To the Editor:
The inner ear contains many different cell types that are necessary for sound detection [Frolenkov et al., 2004; Forge and Wright, 2002]. Because of the small quantity of cells present in the cochlea, classical biochemical and physiological approaches to characterize hearing processes in humans are often not feasible. A genetic approach to identify the molecular players in auditory processes is an alternative strategy and large consanguineous families with inherited hearing impairment have been a key to the mapping and identification of the majority of the mutated genes associated with deafness [Friedman and Griffith, 2003]. Inherited hearing loss is genetically heterogeneous and presently there are about 40 loci for recessively-inherited, nonsyndromic hearing loss (NSHL) reported in peer-reviewed journals, and 20 of the causative genes have been identified through positional cloning efforts [Hereditary Hearing Loss Homepage, http://www.uia.ac.be/dnalab/hhh].
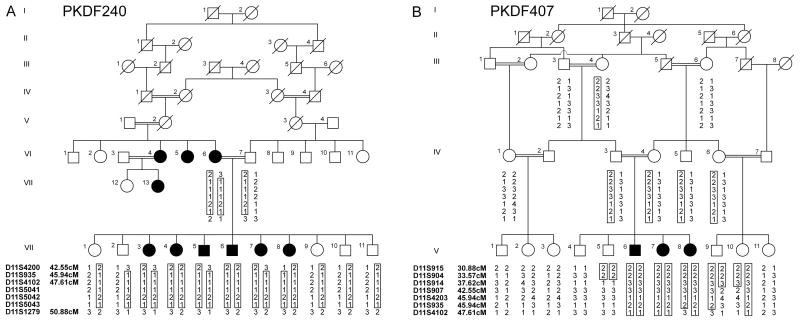
We are undertaking a saturating search for human genes that are necessary for the development of the inner ear and the maintenance of normal hearing. Here we report on mapping of a novel locus DFNB51 to chromosome 11p13-p12 following a whole genome wide scan in two consanguineous families (PKDF240 and PKDF407) segregating recessively inherited, profound congenital deafness (Fig. 1).
Fig. 1.
Pedigrees of families PKDF240 and PKDF407 segregating recessive deafness with haplotypes of markers at 11p13-p12. Filled and clear symbols represent affected and unaffected individuals, respectively. The core haplotypes are boxed representing the ancestral chromosome harboring DFNB51. A Haplotypes of PKDF240 revealed a 8.33 cM interval delimited by markers D11S4200 and D11S1279. Affected individuals VI:6, VII:3, VII:5, VII:7, and VII:8 provided the centromeric break point at marker D11S4200, while affected individuals VI:6 and her offspring provided the telomeric recombination at marker D11S1279. Spouses V:1 and VI:3 are thought to be distantly related cousins; however their relationship could not be confirmed. B Haplotypes of PKDF407 showing a homozygous region of 14.04 cM delimited by markers D11S904 and D11S4102. The STR markers and their relative positions in centiMorgan (cM) according to the Marshfield human genetic map [http://research.marshfieldclinic.org/genetics] are shown on the left side of the each pedigree.
Prior approval was obtained for this study from the IRBs at the National Centre of Excellence in Molecular Biology, Lahore, Pakistan (FWA00001758) and the NIDCD/NINDS at the National Institutes of Health, USA (OH-93-N-016). Signed informed consent was obtained from all participants of families PKDF240 and PKDF407, which were ascertained from rural Sindh and Punjab Provinces, respectively. Due to social customs, family members rarely marry individuals outside of their immediate family. Multiple family members were interviewed to construct the pedigrees and to confirm the relationships in these consanguineous families. Inheritance of deafness in both of the families is consistent with an autosomal recessive trait.
All of the participating members of families PKDF240 and PKDF407 underwent a physical examination. Clinical histories were obtained from all of the individuals to rule out environmental causes of hearing loss such as mumps, rubella, meningitis, ototoxic drugs or chronic otitis media. Pure tone audiometry tests for air conduction were performed at frequencies ranging from 250 to 8000 Hz. Funduscopic examination was conducted to determine gross ocular status and to rule out a retinopathy, while vestibular function was evaluated by using the tandem gait and Romberg tests. All affected individuals of these two families exhibited prelingual bilateral profound sensorineural hearing loss, with no obvious vestibular or ocular abnormalities. No other medical problem was found to co-segregate with the deafness.
Ten milliliters of venous blood samples were collected and genomic DNA was extracted following a standard protocol [Grimberg et al., 1989]. Short tandem repeat (STR) polymorphic markers were typed for the reported deafness loci [Hereditary Hearing Loss Homepage]. After excluding known autosomal recessive deafness loci (DFNB) loci, genome wide scans were initially undertaken on a small number of affected and unaffected members of families PKDF240 and PKDF407. We used panels 1 to 27 of the ABI Prism Linkage Mapping Set, v2.5 (Applied Biosystems) containing 388 fluorescently labeled microsatellite markers spaced at an average interval of 10 cM across the human genome. Markers were amplified by the polymerase chain reaction (PCR) on a Gene Amp PCR system 9700 (Applied Biosystems) and were analyzed on an ABI Prism 3100 Genetic Analyzer. The alleles were assigned by means of Genescan and Genotyper software (Applied Biosystems). For genomic intervals with a positive multipoint lod score, DNA samples from the entire family were retyped for a series of closely spaced markers.
Two-point and multi-point linkage analysis was performed using the MLINK and LINKMAP program, respectively of the FASTLINK implementation of LINKAGE [Schaffer, 1996]. Deafness in families was analyzed as an autosomal recessive trait with full penetrance, and the disease gene allele frequency was set at 0.0001. Meiotic recombination frequencies were considered equal for females and males. Ninety randomly selected normal individuals from the same population were genotyped to determine the allele frequencies for microsatellite markers.
Genome wide linkage analysis of family PKDF240 showed initial evidence of linkage on chromosome 11p13-p12. Additional STR markers were genotyped for all participating family members and haplotype analysis showed a region of homozygosity of 8.33 cM (Fig. 1A), delimited by markers D11S4200 (42.55 cM) and D11S1279 (50.88 cM). The proximal recombination at D11S4200 was given by individuals VI:6, VII:3, VII:5, VII:7, and VII:8 while distal breakpoint of the critical linked region at D11S1279 was provided by individuals VI:6, VII:3, VII:4, VII:5, VII:6, VII:7, and VII:8. A maximum multi-point lod score of 3.8 was obtained at marker D11S4102, while a two-point lod scores (Z max) of 3.5 at recombination fraction θ =0 was obtained at D11S4102 (see Table I).
TABLE I.
Two-Point Lod Scores
| Markers | Marshfield Map Position (cM) | Z max at θ = 0 | |
|---|---|---|---|
| PKDF240 | PKDF407 | ||
| D11S915 | 30.88 | - | -∞ |
| D11S904 | 33.57 | - | -∞ |
| D11S914 | 37.62 | - | 2.18 |
| D11S907 | 42.55 | - | 0.23 |
| D11S4200 | 42.55 | -∞ | - |
| D11S4203 | 45.94 | - | 2.28 |
| D11S935 | 45.94 | 3.38 | 2.50 |
| D11S4102 | 47.61 | 3.50 | -∞ |
| D11S5041 | 3.09 | - | |
| D11S5042 | 3.38 | - | |
| D11S5043 | 0.44 | - | |
| D11S1279 | 50.88 | -∞ | - |
Some markers in the region of homozygosity were not fully informative, thus yielding reduced positive lod scores.
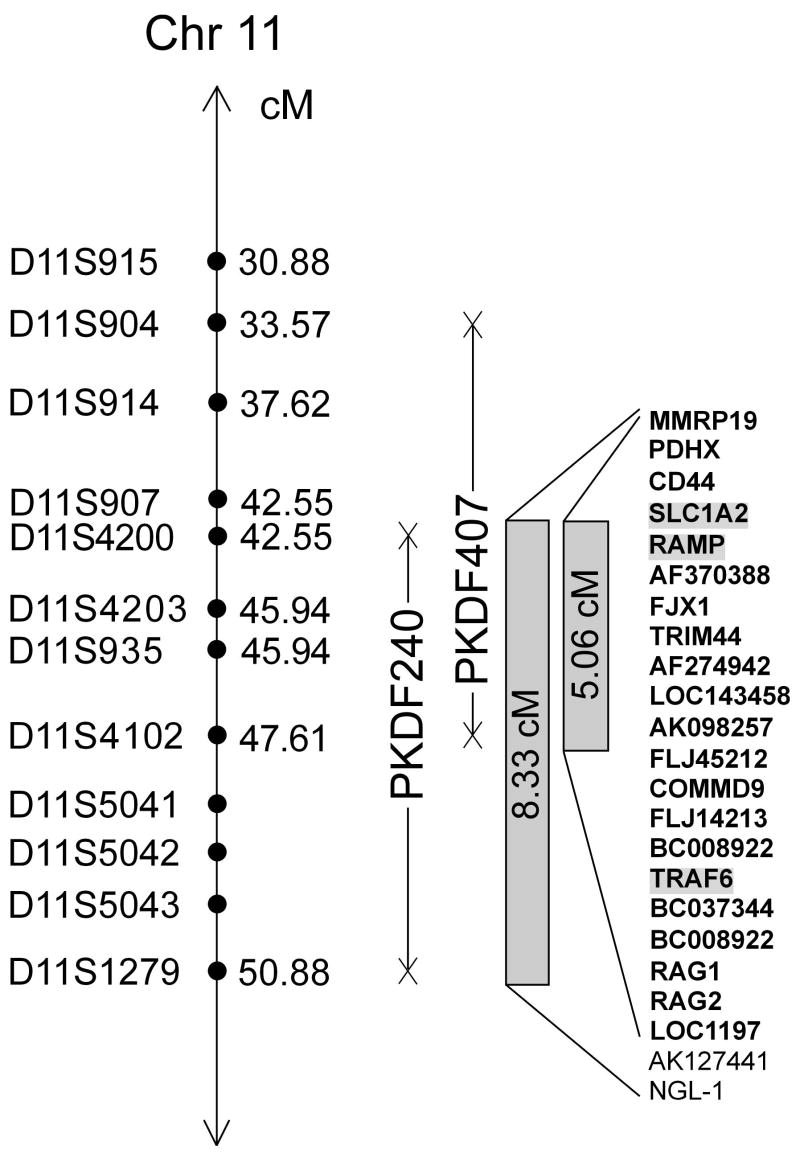
For deafness segregating in family PKDF407, evidence of linkage, although not rising to statistical significance (Z max of 2.5 for D11S935 at =0, see Table I), was found between markers D11S914 (37.62 cM) and D11S935 (45.94 cM) following a genome wide scan. Multi-point linkage analysis yielded a lod score of 2.6 at marker D11S935. After typing additional STR markers in the linkage region, haplotype analysis showed a region of homozygosity of approximately 14.04 cM (Fig. 1B) flanked by markers D11S904 (33.57 cM), a proximal boundary defined by normal hearing individual V:5 and D11S4102 (47.61 cM) a distal recombination given by individual V:8. If we assume the deafness segregating in both families PKDF240 and PKDF407 is caused by allelic mutations, haplotype analysis mapped the gene distal to D11S4200 (42.55 cM) and proximal to D11S4102 (47.61cM), delineating a genetic interval of approximately 5.06 cM (Fig. 2). Most likely the two mutant alleles will not be the same, as the haplotypes in the DFNB51 interval for the two families are different (Fig. 1). An alternative possibility is that there are two closely linked deafness loci.
Fig. 2.
Schematic representation of the DFNB51 interval on chromosome 11p13-p12 showing STR markers (
 ) and meiotic recombinations (X). Solid vertical lines represent the genetic intervals in which affected individuals are homozygous for the STR markers. Based on analyses of family PKDF240, DFNB51 resides in a critical interval of approximately 8.33 cM, delimited by D11S4200 (42.55 cM) and D11S1279 (50.88 cM). There are 23 genes annotated in the DFNB51 interval [UCSC Genome Browser, http://genome.ucsc.edu]. Genes in the 5.06 cM DFNB51 interval are shown in bold font. Genes that are highlighted were sequenced and no mutations were found. Gene symbols were approved by the HUGO Gene Nomenclature Committee [Povey et al., 2001].
) and meiotic recombinations (X). Solid vertical lines represent the genetic intervals in which affected individuals are homozygous for the STR markers. Based on analyses of family PKDF240, DFNB51 resides in a critical interval of approximately 8.33 cM, delimited by D11S4200 (42.55 cM) and D11S1279 (50.88 cM). There are 23 genes annotated in the DFNB51 interval [UCSC Genome Browser, http://genome.ucsc.edu]. Genes in the 5.06 cM DFNB51 interval are shown in bold font. Genes that are highlighted were sequenced and no mutations were found. Gene symbols were approved by the HUGO Gene Nomenclature Committee [Povey et al., 2001].
To date, several NSHL and syndromic hearing loss loci including DFNB2/A11/USH1B, DFNB18/USH1C, DFNB20, DFNB21/A8/A12, DFNB24, and DFNA32, as well as Jervell and Lange-Nielsen syndrome type 1 (JLNS1) have been localized on chromosome 11 [Friedman and Griffith, 2003]. However, all of the above nonsyndromic and syndromic deafness loci and genes are located outside of the DFNB51 genetic interval. Moreover, there is not a reported deaf mouse model mapped to the syntenic region on mouse chromosome 2 corresponding to human chromosome 11p13-p12.
Approximately 21 genes reside in the DFNB51 interval (5.06 cM) including candidates such as CD44, SLC1A2, RAMP, and TRAF6 (Fig. 3). CD44 belongs to a family of transmembrane glycoproteins and is considered to be one of the major hyaluronic acid receptors [Underhill, 1992]. Previous studies have indicated that the intracellular domain of CD44 binds to certain cytoskeletal proteins such as ankyrin, and ezrin, moesin, and radixin [Hirao et al., 1996; Bretscher, 1999; Thorne et al., 2004]. Recently it was reported that radixin deficiency in a mouse knock out of this gene is associated with deafness [Pataky et al., 2004; Kitajiri et al., 2004].
Another interesting candidate is SLC1A2 (GLT1), which encodes a sodium-dependent glutamate/aspartate transporter, and is a member of a super family of transporters [Kanai et al., 2004]. Lethal spontaneous seizures and increased susceptibility to acute cortical injury was observed in a SLC1A2 knock out mouse, but deafness was not reported [Tanaka et al., 1997]. Pendred syndrome, DFNB4, and EVA (Enlarged Vestibular Aqueduct) are allelic disorders caused by mutations of SLC26A4 [Everett et al., 1997; Li et al., 1998; Usami et al., 1999]. Mutations of SLC26A4 contribute to approximately 10% of hereditary deafness in diverse populations including east and south Asians [Park et al., 2003].
RAMP has three major domains, one of which predicts serine protease activity. So far, TMPRSS3 is the only gene encoding a serine protease which when mutated is involved in the etiology of nonsyndromic deafness. Mutations of TMPRSS3 account for 1.8% of the hereditary deafness segregating in 449 families with this disorder that we have ascertained in Pakistan [Scott et al., 2001; Ben-Yosef et al., 2001; Ahmed et al., 2004].
In the linkage interval of 8.33 cM defined by family PKDF240, there is another interesting candidate NGL-1. The human NGL-1 encodes a transmembrane protein (NGL-1) expressed in embryonic and adult brain [Lin et al., 2003]. NGL-1 is reported to bind to two of the three PDZ domains of whirlin [Delprat et al., 2005]. In hair cells of the inner ear whirlin is transported to the tips of stereocilia by myosin XVa [Belyantseva et al., 2005]. Mutations of both WHRN, and MYO15A, encoding whirlin and myosin XVa, respectively, are associated with nonsyndromic, congenital, profound hearing loss [Mburu et al., 2003; Wang et al., 1998; Liburd et al., 2001].
Sequencing of three candidate genes SLC1A2, RAMP, and TRAF6 was accomplished as part of our still on going search for the causative gene at the new locus DFNB51. Coding regions of these genes were sequenced in a deaf and an unaffected individual from both families used to map DFNB51. No potentially causative variants were identified. The localization of DFNB51 is the first step in identifying the underlying deafness gene as well as a gene necessary for normal hearing.
Acknowledgments
We are grateful to the families for their participation in this study. We also thank Dennis Drayna, Karen Friderici, and Andrew Griffith for their critiques of our manuscript and Fazal Qadir for technical help. This study was supported by the Higher Education Commission, Islamabad, Pakistan; Ministry of Science and Technology, Islamabad, Pakistan, and by the International Centre for Genetic Engineering and Biotechnology, Trieste, Italy under project CRP/PAK02-01 (contract no. 02/013). Part of this study in the USA was supported by intramural funds from the National Institute on Deafness and Other Communication Disorders, NIH (1 ZO1 DC000035-07 and 1 ZO1 DC000039-07) to TBF.
References
- Ahmed ZM, Li XC, Powell SD, Riazuddin S, Young TL, Ramzan K, Ahmad Z, Luscombe S, Dhillon K, MacLaren L, Ploplis B, Shotland LI, Ives E, Riazuddin S, Friedman TB, Morell RJ, Wilcox ER. Characterization of a new full length TMPRSS3 isoform and identification of mutant alleles responsible for nonsyndromic recessive deafness in Newfoundland and Pakistan. BMC Med Genet. 2004;5:24. doi: 10.1186/1471-2350-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva IA, Boger ET, Naz S, Frolenkov GI, Sellers JR, Ahmed ZM, Griffith AJ, Friedman TB. Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat Cell Biol. 2005;7:148–156. doi: 10.1038/ncb1219. [DOI] [PubMed] [Google Scholar]
- Ben-Yosef T, Wattenhofer M, Riazuddin S, Ahmed ZM, Scott HS, Kudoh J, Shibuya K, Antonarakis SE, Bonne-Tamir B, Radhakrishna U, Naz S, Ahmed Z, Riazuddin S, Pandya A, Nance WE, Wilcox ER, Friedman TB, Morell RJ. Novel mutations of TMPRSS3 in four DFNB8/B10 families segregating congenital autosomal recessive deafness. J Med Genet. 2001;38:396–400. doi: 10.1136/jmg.38.6.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol. 1999;11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- Delprat B, Michel V, Goodyear R, Yamasaki Y, Michalski N, El-Amraoui A, Perfettini I, Legrain P, Richardson G, Hardelin JP, Petit C. Myosin XVa and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Hum Mol Genet. 2005;14:401–410. doi: 10.1093/hmg/ddi036. [DOI] [PubMed] [Google Scholar]
- Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, Adawi F, Hazani E, Nassir E, Baxevanis AD, Sheffield VC, Green ED. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat Genet. 1997;17:411–422. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- Forge A, Wright T. The molecular architecture of the inner ear. Br Med Bull. 2002;63:524. doi: 10.1093/bmb/63.1.5. [DOI] [PubMed] [Google Scholar]
- Friedman TB, Griffith AJ. Human nonsyndromic ensorineural deafness. Annu Rev Genomics Hum Genet. 2003;4:341–402. doi: 10.1146/annurev.genom.4.070802.110347. [DOI] [PubMed] [Google Scholar]
- Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet. 2004;5:489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Tsukita S. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch. 2004;447:469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- Kitajiri S, Fukumoto K, Hata M, Sasaki H, Katsuno T, Nakagawa T, Ito J, Tsukita S, Tsukita S. Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. J Cell Biol. 2004;166:559–570. doi: 10.1083/jcb.200402007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liburd N, Ghosh M, Riazuddin S, Naz S, Khan S, Ahmed Z, Riazuddin S, Liang Y, Menon PS, Smith T, Smith AC, Chen KS, Lupski JR, Wilcox ER, Potocki L, Friedman TB. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum Genet. 2001;109:535–541. doi: 10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- Lin JC, Ho WH, Gurney A, Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- Li XC, Everett LA, Lalwani AK, Desmukh D, Friedman TB, Green ED, Wilcox ER. A mutation in PDS causes non-syndromic recessive deafness. Nat Genet. 1998;18:215–217. doi: 10.1038/ng0398-215. [DOI] [PubMed] [Google Scholar]
- Mburu P, Mustapha M, Varela A, Weil D, El-Amraoui A, Holme RH, Rump A, Hardisty RE, Blanchard S, Coimbra RS, Perfettini I, Parkinson N, Mallon AM, Glenister P, Rogers MJ, Paige AJ, Moir L, Clay J, Rosenthal A, Liu XZ, Blanco G, Steel KP, Petit C, Brown SD. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet. 2003;34:421–428. doi: 10.1038/ng1208. [DOI] [PubMed] [Google Scholar]
- Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, Kim HN, Moon SK, Abe S, Tukamoto K, Riazuddin S, Kabra M, Erdenetungalag R, Radnaabazar J, Khan S, Pandya A, Usami SI, Nance WE, Wilcox ER, Riazuddin S, Griffith AJ. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003;40:242–248. doi: 10.1136/jmg.40.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataky F, Pironkova R, Hudspeth AJ. Radixin is a constituent of stereocilia in hair cells. Proc Natl Acad Sci U S A. 2004;101:2601–2606. doi: 10.1073/pnas.0308620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povey S, Lovering R, Bruford E, Wright M, Lush M, Wain H. The HUGO Gene Nomenclature Committee (HGNC) Hum Genet. 2001;109:678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- Schaffer AA. Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered. 1996;46:226–235. doi: 10.1159/000154358. [DOI] [PubMed] [Google Scholar]
- Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi SQ, Radhakrishna U, Papasavvas MP, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne-Tamir B, Antonarakis SE. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T, Ichihara N, Kikuchi T, Okuyama S, Kawashima N, Hori S, Takimoto M, Wada K. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- Thorne RF, Legg JW, Isacke CM. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J Cell Sci. 2004;117:373–380. doi: 10.1242/jcs.00954. [DOI] [PubMed] [Google Scholar]
- Underhill C. CD44: the hyaluronan receptor. J Cell Sci. 1992;03:293–298. doi: 10.1242/jcs.103.2.293. [DOI] [PubMed] [Google Scholar]
- Usami S, Abe S, Weston MD, Shinkawa H, Van Camp G, Kimberling WJ. Nonsyndromic hearing loss associated with enlarged vestibular aqueduct is caused by PDS mutations. Hum Genet. 1999;104:188–192. doi: 10.1007/s004390050933. [DOI] [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, Friedman TB. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280:1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]