI due to two major deformations, global deformation which occurs during fixation and removal of the brain from the skull and histological slice specific deformation. The individual deformations for each slice include asymmetrical shrinkage due to processing and tearing due to slicing and mounting on glass slides. Therefore, to map histology and MRI the coregistration procedure needs to reverse these deformations on histology or/and model them on MRI. From the image processing point of view, the global and slice specific deformations can be described by two different mathematical manipulations: linear (affine) and nonlinear transformation (warping), respectively. Direct coregistration between the MRI and histological sections that performs the affine transformation and warping simultaneously is feasible but technologically difficult. We used the blockface images as an intermediate modality to reduce the variables involved in coregistration and warping of MRI and histology. An affine transformation was performed to register MRI to the blockface images to address the global deformation associated with perfusion and removal of the brain from the skull. Deformations from histological processing were corrected using slice by slice warping of the processed histology back to the blockface image. Similar methods have been used in previous studies (Jacobs et al. 1999; Meyer et al. 2006).
Each histological section is subject to different distortions requiring individual 2-D slice-by-slice warping for registration of MRI to histology. These characteristics make the thin plate splines the best candidate among many image warping techniques (Christensen et al. 1997; Davatzikos 1997; Friston et al. 1995; Hill et al. 2001; Kyriacou et al. 1999; Liu et al. 2006; Pelizzari et al. 1989; Pelizzari 1998; Rueckert et al. 1997; Woods et al. 1998a; Woods et al. 1998b; Zitova and Flusser 2003). The thin plate splines has been extensively used in neuroscience studies (Bookstein 1997; Buchsbaum et al. 1998; Corouge et al. 2003; Darvas et al. 2006; Davis et al. 1997; DeQuardo et al. 1999; E. Downhill J, Jr et al. 2000; Jacobs et al. 1999; Kim et al. 1997; Li et al. 2006; Meyer et al. 1997; Meyer et al. 2006; Miller et al. 2002) due to the capability to correct severe distortion and computational efficiency. However, the selection of control points for the thin plate splines is an error prone and time-consuming procedure. Image features were used for control point selection in previous studies (Jacobs et al. 1999, Rohr et al. 1996; Rohr et al. 2001). However, these methods can not guarantee the exact correspondence between the source and target control points which is the underlying assumption of the thin plate splines. To address this problem, Rohr et al (1996, 2001) relaxed the exact interpolation condition by introducing the Euclidean distance between corresponding control points into the bending energy function. In this method, two control points are considered to be correspondent if the distance is within a preset tolerance. The method we present here extracts control points from anatomical contours which minimizes user interaction, allowing distance and, unlike Rohr et al, local curvature into the control point selection procedure. The match matrix (Equation (3)) based control point optimization method used in this study was recently introduced in applications of pattern recognition. To the best of our knowledge, this is the first time this method has been used in medical image coregistration.
Development of techniques for coregistration and overlay of processed histology and MRI enable us to evaluate the accuracy of MRI to monitor the BMM infiltration. These developments are key to the feasibility of a broad range of studies to evaluate imaging, molecular imaging, and spectroscopic imaging with histology in neurological disease models.
Results shown in Table 2 and Figure 8 demonstrate that there is not an exact correspondence between the volume of cells detected by MRI and histology. There are several sources of potential error which may contribute to this effect. The effect of SPIO on the MRI signal extends well beyond the cell border and when large clusters of SPIO labeled cells are present, the spatial extent of the signal disturbance is enhanced. In addition, the resolution of the histology is much greater (25 micron slice thickness) than the MRI (150 micron slice thickness). These are the primary reasons that the volume determined by MRI is 178% greater than the volume determined by histology. However, this is not the only source of error, as only 72% of the volume of the cells determined by histology is within the MRI volume. Other sources of the difference in spatial extent and distribution of cells include small errors in registration and warping, lack of detection of regions with few cells by MRI, and false positives in MRI from wound healing between the initial and second imaging study. Despite sources of error, the overall correlation between the location and amount of cells shows that the MRI technique can be used to assess the dynamics of cell influx. With histological endpoints, the method can be corrected for overestimated cell volumes. Further developments, including increased spatial resolution, improved methods of cell labeling, and improved image processing will improve accuracy. Having the tools to evaluate the accuracy by coregistration with histology will allow quantitative assessments of the accuracy of image acquisition and processing developments in future works.
Taken together, the migration of macrophages across the BBB in this murine HIV model demonstrates readily identified cells in the area of encephalitis. These responses may be aided, in part, by impairments in the BBB seen in this murine model and in human disease (Kadiu et al. 2005; Persidsky and Poluektova 2006). Further studies to track the responses and kinetics of BMM migration to the brain of other diseases which exhibit inflammation including Alzheimer’s disease, Parkinson’s disease, stroke, and injury (Reynolds, et al. 2007) may provide valuable biological insight into the response of BMM and other cell types to brain injury and disease.
Acknowledgements
The authors thank Robin Taylor for critical reading of the manuscript. The work was supported by the Francine and Louis Blumkin Foundation, the Community Neuroscience Pride of Nebraska Initiative, the Carol Swartz, M.D. Neuroscience Laboratory, and the Alan Baer Charitable Trust (to H.E.G.), NIH grants 1T32 NS07488 (to J.A.N., M.D.B. and H.E.G.), 2R37 NS36126, 1 P01 NS043985-01, 5 P01 MH64570-03, and P20 RR15635 (to M.D.B. and H.E.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson SA, Shukaliak-Quandt J, Jordan EK, Arbab AS, Martin R, McFarland H, Frank JA. Magnetic resonance imaging of labeled T-cells in a mouse model of multiple sclerosis. Ann Neurol. 2004;55(5):654–9. doi: 10.1002/ana.20066. [DOI] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, et al. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69(18):1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendszus M, Stoll G. Caught in the act: In vivo mapping of macrophage infiltration in nerve injury by magnetic resonance imaging. J Neurosci. 2003;23(34):10892–6. doi: 10.1523/JNEUROSCI.23-34-10892.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Hiestand P, Kindler-Baumann D, Rudin M, Rausch M. Analysis of lesion development during acute inflammation and remission in a rat model of experimental autoimmune encephalomyelitis by visualization of macrophage infiltration, demyelination and blood-brain barrier damage. NMR Biomed. 2006;19(1):101–7. doi: 10.1002/nbm.1007. [DOI] [PubMed] [Google Scholar]
- Boesen K, Rehm K, Schaper K, Stoltzner S, Woods R, Luders E, Rottenberg D. Quantitative comparison of four brain extraction algorithms. Neuroimage. 2004;22(3):1255–61. doi: 10.1016/j.neuroimage.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Bookstein FL. Landmark methods for forms without landmarks: Morphometrics of group differences in outline shape. Med Image Anal. 1997;1(3):225–43. doi: 10.1016/s1361-8415(97)85012-8. [DOI] [PubMed] [Google Scholar]
- Bookstein FL. Principal warps: Thin-plate splines and the decomposition of deformations. IEEE Trans. Pattern. Anal. Mach. Intell. 1989;11(6):567–85. [Google Scholar]
- Boska MD, Mosley RL, Nawab M, Nelson JA, Zelivyanskaya M, Poluektova L, Uberti M, Dou H, Lewis TB, Gendelman HE. Advances in neuroimaging for HIV-1 associated neurological dysfunction: Clues to the diagnosis, pathogenesis and therapeutic monitoring. Curr HIV Res. 2004;2(1):61–78. doi: 10.2174/1570162043485095. [DOI] [PubMed] [Google Scholar]
- Broyden CG. The Convergence of a Class of Double-Rank Minimization Algorithms. Journal Inst. Math. Applic. 1970;6:76–90. [Google Scholar]
- Buchsbaum MS, Fallon JH, Wei TC, Guich S, Spiegel-Cohen J, Hamilton M, Tang C. A method of basal forebrain anatomical standardization for functional image analysis. Psychiatry Res. 1998;84(23):113–25. doi: 10.1016/s0925-4927(98)00052-3. [DOI] [PubMed] [Google Scholar]
- Chiverton J, Wells K, Lewis E, Chen C, Podda B, Johnson D. Statistical morphological skull stripping of adult and infant MRI data. Comput Biol Med. 2007;37(3):342–57. doi: 10.1016/j.compbiomed.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE Trans Med Imaging. 1997;16(6):864–77. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- Ciborowski P, Gendelman HE. Human immunodeficiency virus-mononuclear phagocyte interactions: Emerging avenues of biomarker discovery, modes of viral persistence and disease pathogenesis. Curr HIV Res. 2006;4(3):279–91. doi: 10.2174/157016206777709474. [DOI] [PubMed] [Google Scholar]
- Corouge I, Hellier P, Gibaud B, Barillot C. Interindividual functional mapping: A nonlinear local approach. Neuroimage. 2003;19(4):1337–48. doi: 10.1016/s1053-8119(03)00158-7. [DOI] [PubMed] [Google Scholar]
- Darvas F, Ermer JJ, Mosher JC, Leahy RM. Generic head models for atlas-based EEG source analysis. Hum Brain Mapp. 2006;27(2):129–43. doi: 10.1002/hbm.20171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput Vis Image Underst. 1997;66(2):207–22. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- Davis MH, Khotanzad A, Flamig DP, Harms SE. A physics-based coordinate transformation for 3-D image matching. IEEE Trans Med Imaging. 1997;16(3):317–28. doi: 10.1109/42.585766. [DOI] [PubMed] [Google Scholar]
- DeQuardo JR, Keshavan MS, Bookstein FL, Bagwell WW, Green WD, Sweeney JA, Haas GL, Tandon R, Schooler NR, Pettegrew JW. Landmark-based morphometric analysis of first-episode schizophrenia. Biol Psychiatry. 1999;45(10):1321–8. doi: 10.1016/s0006-3223(98)00181-4. [DOI] [PubMed] [Google Scholar]
- Dou H, Destache CJ, Morehead JR, Mosley RL, Boska MD, Kingsley J, Gorantla S, Poluektova L, Nelson JA, Chaubal M, et al. Development of a macrophage- based nanoparticle platform for antiretroviral drug delivery. Blood. 2006;108(8):2827–35. doi: 10.1182/blood-2006-03-012534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downhill J,E, Jr, Buchsbaum MS, Wei T, Spiegel-Cohen J, Hazlett EA, Haznedar MM, Silverman J, Siever LJ. Shape and size of the corpus callosum in schizophrenia and schizotypal personality disorder. Schizophr Res. 2000;42(3):193–208. doi: 10.1016/s0920-9964(99)00123-1. [DOI] [PubMed] [Google Scholar]
- Engberink RD, Blezer EL, Hoff EI, van der Pol SM, van der Toorn A, Dijkhuizen RM, de Vries HE. MRI of monocyte infiltration in an animal model of neuroinflammation using SPIO-labeled monocytes or free USPIO. J Cereb Blood Flow Metab. 2007 doi: 10.1038/sj.jcbfm.9600580. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, West JB. The distribution of target registration error in rigid-body point-based registration. IEEE Trans Med Imaging. 2001;20(9):917–27. doi: 10.1109/42.952729. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, West JB, Maurer CR., Jr. Predicting error in rigid-body point-based registration. IEEE Trans Med Imaging. 1998;17(5):694–702. doi: 10.1109/42.736021. [DOI] [PubMed] [Google Scholar]
- Fletcher R. A new approach to variable metric algorithms. Computer Journal. 1970;13:317–322. [Google Scholar]
- Friston KJ, Ashburner J, Poline JB, Frith CD, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Human Brain Mapping. 1995;2:165–89. [Google Scholar]
- Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med. 1988;167(4):1428–41. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JD, Fedor H, Wesselingh SL, McArthur JC. Immunocytochemical quantitation of human immunodeficiency virus in the brain: Correlations with dementia. Ann Neurol. 1995;38(5):755–62. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- Goldfarb D. A family of variable metric updates derived by variational means. Mathematics of Computing. 1970;24:23–26. [Google Scholar]
- Gorantla S, Dou H, Boska M, Destache CJ, Nelson J, Poluektova L, Rabinow BE, Gendelman HE, Mosley RL. Quantitative magnetic resonance and SPECT imaging for macrophage tissue migration and nanoformulated drug delivery. J Leukoc Biol. 2006;80(5):1165–74. doi: 10.1189/jlb.0206110. [DOI] [PubMed] [Google Scholar]
- Hartley SW, Scher AI, Korf ES, White LR, Launer LJ. Analysis and validation of automated skull stripping tools: A validation study based on 296 MR images from the honolulu asia aging study. Neuroimage. 2006;30(4):1179–86. doi: 10.1016/j.neuroimage.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Hill DL, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Phys Med Biol. 2001;46(3):R1–45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Windham JP, Soltanian-Zadeh H, Peck DJ, Knight RA. Registration and warping of magnetic resonance images to histological sections. Med Phys. 1999;26(8):1568–78. doi: 10.1118/1.598671. [DOI] [PubMed] [Google Scholar]
- Kabanov AV, Gendelman HE. Nanomedicine in the diagnosis and therapy of neurodegenerative disorders. Progress in Polymer Science. 2007;32(89):1054–82. doi: 10.1016/j.progpolymsci.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiu I, Glanzer JG, Kipnis J, Gendelman HE, Thomas MP. Mononuclear phagocytes in the pathogenesis of neurodegenerative diseases. Neurotox Res. 2005;8(12):25–50. doi: 10.1007/BF03033818. [DOI] [PubMed] [Google Scholar]
- Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA. HIV-1 infection and AIDS: Consequences for the central nervous system. Cell Death Differ. 2005;12(Suppl 1):878–92. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- Kim B, Boes JL, Frey KA, Meyer CR. Mutual information for automated unwarping of rat brain autoradiographs. Neuroimage. 1997;5(1):31–40. doi: 10.1006/nimg.1996.0251. [DOI] [PubMed] [Google Scholar]
- Kinglsey JD, Dou H, Morehead J, Rabinow B, Gendelman HE, Destache CJ. Nanotechnology: A focus on nanoparticles as drug delivery system. J Neuroimmune Pharmacol. 2006;1:340–50. doi: 10.1007/s11481-006-9032-4. [DOI] [PubMed] [Google Scholar]
- Kleinschnitz C, Bendszus M, Frank M, Solymosi L, Toyka KV, Stoll G. In vivo monitoring of macrophage infiltration in experimental ischemic brain lesions by magnetic resonance imaging. J Cereb Blood Flow Metab. 2003;23(11):1356–61. doi: 10.1097/01.WCB.0000090505.76664.DB. [DOI] [PubMed] [Google Scholar]
- Kleinschnitz C, Schutz A, Nolte I, Horn T, Frank M, Solymosi L, Stoll G, Bendszus M. In vivo detection of developing vessel occlusion in photothrombotic ischemic brain lesions in the rat by iron particle enhanced MRI. J Cereb Blood Flow Metab. 2005;25(11):1548–55. doi: 10.1038/sj.jcbfm.9600151. [DOI] [PubMed] [Google Scholar]
- Kyriacou SK, Davatzikos C, Zinreich SJ, Bryan RN. Nonlinear elastic registration of brain images with tumor pathology using a biomechanical model. IEEE Trans Med Imaging. 1999;18(7):580–92. doi: 10.1109/42.790458. [DOI] [PubMed] [Google Scholar]
- Sang-Mook Lee, Lynn Abbott A., Neil A. Clark, Philip A. Araman. Spline curve matching with sparse knot sets: Applications to deformable shape detection and recognition. The 29th annual conference of the IEEE industrial electronics societyVirginia, USA.2003a. [Google Scholar]
- Lee JM, Yoon U, Nam SH, Kim JH, Kim IY, Kim SI. Evaluation of automated and semi-automated skull-stripping algorithms using similarity index and segmentation error. Comput Biol Med. 2003b;33(6):495–507. doi: 10.1016/s0010-4825(03)00022-2. [DOI] [PubMed] [Google Scholar]
- Li C, Xu C, Gui C, Fox MD. Level set evolution without re-initialization: A new variational formulation. IEEE computer society conference on computer vision and pattern recognition.2005. [Google Scholar]
- Li G, Nikolova S, Bartha R. Registration of in vivo magnetic resonance T(1)- weighted brain images to triphenyltetrazolium chloride stained sections in small animals. J Neurosci Methods. 2006;156(12):368–75. doi: 10.1016/j.jneumeth.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Linker RA, Kroner A, Horn T, Gold R, Maurer M, Bendszus M. Iron particle-enhanced visualization of inflammatory central nervous system lesions by high resolution: Preliminary data in an animal model. AJNR Am J Neuroradiol. 2006;27(6):1225–9. [PMC free article] [PubMed] [Google Scholar]
- Liu Y, D’Arceuil H, He J, Duggan M, Gonzalez G, Pryor J, de Crespigny A. A nonlinear mesh-warping technique for correcting brain deformation after stroke. Magn Reson Imaging. 2006;24(8):1069–75. doi: 10.1016/j.mri.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Meyer CR, Moffat BA, Kuszpit KK, Bland PL, McKeever PE, Johnson TD, Chenevert TL, Rehemtulla A, Ross BD. A methodology for registration of a histological slide and in vivo MRI volume based on optimizing mutual information. Mol Imaging. 2006;5(1):16–23. [PMC free article] [PubMed] [Google Scholar]
- Meyer CR, Boes JL, Kim B, Bland PH, Zasadny KR, Kison PV, Koral K, Frey KA, Wahl RL. Demonstration of accuracy and clinical versatility of mutual information for automatic multimodality image fusion using affine and thin-plate spline warped geometric deformations. Med Image Anal. 1997;1(3):195–206. doi: 10.1016/s1361-8415(97)85010-4. [DOI] [PubMed] [Google Scholar]
- Miller MI, Trouve A, Younes L. On the metrics and euler-lagrange equations of computational anatomy. Annu Rev Biomed Eng. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- Mitchell SC, Bosch JG, Lelieveldt BP, van der Geest RJ, Reiber JH, Sonka M. 3-D active appearance models: Segmentation of cardiac MR and ultrasound images. IEEE Trans Med Imaging. 2002;21(9):1167–78. doi: 10.1109/TMI.2002.804425. [DOI] [PubMed] [Google Scholar]
- Osher S, Sethian JA. Fronts propagating with curvature dependent speed: Algorithms based on hamilton-jacobi formulations. Journal of Computational Physics. 1988;79:12–49. [Google Scholar]
- Oweida AJ, Dunn EA, Foster PJ. Cellular imaging at 1.5 T: Detecting cells in neuroinflammation using active labeling with superparamagnetic iron oxide. Mol Imaging. 2004;3(2):85–95. doi: 10.1162/15353500200404106. [DOI] [PubMed] [Google Scholar]
- Oweida AJ, Dunn EA, Karlik SJ, Dekaban GA, Foster PJ. Iron-oxide labeling of hematogenous macrophages in a model of experimental autoimmune encephalomyelitis and the contribution to signal loss in fast imaging employing steady state acquisition (FIESTA) images. J Magn Reson Imaging. 2007;26(1):144–51. doi: 10.1002/jmri.21005. [DOI] [PubMed] [Google Scholar]
- Pelizzari CA. Image processing in stereotactic planning: Volume visualization and image registration. Med Dosim. 1998;23(3):137–45. doi: 10.1016/s0958-3947(98)00014-4. [DOI] [PubMed] [Google Scholar]
- Pelizzari CA, Chen GT, Spelbring DR, Weichselbaum RR, Chen CT. Accurate three-dimensional registration of CT, PET, and/or MR images of the brain. J Comput Assist Tomogr. 1989;13(1):20–6. doi: 10.1097/00004728-198901000-00004. [DOI] [PubMed] [Google Scholar]
- Persidsky Y, Poluektova L. Immune privilege and HIV-1 persistence in the CNS. Immunol Rev. 2006;213:180–94. doi: 10.1111/j.1600-065X.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- Persidsky Y, Gendelman HE. Mononuclear phagocyte immunity and the neuropathogenesis of HIV-1 infection. J Leukoc Biol. 2003;74(5):691–701. doi: 10.1189/jlb.0503205. [DOI] [PubMed] [Google Scholar]
- Persidsky Y, Limoges J, McComb R, Bock P, Baldwin T, Tyor W, Patil A, Nottet HS, Epstein L, Gelbard H, et al. Human immunodeficiency virus encephalitis in SCID mice. Am J Pathol. 1996;149(3):1027–53. [PMC free article] [PubMed] [Google Scholar]
- Rausch M, Hiestand P, Foster CA, Baumann DR, Cannet C, Rudin M. Predictability of FTY720 efficacy in experimental autoimmune encephalomyelitis by in vivo macrophage tracking: Clinical implications for ultrasmall superparamagnetic iron oxide-enhanced magnetic resonance imaging. J Magn Reson Imaging. 2004;20(1):16–24. doi: 10.1002/jmri.20057. [DOI] [PubMed] [Google Scholar]
- Rex DE, Shattuck DW, Woods RP, Narr KL, Luders E, Rehm K, Stoltzner SE, Rottenberg DA, Toga AW. A meta-algorithm for brain extraction in MRI. Neuroimage. 2004;23(2):625–37. doi: 10.1016/j.neuroimage.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Kadiu I, Kanmogne G, Gendelman H. HIV and the brain. In: Gorman, Ann Cohen Mary, Jack M., editors. Comprehensive textbook of AIDS psychiatry. Oxford University Press; New York, NY: 2007. . In Press p.
- Rohr K, Stiehl HS, Sprengel R, Buzug TM, Weese J, Kuhn MH. Landmark-based elastic registration using approximating thin-plate splines. IEEE Trans Med Imaging. 2001;20(6):526–34. doi: 10.1109/42.929618. [DOI] [PubMed] [Google Scholar]
- Rohr K, Stiehl HS, Sprengel R, Beil W, Buzug TM, Weese J, Kuhn MH. Nonrigid registration of medical images based on anatomical point landmarks and approximating thin-plate splines. Proceedings of the aacheren workshop: Bildverarbeiterung fur die medizinAachen; Germany. 1996. p. 41 p. [Google Scholar]
- Rueckert PE, Geyer S, Amunts K, Schormann T, Schleicher A, Malikovic A, Zilles K.Automatic construction of 3D statistical deformation models using non-rigid registration MIC-CAI. 199777 p
- Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22(3):1060–75. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Shanno DF. Conditioning of Quasi-Newton Methods for Function Minimization. Mathematics of Computing. 1970;24:647–656. [Google Scholar]
- Smirnov P, Lavergne E, Gazeau F, Lewin M, Boissonnas A, Doan BT, Gillet B, Combadiere C, Combadiere B, Clement O. In vivo cellular imaging of lymphocyte trafficking by MRI: A tumor model approach to cell-based anticancer therapy. Magn Reson Med. 2006;56(3):498–508. doi: 10.1002/mrm.20996. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–55. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer JW, Bulterman D, van Zwet TL, Elzenga-Claasen I, van Furth R. Culture of mononuclear phagocytes on a teflon surface to prevent adherence. J Exp Med. 1978;147(1):271–6. doi: 10.1084/jem.147.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998a;22(1):153–65. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. general methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998b;22(1):139–52. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Zelivyanskaya ML, Nelson JA, Poluektova L, Uberti M, Mellon M, Gendelman HE, Boska MD. Tracking superparamagnetic iron oxide labeled monocytes in brain by high-field magnetic resonance imaging. J Neurosci Res. 2003;73(3):284–95. doi: 10.1002/jnr.10693. [DOI] [PubMed] [Google Scholar]
- Zhuang AH, Valentino DJ, Toga AW. Skull-stripping magnetic resonance brain images using a model-based level set. Neuroimage. 2006;32(1):79–92. doi: 10.1016/j.neuroimage.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Zitova B, Flusser J. Image registration methods: A survey. Image and Vision Computing. 2003;21:977–1000. [Google Scholar]