Abstract
Bacterial chemoreceptors form mixed trimers of homodimers that cluster further in the presence of other cytoplasmic components. The physical proximity between receptors is thought to promote conformational coupling that enhances sensitivity, dynamic range, and collaboration between receptors of different types. We investigated conformational coupling between neighboring dimers by co-expressing two types of receptors, only one of which was labeled with yellow fluorescence protein (YFP). The two types of receptors were stimulated independently, and changes in the relative orientation of the labeled receptors were followed by fluorescence anisotropy. Possible coupling via cytoplasmic components of the taxis system was avoided by working with strains lacking those components. We find that binding of ligand to one type of receptor affects the conformation of the other type of receptor, but not in the same way as binding of ligand to that receptor directly. Thus, different receptors are coupled, but not as simply as previously thought.
To follow chemical gradients, cells of Escherichia coli utilize several types of transmembrane receptors, including the Tsr receptor that senses serine and the Tar receptor that senses aspartate1–3. These sensors form mixed trimers of homodimers, even in the absence of other chemotaxis proteins4,5, and together with a coupling protein, CheW, and the kinase, CheA, form large receptor arrays6–8. These arrays were predicted to facilitate receptor collaborative operation9,10. Indeed, synergetic effects between homodimers of different types have been demonstrated both in-vivo11 and in-vitro12. Here we aimed to test directly whether coupling between homodimers can emerge due to direct physical interactions between neighboring dimers in receptor oligomers. To avoid coupling modes involving cytoplasmic proteins, we made measurements in strains in which CheW and CheA are not expressed. Note that while large clusters appear to be the dominant receptor state in the presence of CheW and CheA, it is the trimer-of-dimers structure that is the dominant state in their absence4,5,13. We showed previously that changes in the relative orientation of adjacent homodimers in response to binding of ligand can be detected by labeling receptors with YFP and measuring fluorescence anisotropy14,15. Changes in fluorescence anisotropy were traced to changes in energy transfer, i.e., to homo-FRET, that occur upon ligand binding. When serine (aspartate) is added to trimers of homodimers of Tsr-YFP (Tar-YFP), the YFPs move farther apart. These responses are absent in mutants that fail to form trimers and can be diluted out by expression of unlabeled receptors14,15.
The rationale of the experiments described here is the following: by forming mixed oligomers composed of homodimers of fluorescently labeled Tar and homodimers of unlabeled Tsr, we can selectively stimulate either type of receptor while reading out changes in the relative orientation of the labeled receptors. Thus, we can directly address the following question: does binding of ligand to the Tsr dimer trigger conformational changes in the Tar dimers? This strategy is illustrated in Fig. 1a. Two extremes are possible: (i) In the absence of coupling, the conformational response of Tsr will not affect Tar and the anisotropy level will not change; (ii) if, however, the trimers form a single allosteric unit (tight coupling) the binding of ligand to Tsr can only toggle the whole structure from the active to the inactive conformation, resulting in a change in anisotropy similar to that observed upon binding of ligand to Tar. The actual behavior turns out to be more complex.
Figure 1.
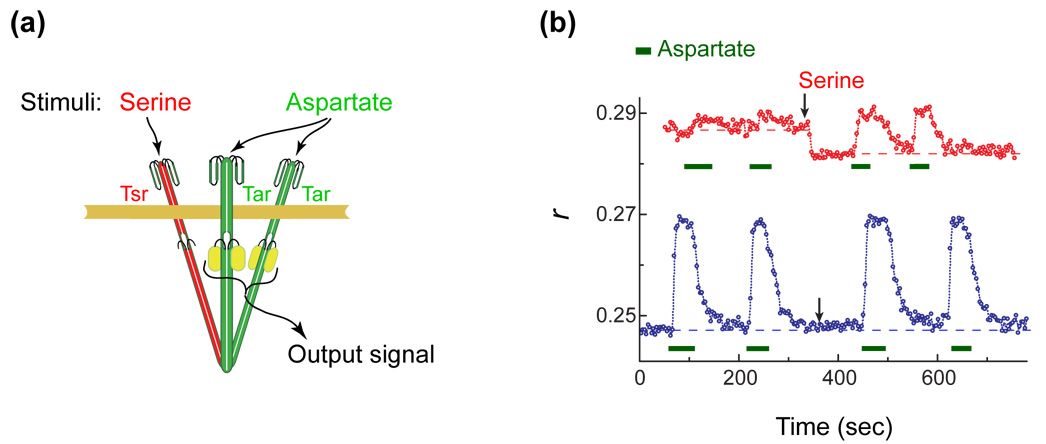
The effect of direct and indirect stimuli on Tar-YFP. (a) The experimental rationale. A mixed trimer is shown containing one Tsr dimer (red) and two Tar-YFP dimers (green). The conformation of the former can be affected directly by binding of serine and the conformation of the latter by binding of aspartate. The relative spacing between the Tar dimers is monitored by measuring the steady-state polarization of the emitted fluorescence, represented here by the fluorescence anisotropy (r). Changes in the fluorescence anisotropy are due to homo-FRET between the YFPs at their C-termini; the anisotropy decreases if the YFPs move more closely together and increases if they move farther apart. The experimental setup for measuring fluorescence polarization was described earlier14. r is defined as (Ipar - Iper) / (Ipar + 2Iper), where Iper was corrected for imperfections of the optical system. Experiments were done at room temperature (22°C). (b) Anisotropy traces measured from flhD cells (VS117; a gift of Victor Sourjik) co-expressing Tar-YFP (induced using 17 µM IPTG from plasmid pAV45: pTrc99A vector carrying tarQQQQ[1-527]-yfpA206K) and different levels of Tsr (induced by different concentrations of sodium salicylate from plasmid pPA114, a gift of Sandy Parkinson). Blue symbols: a low level of Tsr (no sodium salicylate); red symbols: a higher level of Tsr (0.6 µM sodium salicylate). The absolute expression level of Tar was comparable to its native expression level14. At its higher induction (red trace) Tsr expression reached levels roughly four times that of Tar-YFP. Aspartate (500 µM) was added for the time intervals indicated by the green bars. Serine (500 µM) was added at the time indicated by the arrows and left in throughout. Dashed lines indicate the anisotropies observed in the absence of ligand and are a guide to the eye.
We measured the anisotropy in cells without other chemotaxis proteins (flhD cells) expressing different levels of Tsr, together with Tar-YFP, Fig. 1b. At low levels of Tsr expression (lower, blue trace) the addition of serine had little effect, while the anisotropy increased upon addition of aspartate, as observed previously15; the YFPs moved farther apart. At higher levels of Tsr expression (upper, red trace), the baseline anisotropy was higher and increased only marginally upon addition of aspartate, as expected from dilution of the YFP label in mixed trimers. However, serine lowered the anisotropy and increased the response to aspartate, as if binding of serine to Tsr in mixed trimers caused the Tar-YFPs to move closer together. Thus, the response of Tar-YFP homodimers to binding of serine to Tsr dimers in a mixed trimer, albeit small, was opposite of what one might expect.
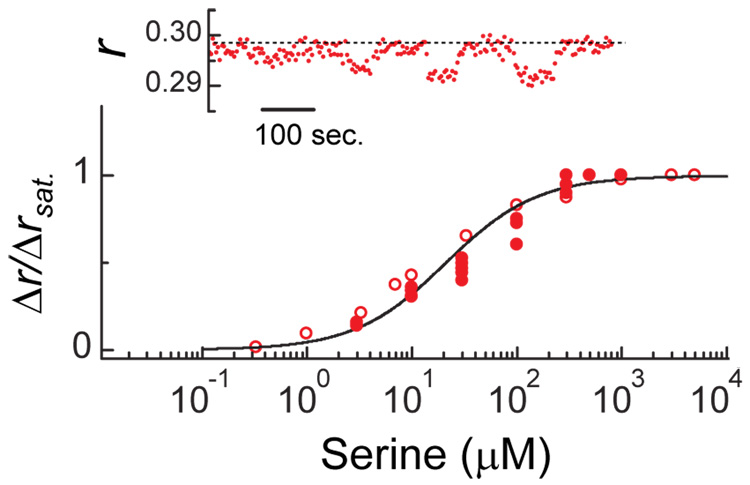
The size of the downward shift in anisotropy (Fig. 1b, red curve) increased with the concentration of serine, as shown in Fig. 2 (inset). The corresponding dose-response curve is shown in the main plot (filled circles) together with a similar dose-response curve measured with a pure population of Tsr-YFP receptors (empty circles) and the curve measured in vitro16 for binding of serine to Tsr (line). The near identity of these curves confirms that the downward shift observed with mixed trimers (Fig. 1b, red curve; and, Fig. 2, inset) is due to the response of the Tsr homodimers to binding of serine, even though it is being read out through a conformational response of the Tar-YFP homodimers. We confirmed that the effect is not unique to Tsr by reversing the roles of Tar and Tsr (data not shown). We also confirmed that the effect appears when Tsr and Tar-YFP are expressed in a strain that is deleted for all other chemotaxis components (UU1581; data not shown). We obtained similar results with constructs where the flexible tail at the C-terminal end of Tar was either present or absent (when YFP was fused to the C-terminus of wildtype Tar or to Tar1-527, missing 27 C-terminal residues15), confirming that the effect does not rely on a possibly large effective volume occupied by the YFPs. Thus, we conclude that binding of ligand to one homodimer, e.g., Tsr, in a mixed trimer affects the conformations of the other homodimers, e.g., Tar. However, it does so in a manner that is qualitatively different from that triggered by direct binding of ligand to Tar. Finally, we find that when both the indirect and direct stimuli are present (e.g. both serine and aspartate) the direct stimulus is dominant (Fig. 1b, red curve).
Figure 2.
The dependence of the downward anisotropy shift of Fig. 1b on the concentration of serine. The inset shows the raw data for one sequence with cells similar to those of Fig. 1b in which the concentration of serine was toggled between 0 and 10, 30, 100, and 300 µM, respectively; the dashed line indicates the anisotropy observed in the absence of ligand and is a guide to the eye. The main plot shows dose-response curves: closed symbols, results obtained from experiments of the kind shown in the inset; open symbols, results obtained from the same strain expressing only Tsr-YFP (induced using 5 µM IPTG from plasmid pAV29: pTrc99A vector carrying tsr–yfpA206K ); line, an in vitro binding curve of serine to Tsr (ref. 16: Kd=20 µM, Hill coefficient=1).
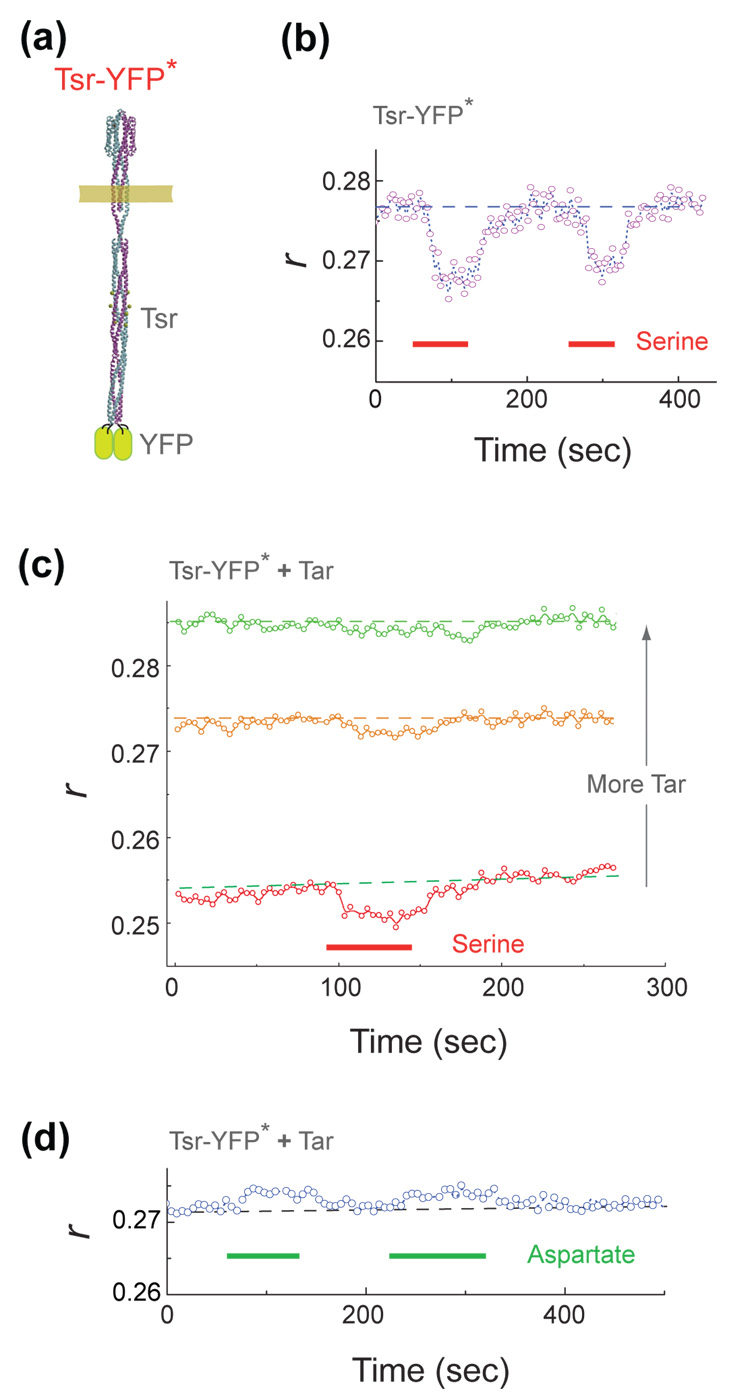
In principle, an alternative interpretation of these results could be that the changes in anisotropy are due to dissociation of receptor oligomers upon ligand binding. Although evidence has been presented elsewhere against such a view (see discussion in ref. 15 and references therein), we further tested this possibility here by varying the location of YFP within the receptor structure. If receptor oligomers dissociate, the efficiency of energy transfer between YFP’s on different dimers should markedly decrease, regardless of where the YFPs are placed. However, if the response is due to conformational changes within the trimers, the sign and/or magnitude of the response should depend upon the position and orientation of the YFP, and thus could be different at different locations. We inserted YFP into the cytoplasmic tip of a full-length Tsr receptor at position 390 (for details see the legend of Fig. 3). This construct is termed Tsr-YFP* and is shown schematically in Fig. 3a. When cells expressing this construct where stimulated with serine, a clear anisotropy response was observed (Fig. 3b) but its sign was opposite of that found with the C-terminal fusion to Tsr. When Tsr-YFP* was co-expressed with increasing levels of unlabeled Tar, the baseline anisotropy increased and the amplitude of the response to addition of serine decreased (Fig. 3c), as expected from dilution of the YFP label within mixed oligomers. When cells expressing Tsr-YFP* with unlabeled Tar at an intermediate level were stimulated by addition of aspartate, an anisotropy response also was observed (Fig. 3d) but again with a sign opposite of that seen with serine (Fig. 3b). Thus, overall, the YFP insertion at the distal cytoplasmic tip of the dimers had a behaviour qualitatively similar to that observed with YFP close to the membrane at the C-terminal end of the receptor, but with smaller amplitude and opposite sign. These findings provide strong evidence for the view that the changes in anisotropy seen in our experiments reflect conformational changes in receptor oligomers rather than dissociation of these oligomers.
Figure 3.
Anisotropy traces with YFP fused to the cytoplasmic tip of a receptor homodimer. (a) The position of YFP in the construct Tsr-YFP*: tsr1-390–yfpA206K –tsr390-end cloned into pTrc99A vector to form plasmid pAV48. (b) An anisotropy trace (see legend of Fig. 1) recorded from flhD cells expressing Tsr-YFP* (induced using 5 µM IPTG). Serine was added for the time intervals indicated by the red bars. (c) Similar traces recorded from cells co-expressing Tsr-YFP* with increasing levels of Tar (induced using increasing concentration of sodium salicylate from plasmid pLC113, a gift of Sandy Parkinson): reading from bottom to top: 0, 0.5, and 1.2 µM sodium salicylate. Cells were stimulated by addition of serine for the time interval indicated by the red bar. (d) Anisotropy traces recorded from cells co-expressing Tsr-YFP* with an intermediate level of Tar (0.7 µM sodium salicylate) and stimulated by addition of aspartate for the time intervals indicated by the green bars. Dashed lines indicate the anisotropies observed in the absence of ligand and are a guide to the eye.
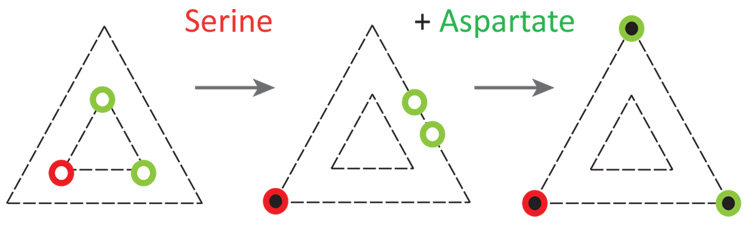
A few qualitative characteristics of trimer responses are illustrated in Fig. 4, which shows the trimer of Fig. 1aviewed from the periplasmic side. In the absence of any ligand the trimer is in the active conformation, with the C-terminal ends of the homodimers close to one another (left image). Ignoring possible structural differences between Tar and Tsr, this conformation would be expected to have a three-fold symmetry. When the ligand binding sites are fully occupied, the trimer switches to the inactive conformation, with the C-terminal ends of the homodimers farther apart (right image). This conformation also has a three-fold symmetry. However, when the ligand binding sites are partially occupied, for example when only serine is bound to Tsr, the symmetry of the trimer is broken, and an asymmetric conformation is possible (middle image). To account for the data presented in Fig. 1b, the C-terminal ends of the Tar homodimers should be closer together than they are in the absence of serine. On the other hand, to account for the monotonic change in anisotropy with ligand concentration observed with homogeneous Tar-YFP trimers or with homogeneous Tsr-YFP trimers (Fig. 2, open symbols), the mean distance between dimers should be larger than observed in the absence of ligand. These constraints are met by the middle image of Fig. 4. It is clear that the actual conformational change of trimers is more complex, as indicated by the differences in sign of the anisotropy responses of the Tsr-YFP and Tsr-YFP* constructs (see also ref. 17); however, the requirement for qualitatively different conformations in the partially or fully bound state should be general. This requirement means, in particular, that trimers on their own do not form tightly coupled ‘two-state’ allosteric units of the sort proposed18–20 to explain recent measurements of kinase activity11,15,21–23. Yet, the conformations of neighbouring dimers are coupled, supporting the view5,24–26 that dimers act collaboratively in trimers to propagate signals to the associated kinase proteins. Additional contributions to receptor coupling could emerge in fully assembled receptor clusters, from direct trimer-trimer interactions or via their association with CheW and CheA.
Figure 4.
Characteristics of trimer responses. A trimer containing one Tsr dimer (red) and two Tar dimers (green) is shown viewed from the periplasmic side. In the absence of any ligand, the trimer has three-fold symmetry (left image). Upon binding of serine to Tsr, the three-fold symmetry is broken (middle image). Upon binding of aspartate to Tar, the three-fold symmetry is restored (right image). The results presented in this paper are explained if the Tar dimers in the middle image are closer together than they are in the left image, while the mean distances between dimers are ranked in the order smallest (left), intermediate (middle), and largest (right), as shown.
Acknowledgements
We thank V. Sourjik and J.S. Parkinson for strains and plasmids, T.S. Shimizu for helpful conversations. This work was supported by a grant from the US NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baker MD, Wolanin PM, Stock JB. Systems biology of bacterial chemotaxis. Curr. Opin. Microbiol. 2006;9:187–192. doi: 10.1016/j.mib.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Berg HC. E. coli in Motion. NY: Springer; 2004. [Google Scholar]
- 3.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 5.Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Kentner D, Thiem S, Hildenbeutel M, Sourjik V. Determinants of chemorecptor cluster formation in Escherichia coli. Mol. Microbiol. 2006;61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 7.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 8.Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 9.Bray D, Duke T. Conformational spread: the propagation of allosteric states in large multiprotein complexes. Annu. Rev. of Biophys. Biomol. Struct. 2004;33:53–73. doi: 10.1146/annurev.biophys.33.110502.132703. [DOI] [PubMed] [Google Scholar]
- 10.Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 11.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 12.Lai RZ, Manson JMB, Bormans AF, Draheim RR, Nguyen NT, Manson MD. Cooperative signaling among bacterial chemoreceptors. Biochemistry. 2005;44:14298–14307. doi: 10.1021/bi050567y. [DOI] [PubMed] [Google Scholar]
- 13.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaknin A, Berg HC. Osmotic stress mechanically perturbs chemoreceptors in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2006;103:592–596. doi: 10.1073/pnas.0510047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J. Mol. Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levit MN, Stock JB. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J. Biol. Chem. 2002;277:36760–36765. doi: 10.1074/jbc.M204325200. [DOI] [PubMed] [Google Scholar]
- 17.Irieda H, Homma M, Homma M, Kawagishi I. Control of chemotactic signal gain via modulation of a pre-formed receptor array. J. Biol. Chem. 2006;281:23880–23886. doi: 10.1074/jbc.M600018200. [DOI] [PubMed] [Google Scholar]
- 18.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc. Natl. Acad. Sci. USA. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mello BA, Tu Y. An allosteric model for heterogeneous receptor complexes: understanding bacterial chemotaxis responses to multiple stimuli. Proc. Natl. Acad. Sci. USA. 2005;102:17354–17359. doi: 10.1073/pnas.0506961102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skoge ML, Endres RG, Wingreen NS. Receptor-receptor coupling in bacterial chemotaxis: evidence for strongly coupled clusters. Biophys. J. 2006;90:4317–4326. doi: 10.1529/biophysj.105.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bornhorst JA, Falke JJ. Evidence that both ligand binding and covalent adaptation drive a two-state equilibrium in the aspartate receptor signaling complex. J. Gen. Physiol. 2001;118:639–710. doi: 10.1085/jgp.118.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 23.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]