Abstract
We describe a chemical method to detect RNA synthesis in cells, based on the biosynthetic incorporation of the uridine analog 5-ethynyluridine (EU) into newly transcribed RNA, on average once every 35 uridine residues in total RNA. EU-labeled cellular RNA is detected quickly and with high sensitivity by using a copper (I)-catalyzed cycloaddition reaction (often referred to as “click” chemistry) with fluorescent azides, followed by microscopic imaging. We demonstrate the use of this method in cultured cells, in which we examine the turnover of bulk RNA after EU pulses of varying lengths. We also use EU to assay transcription rates of various tissues in whole animals, both on sections and by whole-mount staining. We find that total transcription rates vary greatly among different tissues and among different cell types within organs.
Keywords: 5-ethynyluridine, microscopy, azide, alkyne, fluorescence
Two methods have been used to measure rates of total transcription in cells. The first method relies on labeling RNA with radioactive nucleosides followed by tissue autoradiography (1). Autoradiography is very slow, requiring exposure times of weeks to months (1); working with radioactivity is also cumbersome and the spatial resolution of the microscopic images obtained is poor.
The second method measures incorporation into RNA of 5-bromouridine (BrU), delivered to cells either as 5-bromouridine triphosphate (BrUTP) or as BrU. Because cells are impermeable to BrUTP, they must be microinjected with it (2, 3), permeabilized in the presence of BrUTP (4), transfected with BrUTP liposomes (5), scratch labeled (6), or loaded with BrUTP by osmotic shock (7). Aside from the high cost of BrUTP, the methods are labor intensive and not applicable to all cell types or to whole-animal studies. BrU is taken up by cells and incorporated into the pool of nucleotide phosphates by means of the ribonucleoside salvage pathway. Cells are then fixed and BrU incorporation into RNA is detected by immunostaining (8). One of the main disadvantages of BrU detection is that, being an antibody-based method, staining of tissues is limited by antibody diffusion into the specimen. Thus, tissues have to be sectioned and whole-mount examination of RNA synthesis in tissues and organs is practically very limited.
We recently described a chemical method to assay DNA synthesis in vivo by using 5-ethynyl-2′-deoxyuridine (EdU), a thymidine analog that incorporates efficiently into DNA (9). EdU can be detected rapidly and with great sensitivity with fluorescent azides by means of a Sharpless–Meldal copper (I)-catalyzed Huisgen cycloaddition reaction (10, 11), a highly efficient and selective reaction often referred to as a “click” reaction. We have now extended this methodology to devise a chemical method to assay RNA synthesis in vivo.
We show that 5-ethynyluridine (EU) is incorporated into RNA transcripts generated by RNA polymerases I, II, and III in cells. EU-labeled cellular RNA can be detected quickly and with high sensitivity with fluorescent azides. Detection of EU is much faster than an anti-BrU immunostain and allows whole-mount staining of large organ and tissue fragments. Conveniently, EU does not significantly label cellular DNA, thus making it a specific transcriptional label. We demonstrate the use of EU in assaying the turnover of bulk transcripts in cultured cells. Last, we use EU in whole animals to reveal bulk transcriptional patterns in several organs and tissues.
Due to its ease and sensitivity, we anticipate the EU labeling method [particularly through labeling individual RNAs in vitro with 5-ethynyluridine triphosphate (EUTP)] will facilitate high-resolution microscopic analysis of RNA synthesis, transport, localization, and turnover in vivo.
Results and Discussion
EU as a Transcriptional Label.
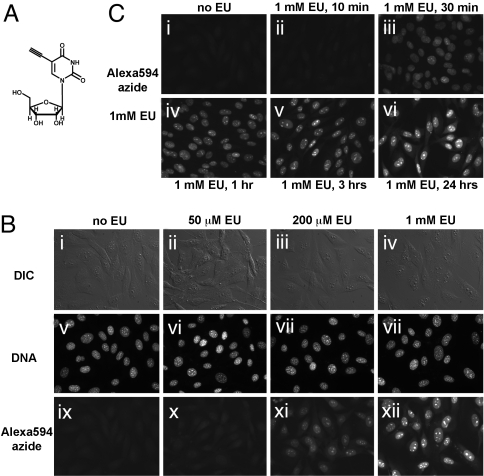
We reasoned that the uridine analog EU (Fig. 1A) should incorporate into cellular RNA. EU-labeled RNA could then be detected by reaction with fluorescent azides, by means of a copper (I)-catalyzed alkyne-azide cycloaddition. EU was synthesized from 5-iodo-uridine. The fluorescent azides used in this study were Alexa594-azide, Alexa568-azide, and tetramethylrhodamine (TMR)-azide.
Fig. 1.
Imaging cellular transcription by using EU. (A) Structure of the uridine analog EU, a biosynthetic RNA label. (B) EU incorporation into RNA in NIH 3T3 cells. Cells were grown without EU (i, v, and ix), with 50 μM EU (ii, vi, and x), 200 μM EU (iii, vii, and xi), or 1 mM EU (iv, viii, and xii) for 20 h. The cells were fixed and reacted with 10 μM Alexa594-azide. EU-labeled cells show strong nuclear and weaker cytoplasmic staining, proportional to the added EU concentration. Note the intense staining of nucleoli. All cells incorporate EU, although some cell-to-cell variability is observed. (C) Rapid uptake and incorporation of EU by cells. NIH 3T3 cells were incubated with 1 mM EU for varying amounts of time, followed by fixation and EU detection. Strong nuclear staining is visible after 30 min (iii), although even after 10 min (ii), a signal above background is observed at longer exposure times (data not shown). EU staining intensity increases quickly in the first 3 h (iv and v) and then more slowly up to 24 h (vi).
We first tested whether EU is incorporated into cellular RNA. NIH 3T3 cells were incubated overnight with increasing concentrations of EU, fixed and stained with a fluorescent azide. As shown in Fig. 1B, the cells showed staining that increased in intensity with increasing EU concentration. Unlabeled cells showed very low background staining (Fig. 1Bi). The staining was very strong in cell nuclei. The nucleoli, sites of abundant ribosomal RNA transcription stained very intensely. Cytoplasmic staining was detectable but weaker than the nuclear signal. Importantly, all cells showed comparable levels of EU staining. The click staining reaction for the EU label was fast and proceeded smoothly with all three fluorescent azides used in this study.
We next asked how long it takes for the cultured cells to incorporate EU. As shown in Fig. 1C, the cells stain strongly after only 30 min of exposure to EU, and the signal increases after 1, 3, and 24 h. Even after incubation with EU for as little as 10 min, a weak but significant signal could be seen in cells at longer camera exposure times (data not shown).
In conclusion, EU is quickly incorporated into cells, and its subcellular staining pattern is consistent with EU labeling newly synthesized, bulk RNA transcripts.
EU Labels Cellular RNA but not DNA.
To be incorporated into RNA, EU has to enter the ribonucleoside salvage pathway and be converted successively to the 5′-ribonucleotide phosphates EUMP, EUDP, and EUTP. Ribonucleotide reductase converts ribonucleotide diphosphates into deoxyribonucleotide diphosphates, which then become dNTPs to be used in DNA synthesis. If EUDP were a substrate for ribonucleotide reductase, it would lead to production of EdUTP, which would be readily incorporated into cellular DNA. This property would limit the utility of EU labeling, because both RNA and DNA would be labeled. Thus, we next asked whether addition of EU to cells in culture labels their DNA.
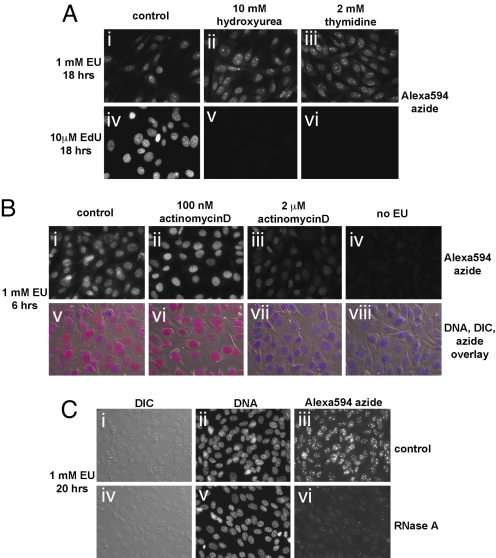
When cells were incubated overnight with EU in the presence of hydroxyurea (Fig. 2Aii) or thymidine (Fig. 2Aiii), two inhibitors of ribonucleotide reductase, the intensity and subcellular pattern of the EU stain, did not change compared with incubation with EU alone (Fig. 2Ai). In a parallel experiment (Fig. 2 Aiv–Avi), the same concentrations of hydroxyurea or thymidine abolished the incorporation of EdU into cellular DNA. We conclude that EU does not significantly label DNA, implying that EU must be a poor substrate for ribonucleotide reductase. This unexpected property of EU allows the specific labeling of cellular RNA, without the complication of concomitant DNA labeling.
Fig. 2.
EU is incorporated into cellular RNA but not DNA. (A) Inhibition of DNA synthesis by hydroxyurea or thymidine does not affect EU incorporation in cells. NIH 3T3 cells incubated with 1 mM EU and 10 mM hydroxyurea (ii) or 2 mM thymidine (iii) show EU staining identical to cells incubated with 1 mM EU alone (i). The same concentrations of hydroxyurea or thymidine strongly inhibit incorporation of EdU (at 10 μM) into DNA (v and vi). (B) Actinomycin D inhibits EU incorporation in cells. At low concentration (100 nM, sufficient to inhibit RNA polymerase I; ii and vi), actinomycin D abolishes the strong EU staining of nucleoli seen after incubation for 6 h in 1 mM EU (compare with i and v). Note that except for the absence of nucleolar staining, cells show normal levels of nuclear EU staining. A high concentration of actinomycin D (2 μM, sufficient to block RNA polymerase II; iii and vii) causes a pronounced inhibition of EU incorporation in cells. (C) EU staining is sensitive to RNaseA. Cells labeled with 1 mM EU for 6 h were permeabilized and lightly fixed followed by treatment with (iv–vi) or without (i–iii) RNaseA. The cells were subsequently stained with 10 μM Alexa594-azide and Hoechst.
To demonstrate that EU labels cellular RNA, we first determined the effect of RNA polymerase inhibition on EU labeling (Fig. 2B). Cells were labeled with EU for 6 h, in the absence or presence of different concentrations of the RNA polymerase inhibitor actinomycin D. Low concentrations of actinomycin D, which block RNA polymerase I, abolished the intense EU nucleolar staining (compare Bii and Bvi with Bi and Bv), whereas leaving intact the EU signal in the rest of the nucleus. This result demonstrates that EU staining of nucleoli indeed represents RNA polymerase I transcripts, most likely pre-rRNA molecules. High concentrations of actinomycin D (which block RNA polymerase II) cause a strong inhibition of EU staining throughout the cell (Fig. 2 Biii and Bvii). We conclude that EU is incorporated into transcripts produced by RNA polymerases I and II.
We also tested the RNase sensitivity of the EU stain. Cells labeled with EU for 6 h were lightly fixed and permeabilized, followed by incubation in the presence or absence of bovine pancreatic RNase A, a single strand-specific RNase. Cells were then stained with Alexa594-azide. RNase A treatment strongly reduces (but does not completely abolish) the intensity of the EU stain (Fig. 2, compare Ciii and Cvi). We speculate that there are several reasons why the RNase treatment does not completely abolish EU staining: (i) some RNA is protected through secondary structure; (ii) some RNA is protected by interaction with proteins; and (iii) some RNA is masked by proteins cross-linked during fixation.
Analysis of EU Incorporation into Cellular RNA.
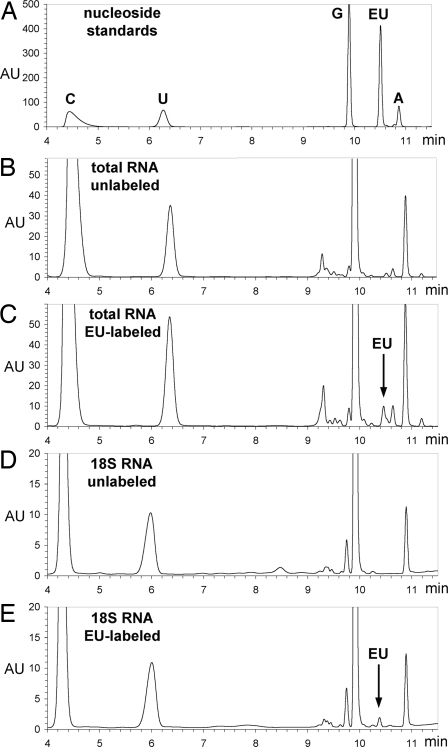
We used HPLC analysis and mass spectrometry to measure directly EU incorporation into cellular RNA. Cultured 293T cells were incubated with or without 1 mM EU for 24 h, followed by total RNA isolation. EU labeling did not affect the yield or electrophoretic appearance of total RNA, as compared with the control (data not shown). Messenger RNA was isolated by polyA selection from total RNA, whereas 28S rRNA, 18S rRNA, and the tRNA-containing fraction were isolated by agarose gel electrophoresis and purification. RNA was then hydrolyzed to nucleoside monophosphates, which were dephosphorylated with calf intestinal phosphatase. The resulting nucleosides were analyzed by reverse-phase HPLC (Fig. 3 B–E). An equimolar mix of pure nucleosides (Fig. 3A) was used to measure retention times for our HPLC method and as standard for measuring the amount of EU incorporated relative to uridine.
Fig. 3.
Analysis of EU incorporation into RNA. (A) HPLC separation of an equimolar mixture of pure cytidine, uridine, guanosine, EU, and adenosine. Absorbtion at 285 nm (in arbitrary units, AU) is plotted against elution time (in minutes). (B and C) HPLC analysis of nucleosides from total RNA isolated from unlabeled (B) and EU-labeled (C) cells. Note the EU peak in C, corresponding to a 2.8% substitution of uridine residues by EU. (D and E) HPLC analysis of nucleosides from unlabeled and EU-labeled, gel-purified 18S ribosomal RNA. The EU peak in E corresponds to a 1.3% substitution of uridine with EU. An identical degree of substitution was measured for the 28S ribosomal RNA (data not shown), consistent with the fact that 18S and 28S rRNAs are both derived from the pre-rRNA 45S precursor transcribed by RNA polymerase I.
As shown in the chromatogram in Fig. 3C, EU is detected among nucleosides isolated from EU-labeled total RNA (but not in unlabeled total RNA, Fig. 3B). We collected the fractions corresponding to the EU peak in Fig. 3C, as well as the fractions of the same retention time in the unlabeled sample in Fig. 3B. Mass spectrometric analysis of these fractions confirmed the presence of EU in the EU-labeled but not in unlabeled total RNA (data not shown; see Materials and Methods).
Assuming equal recovery of uridine and EU during HPLC sample preparation, we calculated that EU labels 2.8% of uridine residues in total RNA, meaning that EU is found once in every 35 uridines. The fraction of uridines replaced by EU is 1.3% in gel-purified 18S rRNA (Fig. 3E), 1.3% in gel-purified 28S rRNA, 3.4% in the gel-purified small RNA fraction that includes tRNAs, 5S and 5.8S rRNA, and 5.7% in polyadenylated mRNA (data not shown). These results demonstrate that EU is incorporated into transcripts produced by all three RNA polymerases, although a lower incorporation is seen in rRNA species synthesized by RNA polymerase I, compared with polyA mRNA (synthesized by RNA polymerase II) and with the tRNA fraction (which contains mostly transcripts made by RNA polymerase III).
Using EU to Study RNA Turnover in Cultured Cells.
We next determined the stability of EU-labeled transcripts by EU pulse–chase in cells. Cultured cells were labeled with EU for either 3 or 24 h, after which they were washed and chased with media without EU. Parallel cultures were fixed at different times during the chase and then stained at the same time with Alexa568-azide.
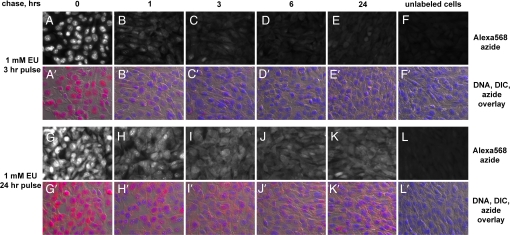
As shown in Fig. 4 A–F and A′–F′), transcripts labeled during a 3-h EU pulse are turned over quickly. At the beginning of the chase, most of the intense EU staining is in the nucleus. The nuclear signal drops dramatically after 1 h and drops even further after 3 and 6 h. However, the cytoplasmic EU signal starts low, shows a small initial increase (compare A and B in Fig. 4) and then decreases to almost background levels after 24 h. Thus, a 3-h EU pulse is too short to significantly label stable RNA species. We speculate that the initial increase in the cytoplasmic signal in the first hour of the chase is due to the export from the nucleus of some of the EU-labeled transcripts.
Fig. 4.
Imaging cellular RNA turnover with EU. NIH 3T3 cells were labeled for 3 h (A–F and A′–F′) or 24 h (G–L and G′–L′) with 1 mM EU. The label was chased with complete media for different periods of time. The cells were fixed and stained with Alexa568-azide and Hoechst. After a 3-h pulse the nuclear EU staining drops quickly in the first hour of the chase (A and B; A′ and B′) and becomes very low after 6 h (D and D′). EU staining of the cytoplasm shows a delayed decline compared with the nuclear signal. Cytoplasmic staining is still visible after 6 h (D and D′), whereas after 24 h (E and E′) it drops very close to background levels (F and F′). After labeling with EU for 24 h, both the nucleus and the cytoplasm are strongly labeled (G and G′). The nuclear signal drops significantly during the chase but does not disappear. Strong cytoplasmic staining persists after a 24-hour chase, indicating the labeling of stable RNA species.
A different picture emerges when cells are chased after a 24-h incubation with EU (Fig. 4 G–L and G′–L′). At the beginning of the chase, the cells show strong staining, both nuclear and cytoplasmic. During the chase, the nuclear staining decreases significantly but does not completely disappear even after 6 and 24 h. The signal in the cytoplasm remains strong even 24 h into the chase, demonstrating that a 24-h EU pulse strongly labels stable RNA species.
Imaging Total Transcription in Animals by Using EU.
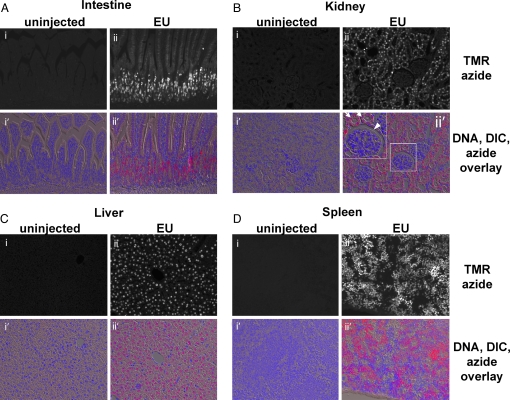
We next asked whether EU is suitable for labeling RNA in whole animals. We injected i.p. 2 mg of EU into a 3-week-old mouse and organs were harvested 5 h later. An uninjected littermate was used as a control. The organs were fixed, embedded in paraffin, and sectioned, followed by staining with TMR-azide and Hoechst staining. The results of staining four different organs (small intestine, kidney, liver, and spleen) are shown in Fig. 5. All four organs surveyed show low background staining on control sections, whereas very intensely stained cells can be seen in all four organs from the EU-injected mouse. The stain in organs was stronger in the nucleus and weaker in the cytoplasm. In the small intestine (Fig. 5A), strong transcriptional activity was detected in the highly proliferative cells of the crypts and at the base of villi, as described in previous studies (1). These earlier studies used tritiated uridine as a label and did not detect transcription in the villar cells; however, the higher sensitivity of EU staining allowed us to detect significant transcription in these cells, although at much lower levels than in crypt cells. On sections through kidney cortex (Fig. 5B), high transcription levels were seen in the cells of kidney tubules, but not in the glomerular capillaries or the capsule, or in the capillaries found throughout the cortex.
Fig. 5.
Imaging total RNA synthesis in vivo by using EU. Tissues from a EU-injected mouse (2 mg EU) and an uninjected control littermate were harvested and fixed 5 h later. Tissue sections were stained with 25 μM TMR-azide and Hoechst. The black-and-white images are fluorescent micrographs of mouse tissues stained with TMR-azide. The color images are overlays of fluorescent azide (red), fluorescent DNA (blue), and DIC micrographs of the sectioned tissues. (A) Villi of the small intestine seen in longitudinal section. Actively transcribing cells are located in the crypts and the base of the villi; EU staining is much weaker but clearly visible in the cells along villi. (B) Kidney. Kidney tubules (the two arrows in the Inset of ii′) show strong EU staining; EU-labeled RNA is strikingly absent from glomeruli (the arrowhead in the Inset of ii′). (C) Liver. All hepatocytes show strong EU staining, whereas staining is absent in cells located at the periphery of liver lobules. (D) Spleen. A large subset of the lymphocytes seen in spleen sections show very intense EU staining; however, EU incorporation is absent from some of the cells, indicating dramatically different levels of bulk transcriptional activity.
In liver sections (Fig. 5C, in which both the control and EU-injected sections show the central vein of a liver lobule), staining is very strong in all hepatocytes but absent from the endothelial cells of the central vein as well as from other cells with an endothelial appearance throughout the section (most likely endothelial cells of bile and blood capillaries).
Last, in sections of spleen (Fig. 5D), EU staining is very intense (the strongest among the four tissues we surveyed). Large patches of cells (presumably lymphocytes) show very strong staining, interspersed with other, smaller areas of the organ, which show little to no EU incorporation. Understanding the physiological basis for these histological observations require further study.
Whole-Mount Staining of EU-Labeled Organs.
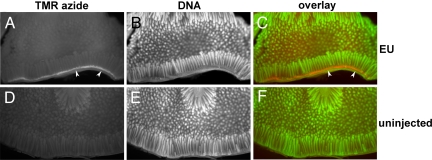
Last, we asked whether EU could be used to detect transcription in large organ fragments by whole-mount azide staining. One centimeter-long pieces of intestine from an EU-injected mouse and from an uninjected control mouse were fixed, cut open longitudinally, and then stained for 1 h with TMR-azide, followed by extensive washing to remove unreacted TMR-azide and counterstaining of DNA with OliGreen. The whole-mount stained intestine was imaged on a dissecting microscope by fluorescence. As shown in Fig. 6, 5 h after injection EU staining is strong in crypt cells and cells at the base of villi (the orange stain in Fig. 6C, indicated by the white arrowheads; compare with the identical pattern seen on the section through the small intestine in Fig. 5A). In contrast, the small intestine from the uninjected control animal shows just a low background signal (Fig. 6 D–F). Thus, EU can be used to metabolically label RNA in whole animals followed by whole-mount imaging of large fragments of tissues and organs at low magnification. We envision that this property will be an important advantage of this technique over conventional immunodetection methods.
Fig. 6.
Whole-mount staining of RNA synthesis in the mouse small intestine by using EU. Fragments of intestine were removed from an uninjected control mouse and from an EU-injected mouse, 5 h after the i.p. injection of 2 mg of EU. The explants were fixed and then stained with 50 μM TMR-azide and OliGreen (to visualize DNA), and were imaged on a fluorescent dissecting microscope. Note the strong EU incorporation in crypt cells and cells located at the base of the villi (the white band in A or the orange band in C, indicated by white arrowheads).
We have so far examined the synthesis and turnover of total RNA in cells, but EU labeling should also be useful for following specific RNAs in cells. Studies of RNA transport and localization have relied on microinjecting cells with RNAs labeled with biotin-UTP or various fluorophore-UTP by in vitro translation. One potential problem with mRNAs labeled in this manner is that the many bulky biotin or fluorophore moieties that decorate the mRNA molecules might inhibit the translation of these mRNAs. Because mRNAs in which all uridine residues are replaced by BrU or 5-fluoro-uridine are efficiently translated (12), we expect that in vitro synthesized mRNAs labeled with EUTP should also be translated well, thus allowing the imaging in cells of a fully functional labeled RNA.
Another unique advantage of EU compared with BrU labeling is the fact that newly transcribed RNAs can be modified covalently with any molecule attached to an azide group. These molecules in turn can react with RNA-interacting proteins. We envision that this feature will facilitate experiments that probe dynamic RNA-protein interactions in vivo.
Materials and Methods
EU Labeling of Cultured Cells.
NIH 3T3 cells were grown on glass coverslips, in DMEM supplemented with 10% bovine calf serum, penicillin, and streptomycin. EU was added to the complete culture medium from a 100 mM stock in DMSO.
After EU labeling, cells were washed with PBS and fixed. Except for the cells used for the RNase experiment, all cells were fixed in 125 mM Pipes, pH 6.8/10 mM EGTA/1 mM magnesium chloride/0.2% Triton X-100/3.7% formaldehyde for 30 min at room temperature. The fixed cells were rinsed once with TBS, followed by click staining by using a fluorescent azide.
To assay the effect of inhibiting DNA synthesis (Fig. 2A), cells were incubated for 18 h with 1 mM EU or 10 μM EdU, in the presence or absence of 10 mM hydroxyurea or 2 mM thymidine. To examine the effect of RNA synthesis inhibition (Fig. 2B), NIH 3T3 cells were grown for 6 h in complete media with 1 mM EU, without actinomycin D or with 100 nM or 2 μM actinomycin D (Sigma). The cells were then rinsed, fixed, and processed for azide staining.
To assay RNaseA sensitivity (Fig. 2C), NIH 3T3 cells grown in the presence of 1 mM EU for 20 h were fixed in PBS with 0.5% formaldehyde and 0.5% Triton X-100 for 30 min at room temperature. The cells were rinsed twice with TBS with 0.2% Triton X-100 (TBST) and then incubated in either TBST alone (control) or TBST with 200 μg/ml RNaseA, for 45 min at room temperature. The cells were then washed with TBS and then stained with 10 μM Alexa594-azide.
EU Detection by Click Chemistry.
All of the steps of EU detection were performed at room temperature. The fixed cells were rinsed with TBS and stained for 30 min at room temperature with 100 mM Tris, pH 8.5/1 mM CuSO4/10–50 μM fluorescent azide/100 mM ascorbic acid (added last from a 0.5 M stock in water). An identical protocol was used to stain EdU-labeled cells. After staining, cells were washed several times with TBS with 0.5% Triton X-100 and then were stained with Hoechst. The cells were imaged by fluorescence microscopy and differential interference contrast microscopy (DIC).
For the experiment in Fig. 3, NIH 3T3 cells labeled with 1 mM EU for 3 or 24 h were washed three times in PBS followed by incubation in complete media. Cells fixed at different times were all stained in parallel with 20 μM Alexa568-azide.
EU Labeling of Mouse Tissues.
A 3 week-old mouse was injected i.p. with 100 μl of a 20 mg/ml solution of EU in PBS. Tissues were harvested 5 h after injection and were fixed in formalin overnight. Tissues from an uninjected littermate were used as negative controls. Pieces of the small intestine, kidney, liver, and spleen were embedded in paraffin and sectioned. After paraffin removal and rehydration, the sections were stained with 25 μM TMR-azide for 30 min, as described for staining cultured cells. Sections were counterstained with Hoechst and examined by fluorescence microscopy and DIC.
For whole-mount staining of EU-labeled small intestine, 1-cm long pieces of fixed small intestine were stained for 1 h with 50 μM TMR-azide. The small intestine pieces were then washed extensively with 20 mM Hepes, pH 7.5/500 mM NaCl/1% Triton X-100 by end-over-end rotation, until the background staining of the control intestine became very low (≈24 h). The intestine pieces were stained with OliGreen (Molecular Probes) to reveal their DNA and were imaged on a fluorescent dissecting microscope equipped with filters for fluorescein and rhodamine.
RNA Isolation and Hydrolysis.
Cultured 293T cells were incubated for 24 h in complete media with 1% DMSO (unlabeled control) or 1 mM EU (added from a 100 mM stock in DMSO). The cells were washed with PBS and total RNA was isolated by using RNazol-B (Tel-Test). We separated 100 μg of total RNA (unlabeled or EU-labeled) by electrophoresis on a 1% low melting agarose gel. Bands corresponding to 28S rRNA, 18S rRNA, and tRNA were excised, melted and incubated with beta-agarase (New England Biolabs) according to the instructions of the manufacturer. After phenol-chloroform extraction, the RNA was recovered by isopropanol precipitation. PolyA mRNA was purified from total RNA by using an Oligotex mRNA isolation kit (Qiagen).
Between 5 and 100 μg of the different RNA species purified as described above were hydrolyzed to nucleoside monophosphates with 50 mM potassium hydroxide for 1 h at 100°C. The reaction mixture was adjusted to 50 mM Tris, pH 8/40 mM KCl/60 mM NaCl/10 mM MgCl2 and incubated with 30 units of calf intestinal phosphatase (New England Biolabs) at 37°C for 2 h to hydrolyze the nucleoside monophosphates. The mixture was evaporated under reduced pressure and the dry residue was extracted with 80% ethanol by vortexing for 10 min at 37°C. After centrifugation to pellet insoluble materials, the supernatant was recovered and dried under reduced pressure. The dry residue was dissolved in water, filtered, and analyzed by HPLC.
HPLC and Mass Spectrometric Analysis of Nucleosides.
Nucleosides were analyzed by using a 9.4 × 150 mm Zorbax C-18 column on an Agilent 1200 HPLC instrument equipped with a diode array detector. The conditions used to separate nucleosides were as follows: 5 min constant 1% acetonitrile in water, followed by 8 min of a linear gradient from 1% acetonitrile in water to 31% acetonitrile in water, at a flow rate of 3 ml/min. Pure standards for adenosine, cytidine, guanosine, and uridine were from Sigma. EU standard was obtained by preparative HPLC purification. Under the conditions above, the retention times were 4.4 min for cytidine, 6.3 min for uridine, 9.9 min for guanosine, 10.4 for EU, and 10.88 for adenosine. Nucleoside separation was monitored by absorbance at both 285 and 220 nm. By using pure EU and uridine, we determined that the extinction coefficient of EU is 3.28 times larger than that of uridine at 285 nm. This value was used to calculate the amount of EU relative to uridine in EU-labeled RNA samples, from the intensities of the EU and uridine absorbtion peaks at 285 nm.
Nucleoside samples from both control and EU-labeled cells were separated by HPLC by using the chromatographic conditions described above. Peaks corresponding to various nucleosides were identified based on their characteristic retention times. To confirm their identity, fractions corresponding to distinct peaks were collected, dried under reduced pressure, dissolved in water, and analyzed by LC/MS on an Agilent 6130 Quadrupole LC/MS system, by using a 4.6 × 100 mm Luna C-18 column. The conditions used for LC/MS analysis were as follows: 1 ml/min flow rate, 5 min of 100% water plus 0.1% formic acid, followed by 2 min of a linear gradient to 10% acetonitrile plus 0.1% formic acid, followed by another 5 min of a linear gradient to 30% acetonitrile plus 0.1% formic acid.
Chemicals.
All chemicals were purchased from Sigma-Aldrich and were used without further purification. NMR spectra were recorded on a 600 MHz Varian Oxford NMR A5600.
EU was synthesized from 5-Iodo-uridine (IU) and trimethyl-silyl-acetylene by means of a Sonogashira coupling reaction. Briefly, 2 g of IU (5.4 mmol) were reacted with 3 ml of trimethylsilyl-acetylene (21 mmole) essentially as described in ref. 13 for the synthesis of 5-ethynyl-deoxyuridine. The product was purified by flash chromatography on silica and the trimethylsilyl group was removed with potassium carbonate in methanol, yielding EU. EU was purified by flash chromatography on silica. Aliquots of the light tan solid thus obtained were further purified by preparative HPLC on a C18 reverse-phase column, affording EU as white crystals.
EU: white crystals, molecular weight: 268.22, ES-API LC/MS: M = 268 (m/z), [M+H]+ = 269.0, [M+Na]+ = 291.0, [2M+Na]+ = 559.1, (1)H NMR (600 MHz, DMSO-d6): δ 8.36 (s, 1H, H-6), 5.72 (d, 1H, J = 4.4 Hz, H-1′), 5.50 (b, 1H, HO-2′), 5.25 (b, 2H, HO-3′, HO-5′), 4.07 (s, 1H, H-8), 4.02 (t, 1H, J = 4.7 Hz, H-2′), 3.97 (t, 1H, J = 4.7, H-3′), 3.84 (m, 1H, H-4′), 3.66–3.56 (ddd, 2H, J5′a,5′b = 12.0 Hz, J5′,OH = 2.64 Hz, J5′,4′ = 2.35 Hz, H-5′), 3.30 (s, H2O), 2.50 (s, (CD3)2SO), (13)C NMR (600 MHz, DMSO-d6): δ 162.4 (C-4), 150.4 (C-2), 145.3 (C-6), 98.3 (C-5), 89.1 (C-1′), 85.4 (C-4′), 84.3 (C-7), 77.1 (C-8), 74.7 (C-2′), 70.0 (C-3′), 60.9 (C-5′).
The three fluorescent azides used in this study were TMR-azide, Alexa594-azide, and Alexa568-azide. The synthesis of TMR-azide was described elsewhere (9). Alexa594-azide was obtained by reacting the succinimidyl ester of Alexa594 carboxylic acid (Molecular Probes) with 2-amino-2′-[6-azido-hexanamide]ethylene glycol diethyl ether (9), in dry DMSO according to the instructions of the manufacturer. Alexa568-azide was obtained by reacting the succinimidyl ester of Alexa568 carboxylic acid (Molecular Probes) with O-(2-aminoethyl)-O′-(2-azidoethyl)-pentaethylene glycol (Fluka) in dry DMSO according to the instructions of the manufacturer.
Acknowledgments.
We thank Frank McKeon for mouse injections and dissections, and Hongmei Mou for help with tissue sectioning. A.S. was supported by the Rita Allen Foundation, the Beckman Foundation, and the Sontag Foundation. C.Y.J. is a recipient of a National Science Foundation predoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
References
- 1.Uddin M, et al. Radioautographic visualization of differences in the pattern of [3H]uridine and [3H]orotic acid incorporation into the RNA of migrating columnar cells in the rat small intestine. J Cell Biol. 1984;98:1619–1629. doi: 10.1083/jcb.98.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wansink DG, et al. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cmarko D, et al. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei X, et al. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haukenes G, et al. Labeling of RNA transcripts of eukaryotic cells in culture with BrUTP using a liposome transfection reagent (DOTAP) BioTechniques. 1997;22:308–312. doi: 10.2144/97222st03. [DOI] [PubMed] [Google Scholar]
- 6.Sadoni N, Zink D. Nascent RNA synthesis in the context of chromatin architecture. Chromosome Res. 2004;12:439–451. doi: 10.1023/B:CHRO.0000034739.96307.8d. [DOI] [PubMed] [Google Scholar]
- 7.Koberna K, et al. Nuclear organization studied with the help of a hypotonic shift: Its use permits hydrophilic molecules to enter into living cells. Chromosoma. 1999;108:325–335. doi: 10.1007/s004120050384. [DOI] [PubMed] [Google Scholar]
- 8.Halicka HD, et al. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res. 2000;260:248–256. doi: 10.1006/excr.2000.5027. [DOI] [PubMed] [Google Scholar]
- 9.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tornoe CW, et al. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 11.Rostovtsev VV, et al. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Schmittgen TD, et al. Effect of 5-fluoro- and 5-bromouracil substitution on the translation of human thymidylate synthase mRNA. J Biol Chem. 1994;269:16269–16275. [PubMed] [Google Scholar]
- 13.Yu CS, Oberdorfer F. Synthesis of (E)-5-[2-(tri-n-butylstannyl)vinyl] substituted 2′-deoxyuridine derivatives for use in halogenation and radiohalogenation reactions. Synlett. 2000:86–88. [Google Scholar]