Summary
A naturally occurring unusual amino acid, hypusine [Nε-(4-amino-2-hydroxybutyl)-lysine] is a component of a single cellular protein, eukaryotic translation initiation factor 5A (eIF5A). It is a modified lysine with structural contribution from the polyamine spermidine. Hypusine is formed in a novel posttranslational modification that involves two enzymes, deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH). eIF5A and deoxyhypusine/hypusine modification are essential for growth of eukaryotic cells. The hypusine synthetic pathway has evolved in eukaryotes and eIF5A, DHS and DOHH are highly conserved, suggesting maintenance of a fundamental cellular function of eIF5A through evolution. The unique feature of the hypusine modification is the strict specificity of the enzymes toward its substrate protein, eIF5A. Moreover, DHS exhibits a narrow specificity toward spermidine. In view of the extraordinary specificity and the requirement for hypusine-containing eIF5A for mammalian cell proliferation, eIF5A and the hypusine biosynthetic enzymes present new potential targets for intervention in aberrant cell proliferation.
Keywords: Hypusine, eIF5A, Posttranslational modification, Deoxyhypusine synthase, Deoxyhypusine hydroxylase, Polyamine
Introduction
Hypusine [Nε-(4-amino-2-hydroxybutyl)-lysine, or (2S, 9R)-2-11-diamino-9-hydroxy-7-azaundecanoic acid] was discovered from bovine brain extracts in 1971 by Shiba (Shiba et al., 1971) in their search for unusually basic, natural amino acids. Later hypusine was found in various animal tissues as a protein component as well as a free amino acid. After determination of its chemical structure, this amino acid was named hypusine, based on its two structural components, hydroxyputrescine and lysine (Shiba et al., 1971). However, the origin of the 4-amino-2-hydroxybutyl moiety, the mechanism of its synthesis and the source of hypusine were unknown. In 1981, we discovered that the aminobutyl side chain of hypusine is derived from spermidine and that hypusine synthesis occurs posttranslationally only in one cellular protein (Park et al., 1981). We later identified this hypusine-containing protein as eukaryotic translation initiation factor 5A (eIF5A, old nomenclature, eIF4D) (Cooper et al., 1983). There is no known pathway of its synthesis as a free amino acid. Instead, hypusine is formed exclusively by posttranslational modification involving two enzymatic steps (Fig. 1) (Park et al., 1982, 1996; Murphey and Gerner, 1987; Park, 2006). In the first step, deoxyhypusine synthase (DHS) catalyzes the transfer of the aminobutyl moiety of spermidine to one specific lysine residue of eIF5A precursor to form an intermediate, deoxyhypusine [Nε-(4-aminobutyl)-lysine] residue. This intermediate is subsequently hydroxylated by deoxyhypusine hydroxylase (DOHH) (Abbruzzese et al., 1986) to complete hypusine synthesis and eIF5A activation. In 1995, we and others reported purification and cloning of the first step enzyme, deoxyhypusine synthase (Joe et al., 1995; Tao and Chen, 1995). The identity of the second step enzyme, deoxyhypusine hydroxylase, remained elusive, until our recent identification of the previously reported LIA1 (ligand of eIF5A) (Thompson et al., 2003) as the DOHH gene, its cloning and characterization (Kim et al., 2006; Park et al., 2006). eIF5A and its deoxyhypusine/hypusine modification are vital for eukaryotic cell proliferation (Gerner et al., 1986; Byers et al., 1992; Chen and Liu, 1997; Chattopadhyay et al., 2003; Park, 2006). Availability of both enzymes of the hypusine synthesis pathway has opened new avenues for exploration of this critical cellular pathway as a target for intervention in hyperproliferative diseases. In a recent minireview, we have discussed the structures of eIF5A, deoxyhypusine synthase and deoxyhypusine hydroxylase and the enzymatic reaction mechanisms (Park, 2006). Therefore, in the current paper, we will cover areas not emphasized in the previous review.
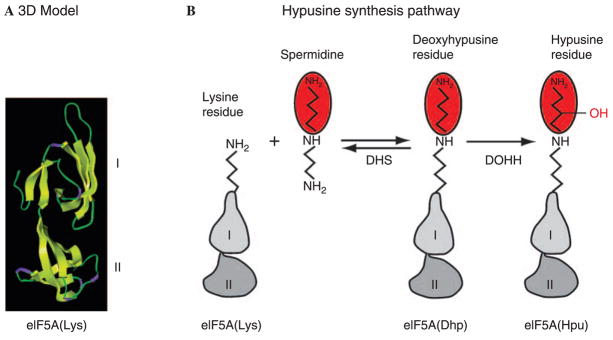
Fig. 1.
Model structure of eIF5A (A) and the hypusine synthesis pathway (B). eIF5A is comprised of two domains with β-sheet structures (based on Facchiano et al., 2001). Domain I (N-terminal domain) and II (C-terminal domain) are connected by a hinge (around Pro82 for the human protein). Hypusine synthesis occurs at one specific lysine residue (Lys50 for human protein) located on an exposed loop of the N-terminal domain. Side chain structures of lysine, deoxyhypusine and hypusine are shown. Deoxyhypusine synthase (DHS) reaction is reversible, whereas the deoxyhypusine hydroxylase (DOHH) reaction is not
Evolutionary progression of the hypusine synthesis pathway
The great mystery of the hypusine modification is that nature has evolved a unique posttranslational mechanism involving two new enzymes just to modify one cellular protein. The emergence of the hypusine biosynthetic pathway and its requirement for cell viability and growth show an interesting development in the course of evolution. Hypusine, eIF5A protein, DHS and DOHH occur in all eukaryotes. Archaea contains an eIF5A homolog protein, termed archaeal initiation factor 5A (aIF5A), and DHS, but a DOHH homolog has not yet been detected in a proteome or genome of this kingdom (Park et al., 2006). Despite the lack of evidence for DOHH, deoxyhypusine, hypusine or both, were reported to occur in archaea species (Bartig et al., 1990). In those species supposedly containing hypusine, it is curious how hypusine can be produced. In contrast to eukaryotes and archaea, there is no evidence for the occurrence of deoxyhypusine or hypusine in eubacteria. Although a distant ortholog of eIF5A, elongation factor P (EF-P), which is an essential bacterial protein that stimulates the peptidyl transferase activity of ribosomes, exists in eubacteria, DHS is not found in most commonly studied bacteria. However, a detailed phylogenetic analysis suggests that several bacterial species contain DHS cognate genes presumably transferred from archaea by horizontal gene transfer (Brochier et al., 2004). It is not known whether EF-P undergoes deoxyhypusine modification in those bacteria that contain a DHS cognate, although it seems unlikely (Brochier et al., 2004). These findings provide evidence that eIF5A and the two modification enzymes have evolved sequentially, but in an independent manner without a tight co-evolutionary linkage between them.
A significant amino acid sequence identity is observed among the eukaryotic protein, eIF5A, and its orthologs, aIF5A and EF-P. Of greater significance is the structural conservation between EF-P and aIF5A (Hanawa-Suetsugu et al., 2004). Crystal structures have been determined for three aIF5A proteins and two Leishmania eIF5A proteins that exhibit only minor differences (PDB 1EIF, 1IZ6, 1BKB, 1X60, 1XTD). Other eukaryotic proteins are also predicted to have a similar structure (Facchiano et al., 2001; Costa-Neto et al., 2006) consisting of two domains: a basic N-terminal domain with the lysine residue that undergoes deoxyhypusine/hypusine modification in an exposed loop and an acidic C-terminal domain that resembles oligonucleotide-binding fold (Fig. 1A). The structure of aIF5A is superimposable on that of the first two (of three) domains of EF-P (Hanawa-Suetsugu et al., 2004). In addition to the similarity of activities of EF-P and eIF5As in in vitro assays, the structural similarities further suggest functional conservation between the eubacterial, archaeal and eukaryotic proteins.
The requirement for eIF5A and the deoxyhypusine/hypusine modification for eukaryotic cell proliferation has been established by genetic manipulations. Inactivation of the aerobic eIF5A gene (TIF51A) (Wöhl et al., 1993) the two eIF5A genes (TIF51A and TIF51B) (Schnier et al., 1991) or the single deoxyhypusine synthase gene (Sasaki et al., 1996; Park, 2006) in Saccharomyces cerevisiae is lethal. In contrast, DOHH null strain of S. cerevisiae is viable (Thompson et al., 2003; Weir and Yaffe, 2004), although the growth rate is slightly lower in the mutant than in the parent strain (Park et al., 2006). This finding indicates that the deoxyhypusine-containing form of eIF5A can perform the basic cellular function(s) of eIF5A necessary for the survival of this budding yeast. In case of the fission yeast, S. pombe, a mutation (E66K) in DOHH caused altered morphology, abnormal mitochondrial distribution and temperature sensitive growth, suggesting a structural function of DOHH or of mature, hydroxylated form of eIF5A in this organism (Weir and Yaffe, 2004). In contrast to S. cerevisiae, suppression of expression of DOHH results in the recessive embryonic lethal phenotypes in C. elegans (Sugimoto, 2004) or Drosophila melanogaster (Spradling et al., 1999) indicating the requirement for fully modified, hypusine-containing eIF5A in higher multicellular eukaryotes. Both human eIF5A isoforms (isoforms 1 and 2) can support the growth of eIF5A null strain of S. cerevisiae (Clement et al., 2003), indicating that the fundamental function of eIF5A has been conserved from yeast to human. The finding that this hydroxylation is vital in the higher eukaryotes suggests either a specialized eIF5A function and/or a critical hydroxylation-dependent regulation mechanism involving eIF5A. It is also conceivable that eIF5A in higher eukaryotes is multi-functional and performs a hydroxylation-dependent function in addition to a hydroxylation-independent basic function common to yeast and human.
Sequence conservation of eIF5A, DHS and DOHH in eukaryotes
eIF5A and the hypusine biosynthetic enzymes, DHS and DOHH occur in all eukaryotes and are highly conserved as illustrated in Fig. 2A–C. The high sequence conservation of each of the three proteins may have been dictated by the structural requirement of eIF5A for its interaction with cellular effector molecules as well as with the modification enzymes. Two or more genes encoding eIF5A isoforms have been identified in various eukaryotes including fungi, plants, vertebrates and mammals. In yeast, two eIF5A genes, TIF51A (aerobic gene) and TIF51B (anaerobic gene) are reciprocally regulated by oxygen, while the two isoform proteins are functionally indistinguishable. In fish, amphibians and chicken, the two eIF5A genes appear to be co-expressed (Clement et al., 2003). In contrast, most mammalian cells and tissues constitutively express only the predominant isoform, eIF5A-1. Although eIF5A-2 mRNA appears to be expressed in specific tissues like brain and testis (Jenkins et al., 2001), the second isoform protein is hardly detectable in normal cells or tissues (Clement et al., 2003). Overexpression of eIF5A-2 gene in certain ovarian cancer tissues and cells was observed, leading to a hypothetical role of eIF5A-2 as an oncogene (Guan et al., 2001). However, it is unknown whether eIF5A-2 has a specialized function distinct from that of eIF5A-1 in mammals.
Fig. 2.
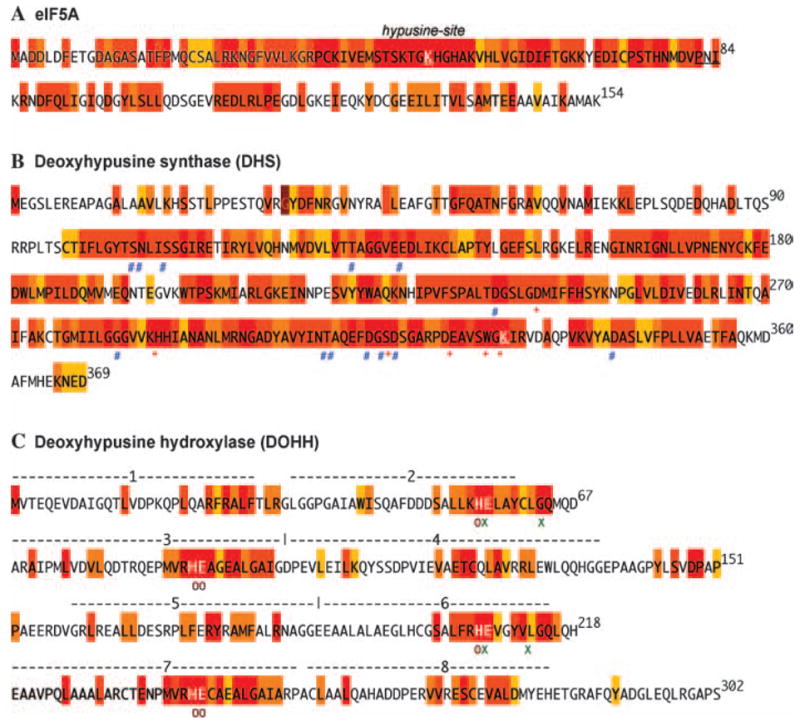
Amino acid sequence conservation of eIF5A, DHS and DOHH in eukaryotes. In each case (eIF5A-1, DHS and DOHH), the human sequence is shown. The degree of conservation is indicated by color coding: red, 100% identity; dark orange to yellow, conservative replacements (e.g. F, Y; D, E; K, R, L, V, I, M; T, S; A, G, C, S) with >80 to >50% sequence identity; white, no significant sequence identity. The diagram is based on eukaryotic sequences (40 for eIF5A, 36 for DHS and 14 for DOHH) chosen from a range of eukaryotic phyla and species. Critical amino acids for binding of substrates are shown by symbols under the residues
As shown in Fig. 2A, eIF5A is highly conserved in eukaryotes (with amino sequence identity of 63% between the human eIF5A-1 and S. cerevisiae eIF5A (TIF51A gene product). It is comprised of two domains, the N-terminal (aa 1–81) and C-terminal (aa 85–154) domains. The sequence identity is especially high in the region surrounding the hypusine modification site (Lys50 for the human protein) in the basic N-terminal domain. This finding suggests an importance of this site for the interaction of eIF5A with the modification enzymes or with its binding partners (proteins or RNA). Indeed, alanine substitution of the strictly conserved amino acids surrounding the modification site causes the loss of its substrate activity towards the modification enzymes or abolishes its ability to support the growth of yeast (G. Jeon, V. Oliveira, M. H. Park, unpublished results). The C-terminal domain, which is less highly conserved than the N-terminal domain, contains a putative oligonucleotide binding fold and has been proposed to be important for interaction with downstream effector molecules (Chen and Liu, 1997). Although the N-terminal domain alone is sufficient as a substrate for DHS and for DOHH as well (as will be discussed later, Fig. 4), both the N- and C-terminal core domains are required for its activity in supporting the growth of yeast (G. Jeon, V. Oliveira, M. H. Park, unpublished results).
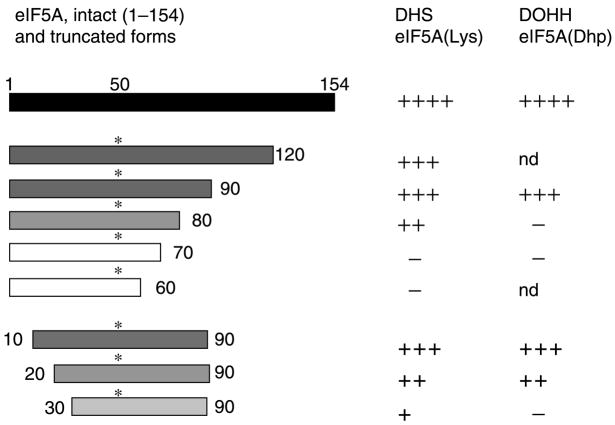
Fig. 4.
Structural requirements for eIF5A polypeptides as substrates for DHS and DOHH. Truncated polypeptides of eIF5A(Lys) and eI-F5A(Dhp) were used as substrates for DHS (Joe and Park, 1994) and DOHH (Kang et al., 2007), respectively. The relative effectiveness of truncated peptides as substrates, in comparison to the intact protein (1–154), is indicated as + + ++, + + +, ++, +, or −, where − denotes little or no substrate activity. nd Not determined
DHS is a tetrameric enzyme composed of four identical subunits of ~40 kDa (Umland et al., 2004; Park, 2006). In most eukaryotes, a single gene has been found for DHS, while a homolog gene encoding homospermidine synthase, that presumably was derived from the DHS gene, also exists in certain plants (Ober et al., 2003). Like eIF5A, DHS is highly conserved (Fig. 2B). The amino acid identity between the S. cerevisiae and human enzyme is 68%. The active site amino acid residues involved in the binding of its amine substrate spermidine (red*), and the cofactor NAD (blue #) have been identified from the crystal structure (Umland et al., 2004) and their roles were confirmed by site-directed mutagenesis (Lee et al., 2001). The active site lysine (K329) (Wolff et al., 1997), and the four of the five residues (Asp243, His288, Asp316, Glu323 and Trp327 Fig. 3) critical for binding of spermidine, are strictly conserved. Alanine substitution of any single amino acid of these residues abolishes spermidine binding and DHS activity. The amino acid residues predicted to be involved in the binding of NAD (indicated by #) are also highly conserved. All the alanine substitution mutants of the spermidine binding site and NAD binding site and K329A were all capable of forming stable complexes with eIF5A(Lys), as evidenced by gel mobility shifts of the complexes (Lee et al., 2001), indicating that these residues are not critically involved in the binding of the substrate protein. In spite of evidence for the formation of a stable complex between DHS and eIF5A(Lys) or eIF5A(Dhp), crystal structures have not been determined for these complexes. The amino acid residues of both proteins involved in the binding have yet to be identified.
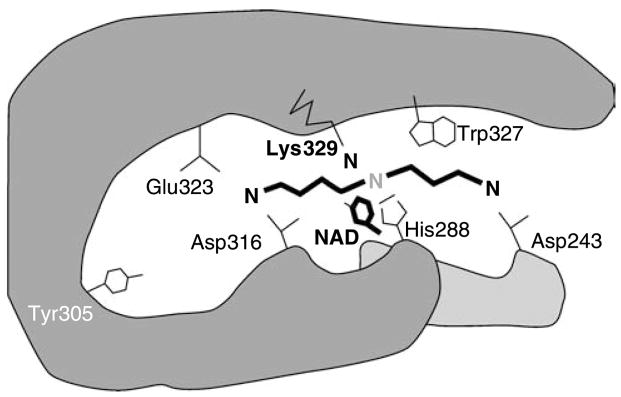
Fig. 3.
Proposed mode of spermidine binding at the active site of human deoxyhypusine synthase. The amino acid residues of the spermidine binding site of human DHS are shown. Upon NAD-dependent dehydrogenation of spermidine between the secondary nitrogen and carbon 5, the butylimine moiety is transferred to the ε-amino group of Lys329 to form a covalent enzyme intermediate. Subsequently, the butylimine moiety is transferred from the enzyme intermediate to the substrate protein. Spermidine is predicted to be anchored at its terminal amino groups by the acidic amino acids, Glu323, Asp316, and Asp243. Hydrophobic amino acid Trp327 is presumed to interact with the methylene chain of spermidine. The diagram was constructed based on the crystal structures of human DHS and is consistent with the results of mechanism and site-directed mutagenesis studies. Modified from Lee et al. (2001)
DOHH is also highly conserved (Fig. 2C) with 47% amino sequence identity between the S. cerevisiae and the human enzymes. A single DOHH homolog gene exists in most eukaryotes (Park et al., 2006). DOHH is a super helical protein with eight tandem HEAT-repeats (alpha helical hairpin structure composed of ~50 amino acids) numbered 1–8 (Park et al., 2006). It is a metalloenzyme with two iron atoms coordinated by four strictly conserved His-Glu motifs (per enzyme monomer). The composition and structure of a native DOHH enzyme are as yet unknown and the cloned DOHH may present a catalytic subunit or a monomeric protein. DOHH monomer is a dyad of symmetrical N- (HEAT-repeats 1–4) and C-terminal (HEAT-repeats 5–8) domains connected by a variable loop. Although symmetrical, neither the N- nor the C-terminal domain alone shows any enzymatic activity. Both domains are required for binding of iron, and the substrate protein, eIF5A(Dhp), and for deoxyhypusine hydroxylation. The amino acid residues identified to be critical for binding of iron (His56, His89, Glu90, His207, His240 and Glu241 indicated by red ○) (Kim et al., 2006) and for eIF5A(Dhp) binding (Glu57, Glu208, and Gly63 and Gly214 indicated by green ×) (Kang et al., 2007), are strictly conserved, indicating their importance in DOHH catalysis. A number of other strictly conserved amino acid residues are also noted, but how these other amino acid residues may contribute to DOHH structure or function (enzymatic or structural), has yet to be determined.
Specificity of deoxyhypusine synthase: spermidine analogs as inhibitors and/or substrates of DHS
The first step in the DHS reaction is a NAD-dependent dehydrogenation of spermidine with subsequent transfer of the aminobutyl moiety from spermidine to eIF5A(Lys) (Park, 2006). This enzyme exhibits a narrow specificity toward spermidine, NAD and the protein substrate. Initially, spermidine binding site of DHS was probed using the partially purified enzyme from rat testis and a series of diamines, polyamines and spermidine analogs as inhibitors (Table 1A) (Jakus et al., 1993). Of these compounds, 1,7-diaminoheptane and 1,8-diaminooctane were the most effective inhibitors with Ki values (5.6 and 6.2 μM, respectively) similar to the Km for spermidine (4.5 μM). Alkylation of either of the two terminal amino groups of spermidine markedly reduces its affinity for DHS, indicating that the two-terminal amino groups spaced by 7–8 methylenes are important for binding. Caldine, homospermidine and aminopropylcadaverine were less effective than the two diamines, 1,7-diaminoheptane and 1,8-diaminooctane, indicating that the secondary amino group of the polyamines does not contribute to their binding to the enzyme active site. Therefore, inhibition of DHS by 1,3-diaminopropane or putrescine is not likely due to their competition with spermidine for binding to the enzyme. Rather, this inhibition may result from the enhancement of the reversal of deoxyhypusine synthesis reaction by these two amines, which act as alternate acceptors of the aminobutyl moiety from the enzyme-imine intermediate (Park et al., 2003). Attachment of various bulky groups at the terminal amino group or at the secondary amino group of spermidine reduced the inhibitory activity, indicating a narrow channel of spermidine binding site. One exception was monoguanylation of the terminal amino groups, which increases the basicity at one amino terminal. Monoguanyl derivatives of spermidine, 1,7-diaminoheptane and 1,8-diaminooctane were much more effective than the parent counterparts in the inhibition of DHS. The most effective inhibitors of DHS, thus far identified, are N1-guanyl-1,7-diaminoheptane (GC7) and its methyl derivative (7-methyl GC7, or 7-amino-1-guanidinooctane) (Ki value for GC7, 0.01 μM, compared to Km for spermidine, 4.5 μM). Consistent with the potency of the inhibition of DHS in vitro, GC7 also effectively inhibits hypusine synthesis in cultured mammalian cells (Joe and Park, 1994). GC7 and its analogs exerted strong antiproliferative effects in CHO cells and in various human cancer cell lines (Joe and Park, 1994). Although a close correlation was observed between the inhibition of hypusine synthesis and inhibition of cell growth by GC7 and its analogs, the growth inhibitory effects may not be entirely attributable to the reduction of hypusine-containing eIF5A.
Table 1.
The inhibitory (IC50 and Ki) and substrate activity of diamines and spermidine analogs toward deoxyhypusine synthase. In B, the structure of the product formed with eIF5A(Lys) as acceptor, and growth effects are shown. Modified from Jakus et al. (1993) (A) and Park et al. (2003) (B). IC50 is the concentration to achieve 50% inhibition under the stated experimental conditions
| A. Comparison of amine and guanyl analogs of spermidine as inhibitors of DHS
| ||||
|---|---|---|---|---|
| Compound | Structure | IC50 (μM) | Ki (μM) | |
| 1,2-diaminoethane |
|
–a | ||
| 1.3-diaminopropane |
|
6.3 | 4.0 ± 0.2 | |
| putrescine |
|
91 | ||
| cadaverine |
|
–a | ||
| 1,6-diaminohexane |
|
171 | ||
| 1,7-diaminoheptane |
|
8.1 | 5.6 ± 0.6 | |
| 1,8-diaminooctane |
|
9.1 | 6.2 ± 1.3 | |
| 1,9-diaminononane |
|
216 | 159 ± 16 | |
| caldine |
|
41.2 | 28 ± 4 | |
| spermidine |
|
7.8b | 4.5 ± 0.7c | |
| N-(3-aminopropyl)-cadaverine |
|
118 | 83 ± 7 | |
| spermine |
|
–a | ||
| agmatine |
|
71 | ||
| N1-guanyl-1,7-diaminoheptane |
|
0.017 | 0.0097 ± 0.0005 | |
| N1-guanyl-1,8-diaminooctane |
|
0.37 | 0.24 ± 0.02 | |
| N1-guanylcaldine |
|
1.2 | 0.74 ± 0.03 | |
| N1-guanylspermidine |
|
0.57 | 0.33 ± 0.05 | |
| N8-guanylspermidine |
|
0.24 | 0.15 ± 0.04 | |
| hirudonine |
|
7.45 | 4.89 ± 1.32 | |
|
B. Spermidine analogs as donor substrates for deoxyhypusine synthase
| ||||
| Compound (donor amine)d | Product (with eIF5A(Lys)) | IC50 (μM)
|
Growth supporti | |
|
| ||||
| Structureg | Name | |||
| Spermidine (Spd) (3,4) |
|
deoxyhypusine | 7.8 | + |
| Homospermidine (4,4) |
|
deoxyhypusine | 20 | n.d.j |
| Caldine (3,3) | – | – | 41.2 | n.d. |
| N-(3-aminopropyl)-cadaverine(3,5) |
|
homodeoxyhypusine | 118 | + |
| Compound (donor amine)d | Product (with eIF5A(Lys) | IC50 (mM) | Growth supporti | |
| Structureg | Name | |||
| cis = Spde |
|
cis =deoxyhypusinee | <10 | + |
| trans = Spdf |
|
trans =deoxyhypusinef | <3 | – |
| 1-MeSpd |
|
deoxyhypusine | 184 | + |
| 8-MeSpd |
|
Methyldeoxyhypusineh | –a | n.d. |
| N1EtSpd |
|
deoxyhypusine | –a | n.d. |
| N8EtSpd |
|
N-ethyldeoxyhypusineh | –a | n.d. |
| 5,5′-Me2Spd | – | – | –a | – |
Less than 50% inhibition at 1 mM
Apparent “inhibition” by unlabeled spermidine
Km for spermidine
Numbers in parentheses indicate number of methylene groups between amino groups
cis = Spd, cis unsaturated spermidine, N-(3-aminopropyl)1,4-diamino-cis-but-2-ene; cis = deoxyhypusine, Nε (4-amino-cis-but-2-ene)lysine
trans = Spd, trans unsaturated spermidine, N-(3-aminopropyl)1,4-diamino-trans-but-2-ene; trans = deoxyhypusine, Nε (4-amino-trans-but-2-ene)lysine
LysNH indicates the lysine residue of eIF5A with its epsilon amino group, which becomes a secondary amino group in deoxyhypusine and its derivatives
Detectable, but ≤3% of the amount of product from spermidine
Ability to support growth of spermidine-depleted mammalian cells (Byers et al., 1992, 1994; Park et al., 2003)
n.d. Not determined
The narrow spermidine binding site of DHS predicted from inhibitor studies using spermidine analogs was validated by determination of crystal structures of human recombinant deoxyhypusine synthase in a complex with NAD, and of a DHS ternary complex with NAD and GC7 (Umland et al., 2004). Four active sites were identified at the dimer–dimer interface of this tetrameric enzyme. The mode of binding of GC7 at this active site is consistent with the proposed mode of spermidine binding with the active site lysine residue (K329) positioned close to the secondary amino group of spermidine and Asp243, Asp316 and Glu323 anchoring its terminal amino groups (Fig. 3).
The absolute requirement for spermidine for survival and growth of S. cerevisiae hinges on the specificity of DHS toward spermidine and the cellular necessity for the modified eIF5A protein. Other natural amines putrescine or spermine cannot themselves fulfill this substrate function for hypusine modification, neither can they support the growth of eukaryotes in the absence of their conversion to spermidine (Chattopadhyay et al., 2003). Byers et al. reported evidence that cytostasis in the polyamine-depleted L1210 cells (by a treatment with an inhibitor of S-adenosyl-methionine decarboxylase 5′-5′ Deoxyadenosine (AbeAdo) was ultimately due to deprivation of hypusine-containing eIF5A (Byers et al., 1992, 1994). Whereas a number of polyamine analogs could substitute for natural polyamines in the polycationic requirement of cells, only a few compounds closely related to spermidine could substitute for spermidine as a substrate for DHS and in supporting cell proliferation (Table 1B). Of these spermidine analogs, all the compounds except 5,5′-dimethyl-spermidine were capable of forming covalent enzyme–imine intermediates (Park et al., 2003). Those that formed the enzyme-intermediates were able to form deoxyhypusine or its homologs, except caldine. It is noteworthy that there is no known spermidine analog that can support the growth of spermidine-depleted cells, without being a substrate for DHS. Aminopropyl cadaverine, cis unsaturated spermidine and 1-methylspermidine were reported to substitute for spermidine in supporting mammalian cell growth (Byers et al., 1992, 1994). However, trans unsaturated spermidine failed to support growth of spermidine-depleted cells, in spite of its ability to form a deoxyhypusine analog. This indication that eIF5A containing a variant deoxyhypusine analog side chain is biologically inactive, reflects an intricate structural requirement of hypusine side chain in the activity of eIF5A.
Specificity of DHS and DOHH toward their respective protein substrates
Perhaps the most unique characteristic of hypusine synthesis is its strict specificity toward the protein substrate. Of all the cellular proteins, only eIF5A isoform proteins undergo this posttranslational modification. This is readily demonstrated by culture of eukaryotic cells in the presence of [3H]spermidine and separation of cellular proteins by two dimensional gel electrophoresis (Park et al., 1981). One major spot of radiolabeled eIF5A protein is detected in most mammalian cells. In certain species like chicken embryo, which expresses two isoforms concomitantly, two radiolabeled eIF5A isoforms were detected. No other cellular protein containing hypusine has thus far been identified. Therefore, it is apparent that the modification enzyme binds and modifies only the eIF5A precursor protein. Deoxyhypusine synthase does not modify free lysine or short peptides with amino acid sequences corresponding to the modification site and deoxyhypusine hydroxylase does not hydroxylate free deoxyhypusine either. In order to determine the structural requirements of eIF5A substrate peptides for modification by DHS (Joe and Park, 1994) and DOHH (Kang et al., 2007), truncated eIF5A peptides were tested. As summarized in Fig. 4, both enzymes display similar structural requirements for their respective substrates, eIF5A(Lys) and eIF5A(Dhp). The amino terminal half (aa 1–83) of eIF5A seems to be sufficient for modification by both enzymes, since truncated peptide substrates (aa 1–90) were nearly as effective as the intact protein. However, further truncation from the C-terminal caused a significant loss of substrate activity, as shown for aa 1–80 and aa 1–70. In the case of N-terminal truncation, the deletion of 10 amino acids from the N-terminal did not affect the substrate activity (aa 10–90), whereas truncation of 20 or 30 amino acids caused a significant reduction (aa 20–90, and aa 30–90). From the predicted structure of human eIF5A (Facchiano et al., 2001), the N-terminal domain of human eIF5A contains a β-sheet core encompassing Ser15-Pro82. In this regard, preservation of an intact β-sheet core structure of the N-terminal domain may be important for these peptides to serve as substrates for DHS as well as for DOHH.
Although both enzymes exhibit a similar dependence on the eIF5A polypeptide length, there are fine differences to be noted between eIF5A/DHS and eIF5A/DOHH interactions. DHS forms stable complexes with the substrate, eIF5A(Lys), as well as with the product of the reaction, eIF5A(Dhp), suggesting that it binds both proteins tightly and that eIF5A/DHS binding does not depend on the modification status (Lys50, Dhp50 or Hpu50). This high affinity of DHS for both eIF5A(Lys) and eIF5A(Dhp) is consistent with the reversibility of the DHS reaction. In contrast, DOHH shows a strong preference for eIF5A(Dhp) over eIF5A(Lys) or eIF5A(Hpu) for binding, indicating that the substrate binding to DOHH depends on the deoxyhypusine side chain of eIF5A(Dhp) (Kang et al., 2007). Acidic amino acid residues at the active site of DOHH that are critical for anchoring the deoxyhypusine side chain of eIF5A(Dhp) have been identified by site-directed mutagenesis (Kang et al., 2007). However, there is no experimental evidence as yet concerning the sites of interaction between eIF5A(Lys) and DHS and their mode of binding. We are characterizing a number of mutant proteins of eIF5A and DHS to identify amino acid residues directly involved in eIF5A/DHS binding. The basis of the exclusive specificity of hypusine modification will be revealed upon determination of crystal structures of eIF5A(Lys)/DHS and eIF5A(Dhp)/DOHH complexes.
Concluding remarks
The essential nature and unique specificity of hypusine synthesis and the high conservation of eIF5A and its modification enzymes, DHS and DOHH, attest to the importance of this posttranslational modification. Although remarkable progress has been made in elucidating the biosynthetic pathway of hypusine, challenging questions remain to be answered on the physiological function of eIF5A and on how the deoxyhypusine/hypusine modification activates eIF5A. For example, addition of the 4-aminobutyl moiety or the 4-amino-2-hydroxybutyl moiety to eIF5A(Lys) may cause a conformational change in the eIF5A protein to promote its interaction with downstream effector molecules. Alternatively, the deoxyhypusine/hypusine site may serve as a focal contact point for downstream effectors, as exemplified in case of eIF5A(Dhp)/DOHH binding. It is also possible that eIF5A is regulated by further post-translational modification at the hypusine residue, such as phosphorylation or glycosylation, although no biochemical evidence for such additional modification has yet been obtained. In spite of the indispensable nature of the hypusine/deoxyhypusine modification in eukaryotic cell proliferation, the precise cellular function of eIF5A remains to be elucidated. eIF5A stimulates methionyl-puromycin synthesis in a model assay for translation initiation and partially associates with actively translating ribosomes (Jao and Chen, 2006; Zanelli et al., 2006). Thus, eIF5A is evidently involved in translational control, stimulating translation either at the first peptide bond formation or at the elongation step, similar to the role of bacterial EF-P. eIF5A has also been proposed to be a specific initiation factor for a subset of mRNAs (Kang and Hershey, 1994; Hanauske-Abel et al., 1995). From differential display analysis of eIF5A-associated mRNAs, a number of mRNAs that are potential targets of eIF5A regulation have been reported (Xu et al., 2004). Genetic studies using S. cerevisiae harboring eIF5A temperature-sensitive mutants suggest a direct or indirect role of eIF5A in cell wall integrity, mRNA decay, actin polarization, cell cycle progression and anti-apoptotic protection (Zuk and Jacobson, 1998; Valentini et al., 2002; Zanelli and Valentini, 2005; Chatterjee et al., 2006; Schrader et al., 2006). Future studies will be directed toward identification of eIF5A binding partners (proteins and mRNA) and understanding how downstream effector molecules are involved in the expression of these pleiotropic phenotypes in the temperature-sensitive eIF5A mutant strains.
Acknowledgments
The research in the authors’ laboratory was supported by the Intramural Research Program of National Institutes of Health (NIDCR).
Abbreviations
- aIF5A
archaeal initiation factor 5A
- DHS
deoxyhypusine synthase
- DOHH
deoxyhypusine hydroxylase
- EF-P
elongation factor P
- eIF5A
eukaryotic translation initiation factor 5A
- eIF5A-1
primary isoform of eIF5A
- eIF5A-2
secondary isoform of eIF5A
- eIF5A(Dhp)
eIF5A intermediate containing deoxyhypusine
- eIF5A(Hpu)
eIF5A active form containing hypusine
- eIF5A(Lys)
eIF5A precursor
- GC7
N1-guanyl-1,7-diaminoheptane
- HEAT-repeat
alpha-helical structural motif characteristic of Huntingtin, elongation factor 3E, a subunit of protein phosphatase 2A, and the target of rapamycin
- NAD
nicotinamide adenine dinucleotide
Footnotes
Only representative studies are cited; consult the reviews (Park et al., 1996; Chen and Liu, 1997; Park, 2006) for more citations.
References*
- Abbruzzese A, Park MH, Folk JE. Deoxyhypusine hydroxylase from rat testis: partial purification and characterization. J Biol Chem. 1986;261:3085–3089. [PubMed] [Google Scholar]
- Bartig D, Schumann H, Klink F. The unique posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the archebacterial kingdom. System Appl Microbiol. 1990;13:112–116. [Google Scholar]
- Brochier C, Lopez-Garcia P, Moreira D. Horizontal gene transfer and archaeal origin of deoxyhypusine synthase homologous genes in bacteria. Gene. 2004;330:169–176. doi: 10.1016/j.gene.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Byers TL, Ganem B, Pegg AE. Cytostasis induced in l1210 murine leukaemia cells by the S-adenosyl-L-methionine decarboxylase inhibitor 5′-([(Z)-4-amino-2-butenyl]methylamino)-5′-deoxyadenosine may be due to hypusine depletion. Biochem J. 1992;287:717–724. doi: 10.1042/bj2870717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers TL, Lakanen JR, Coward JK, Pegg AE. The role of hypusine depletion in cytostasis induced by S-adenosyl-L-methionine decarboxylase inhibition: new evidence provided by L-methylspermidine and 1,12-dimethylspermine. Biochem J. 1994;303:363–368. doi: 10.1042/bj3030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I, Gross SR, Kinzy TG, Chen KY. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol Genet Genomics. 2006;275:264–276. doi: 10.1007/s00438-005-0086-4. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H. Spermidine, but not spermine, is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the FMS1-amine oxidase. Proc Natl Acad Sci USA. 2003;100:13869–13874. doi: 10.1073/pnas.1835918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Liu AY. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol Signals. 1997;6:105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- Clement PC, Henderson CA, Jenkins ZA, Smit-McBride Z, Wolff EC, Hershey JWB, Park MH, Johansson HE. Identification and characterization of eukaryotic initiation factor 5A-2. Eur J Biochem. 2003;147:4254–4263. doi: 10.1046/j.1432-1033.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein Hy+ as translation initiation factor eIF-4D. Proc Natl Acad Sci USA. 1983;80:1854–1857. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Neto CM, Parreiras ESLT, Ruller R, Oliveira EB, Miranda A, Oliveira L, Ward RJ. Molecular modeling of the human eukaryotic translation initiation factor 5A (eIF5A) based on spectroscopic and computational analyses. Biochem Biophys Res Commun. 2006;347:634–640. doi: 10.1016/j.bbrc.2006.06.119. [DOI] [PubMed] [Google Scholar]
- Facchiano AM, Stiuso P, Chiusano ML, Caraglia M, Giuberti G, Marra M, Abbruzzese A, Colonna G. Homology modelling of the human eukaryotic initiation factor 5A (eIF5A) Protein Eng. 2001;14:881–890. doi: 10.1093/protein/14.11.881. [DOI] [PubMed] [Google Scholar]
- Gerner EW, Mamont PS, Bernhardt A, Siat M. Post-translational modification of the protein-synthesis initiation factor eIF-4D by spermidine in rat hepatoma cells. Biochem J. 1986;239:379–386. doi: 10.1042/bj2390379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 2001;61:3806–3809. [PubMed] [Google Scholar]
- Hanauske-Abel HM, Slowinska B, Zagulska S, Wilson RC, Staiano-Coico L, Hanauske AR, McCaffrey T, Szabo P. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary. Proposal of a role for eIF5A in onset of DNA replication. FEBS Lett. 1995;366:92–98. doi: 10.1016/0014-5793(95)00493-s. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci USA. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakus J, Wolff EC, Park MH, Folk JE. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono-guanylated diamines and polyamines. J Biol Chem. 1993;268:13151–13159. [PubMed] [Google Scholar]
- Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97:583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- Jenkins ZA, Haag PG, Johansson HE. Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics. 2001;71:101–109. doi: 10.1006/geno.2000.6418. [DOI] [PubMed] [Google Scholar]
- Joe YA, Park MH. Structural features of the eIF5A precursor required for posttranslational synthesis of deoxyhypusine. J Biol Chem. 1994;269:25916–25921. [PubMed] [Google Scholar]
- Joe YA, Wolff EC, Park MH. Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins. J Biol Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- Kang HA, Hershey JW. Effect of initiation factor 5A depletion on protein synthesis and of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- Kang KR, Kim YS, Wolff EC, Park MH. Specificity of the deoxyhypusine hydroxylase-eIF5A interaction: identification of amino acid residues of the enzyme required for binding of it substrate, deoxyhypusine-containing eIF5A. J Biol Chem. 2007;282:8300–8308. doi: 10.1074/jbc.M607495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kang KR, Wolff EC, Bell JK, McPhie P, Park MH. Deoxyhypusine hydroxylase is an Fe(II)-dependent, HEAT-repeat enzyme. Identification of amino acid residues critical for Fe(II) binding and catalysis. J Biol Chem. 2006;281:13217–13225. doi: 10.1074/jbc.M601081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Um PY, Park MH. Structure-function studies of human deoxyhypusine synthase: identification of amino acid residues critical for the binding of spermidine and NAD. Biochem J. 2001;355:841–849. doi: 10.1042/bj3550841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey RJ, Gerner EW. Hypusine formation in protein by a two-step process in cell lysates. J Biol Chem. 1987;262:15033–15036. [PubMed] [Google Scholar]
- Ober D, Harms R, Witte L, Hartmann T. Molecular evolution by change of function. Alkaloid-specific homospermidine synthase retained all properties of deoxyhypusine synthase except binding the eIF5A precursor protein. J Biol Chem. 2003;278:12805–12812. doi: 10.1074/jbc.M207112200. [DOI] [PubMed] [Google Scholar]
- Park J-H, Wolff EC, Folk JE, Park MH. Reversal of the deoxyhypusine synthesis reaction. Generation of spermidine or homospermidine from deoxyhypusine by deoxyhypusine synthase. J Biol Chem. 2003;278:32683–32691. doi: 10.1074/jbc.M304247200. [DOI] [PubMed] [Google Scholar]
- Park J-H, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc Natl Acad Sci USA. 2006;103:51–56. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci USA. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE. The biosynthesis of protein-bound hypusine (N-epsilon-(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N-epsilon-(4-aminobutyl)lysine) J Biol Chem. 1982;257:7217–7222. [PubMed] [Google Scholar]
- Park MH, Joe YA, Kang KR, Lee YB, Wolff EC. The polyamine-derived amino acid hypusine: its post-translational formation in eIF5A and its role in cell proliferation. Amino Acids. 1996;10:109–121. doi: 10.1007/BF00806584. [DOI] [PubMed] [Google Scholar]
- Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem (Tokyo) 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Abid MR, Miyazaki M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;384:151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem. 2006;281:35336–35346. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- Shiba T, Mizote H, Kaneko T, Nakajima T, Kakimoto Y. Hypusine, a new amino acid occurring in bovine brain. Isolation and structural determination. Biochim Biophys Acta. 1971;244:523–531. doi: 10.1016/0304-4165(71)90069-9. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project Gene Disruption Project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A. High-throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation. 2004;72:81–91. doi: 10.1111/j.1432-0436.2004.07202004.x. [DOI] [PubMed] [Google Scholar]
- Tao Y, Chen KY. Molecular cloning and functional expression of Neurospora deoxyhypusine synthase cDNA and identification of yeast deoxyhypusine synthase cDNA. J Biol Chem. 1995;270:23984–23987. doi: 10.1074/jbc.270.41.23984. [DOI] [PubMed] [Google Scholar]
- Thompson GM, Cano VS, Valentini SR. Mapping eIF5A binding sites for Dys1 and Lia1: in vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 2003;555:464–468. doi: 10.1016/s0014-5793(03)01305-x. [DOI] [PubMed] [Google Scholar]
- Umland TC, Wolff EC, Park MH, Davies DR. A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme NAD inhibitor ternary complex. J Biol Chem. 2004;279:28697–28705. doi: 10.1074/jbc.M404095200. [DOI] [PubMed] [Google Scholar]
- Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Yaffe MP. Mmd1p, a novel, conserved protein essential for normal mitochondrial morphology and distribution in the fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2004;15:1656–1665. doi: 10.1091/mbc.E03-06-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhl T, Klier H, Ammer H. The Hyp2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. Mol Gen Genet. 1993;241:305–311. doi: 10.1007/BF00284682. [DOI] [PubMed] [Google Scholar]
- Wolff EC, Folk JE, Park MH. Enzyme-substrate intermediate formation at lysine 329 of human deoxyhypusine synthase. J Biol Chem. 1997;272:15865–15871. doi: 10.1074/jbc.272.25.15865. [DOI] [PubMed] [Google Scholar]
- Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem J. 2004;384:585–590. doi: 10.1042/BJ20041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- Zuk D, Jacobson A. A single amino acid substitution in yeast eIF5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]