Abstract
Hybridization between populations can disrupt gene expression, frequently resulting in deleterious hybrid phenotypes. Reduced fitness in interpopulation hybrids of the marine copepod Tigriopus californicus has been traced to interactions between the nuclear and mitochondrial genomes. Here, we determine transcript levels of four to six genes involved in the mitochondrial oxidative phosphorylation pathway for a series of parental and inbred hybrid lines using RT-qPCR. Both nuclear and mitochondrial-encoded genes are included in the analysis. Although all genes studied are up-regulated under salinity stress, only expression of genes located on the mtDNA differed among lines. Because mitochondrial genes are transcribed by a dedicated RNA polymerase encoded in the nuclear genome, we compare transcript levels among hybrid lines with different combinations of mitochondrial RNA polymerase and mtDNA genotypes. Lines bearing certain mtDNA-mitochondrial RNA polymerase genotypic combinations show a diminished capacity to up-regulate mitochondrial genes in response to hypoosmotic stress. Effects on the transcriptional profile depend on the specific interpopulation cross and are correlated with viability effects. We hypothesize that disruption of the mitochondrial transcriptional system in F2 hybrids may play a central role in hybrid breakdown.
Keywords: hybrid breakdown, mitochondria, Tigriopus californicus, transcription
Geographically isolated populations gradually diverge because of the combined forces of mutation, selection, and genetic drift. This genetic divergence may ultimately result in reproductive isolation and the formation of new species. During the course of this process, hybrids between populations experience reductions of fitness, often initially restricted to the F2 and later generations (reviewed in ref. 1). Although the widely cited Dobzhansky-Muller model (2, 3) provides a general mechanism for F2 hybrid breakdown, detailed understanding of gene–gene interactions underlying the phenomenon remains elusive. Such interactions may occur at both the structural level of protein–protein interaction and at the regulatory level, typically involving protein–nucleic acid interaction. Expression of many genes is disrupted in hybrids and can have strongly deleterious effects (4–6). Epistatic interactions are ubiquitous in all regulatory networks (7), and their dysfunction in hybrids is often envisioned as a product of altered interaction strength between transcription factors and their binding sites evolving under a Dobzhansky-Muller or compensatory evolution model (8–11). However, identification of specific transcription factors, RNA polymerases, or cis-regulatory motifs underlying these observations has proven difficult.
No studies to date have directly addressed the impact of hybridization on the expression of mitochondrial genes, which rely on a wholly different transcriptional system than nuclear genes. The animal mitochondrial genome typically encodes 13 polypeptides, each of which is an integral component of the oxidative phosphorylation pathway (OXPHOS) (12). Normal OXPHOS operation relies on the functional interaction and coordinated expression of nuclear and mitochondrial genes (13). In contrast to the far larger nuclear genome, the mitochondrial genome is extremely compact, with only a single region thought to contain the regulatory features for mitochondrial gene expression in many taxa (14). The mitochondrial genome is transcribed by a dedicated, nuclear-encoded RNA polymerase (mtRPOL), which functions independently of the transcription network of nuclear-encoded OXPHOS genes. In contrast to the large, multimeric nuclear RNA polymerase complexes, mtRPOL is a simple, phage-derived polymerase consisting of a single-subunit core polymerase, mitochondrial transcription factor A (TFAM), and one of two mitochondrial transcription factor B paralogues (TFB1M and TFB2M) (15, 16). Importantly, mtRPOL, and not one of the transcription factors, appears to be responsible for promoter binding affinity and specificity (17, 18).
The marine copepod Tigriopus californicus inhabits rock pools in the high intertidal zone along the Pacific coast of North America from southern Alaska to central Baja California, Mexico. Both nuclear and mitochondrial DNA sequences are sharply divergent across populations with no evidence of gene flow among geographically isolated populations (19, 20). Recent work has demonstrated a ubiquitous pattern of F2 hybrid breakdown in laboratory crosses between genetically divergent populations (21–23). Back-crossing approaches have mapped the effect to the cytoplasm (24, 25) and suggest that hybridization disrupts coadapted nuclear-mitochondrial gene complexes evolving within the study populations. In support of this hypothesis, mitochondrial function is reduced in interpopulation hybrids, as are the activities of those mitochondrial enzyme complexes requiring integration of nuclear and mitochondrial gene products (26). Some of this effect may be attributed to known deleterious protein–protein interactions present in hybrids (27–29). However, the pervasive nature of breakdown in all enzyme complexes containing both nuclear and mitochondrial gene products (Complexes I, III, IV, and V), and its absence in the single, wholly nuclear-encoded enzyme complex (Complex II) suggests that regulation of mtDNA transcription may play a role. Although regulatory components of this system are largely unexplored, extensive mtDNA control region variation and numerous amino acid substitutions between population-specific alleles of mtRPOL have been found and the three populations of T. californicus studied here [Abalone Cove (AB), Santa Cruz North (SCN), and San Diego (SD)], each has numerous fixed differences in both mtDNA and mtRPOL (refs. 30, 31; GenBank accession nos. ABC17814-ABC17826).
Here, we investigate the impact of mtRPOL genotype on the expression of mitochondrial-encoded OXPHOS genes and associated, but independently transcribed nuclear-encoded OXPHOS genes in hybrids. Because of the relative simplicity of the mitochondrial transcription machinery, interpopulation hybrids can be generated that are homozygous for either the maternal mtRPOL genotype (matched mtRPOL-mtDNA genotype) or paternal mtRPOL genotype (mismatched mtRPOL-mtDNA genotype). We hypothesize that hybrids with matched mtRPOL-mtDNA combinations will have OXPHOS transcriptional profiles similar to parental animals, whereas transcriptional profiles of mismatched mtRPOL-mtDNA genotype hybrids will be altered, particularly for the expression of mitochondrial genes. Using this approach, we can evaluate the level of coadaptation in the mitochondrial transcriptional network within independent lineages and determine how it is affected by hybridization.
Results
Effect of Salinity Stress on OXPHOS Transcriptional Profiles.
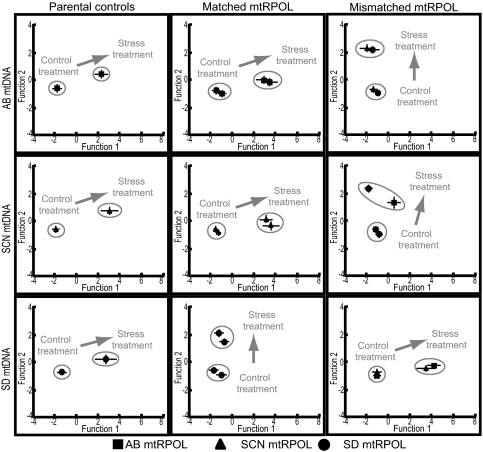
Previous work indicated that hypoosmotic stress results in a sharp increase in oxygen consumption in T. californicus (32). To evaluate the effect of hypoosmotic stress on OXPHOS transcriptional regulation, we used a discriminate function analysis incorporating gene expression data for all nuclear and mitochondrial genes studied, for each cross and reciprocal cross, with all mtRPOL-mtDNA genotypic combinations. The parameters of the primary discriminate functions within each subset of data describe the transcriptional profile, based on a set of four or six genes, depending on the cross, for that subset. Results are summarized in Fig. 1 and supporting information (SI) Tables S1 and S2. No significant transcriptional profile differences were found among genotypes within the control treatment. However, strong differences were found between the control and stress treatments and more complex differences were uncovered among genotypes within the stress treatment (Table S2). The primary discriminate function in Fig. 1 (Function 1) was weighted most strongly with mitochondrial-encoded genes, suggesting that expression of mitochondrial genes contributed more to differences among genotypic classes than did expression of nuclear genes (Table S1).
Fig. 1.
Salinity stress affects OXPHOS transcriptional profile in interpopulation hybrids. Discriminant function analysis results showing effects of salinity stress on transcriptional profile in hybrids and parental animals. Data are grouped by mtDNA genotype and parental, matched, or mismatched hybrid mtRPOL genotypes. Mean and standard errors shown for each treatment and mtRPOL genotype class. AB = Abalone Cove; SCN = Santa Cruz; SD = San Diego.
For hybrids bearing either the AB or SCN mtDNA genotypes, control and stress treatment transcriptional profiles of matched mtRPOL-mtDNA hybrids were similar to those of parental controls on both primary discriminant function axes. Mismatched mtRPOL-mtDNA hybrids were significantly different from either parental controls or matched mtRPOL-mtDNA hybrids; transcriptional profiles of mismatched hybrids subjected to hypoosmotic stress were more similar to the control treatment groups than to the other genotypes subjected to stress (Fig. 1; Table S2). Hybrids with the SD mtDNA genotype showed a strikingly different pattern in which matched mtRPOL-mtDNA hybrids were significantly different from both parental controls and mismatched mtRPOL-mtDNA hybrids and more similar to the control treatment groups (Fig. 1; Table S2).
Effect of mtRPOL Genotype on OXPHOS Transcriptional Profiles.
The effect of mtRPOL genotype on transcriptional profile was apparent only under hypoosmotic stress conditions; here, we focus only on these stress treatment data. Gene expression for each cross and each gene assayed is presented in Fig. 2. Mitochondrial gene transcript levels were typically 2-fold higher than associated OXPHOS nuclear-encoded genes, and expression was much more variable across genotypes for mitochondrial genes. ANOVA results show that five of six crosses had significant differences between genotypic groups for all mitochondrial genes studied, whereas no nuclear gene in any cross showed significant expression differences between genotypic classes (Table 1). This suggests that the expression of mitochondrial, rather than nuclear, genes is the primary cause of transcriptional profile differentiation based on mtRPOL-mtDNA genotype combinations in hybrids under hypoosmotic stress.
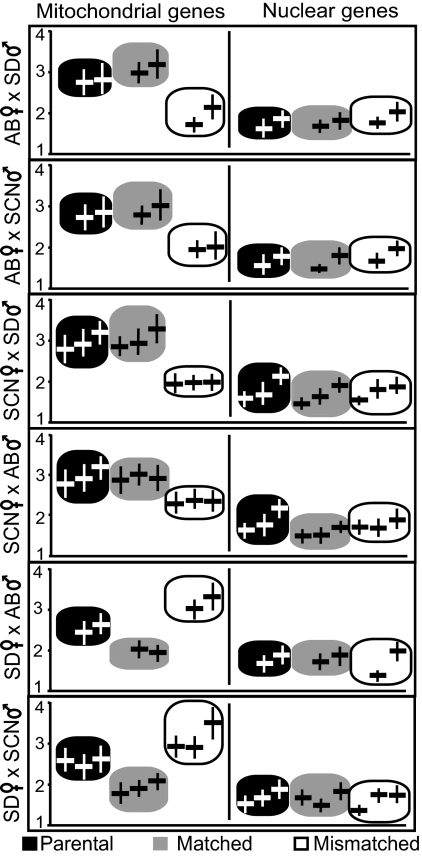
Fig. 2.
Transcript levels among mtDNA-mtRPOL genotype classes for mitochondrial and nuclear genes. Mean gene expression measures in hypoosmotic stress treatment only shown for each gene and cross with respective standard error (all values = cDNA copy number x105). Mitochondrial gene expression values (left column) are presented, left to right, as COI, CytB, and ATP6; nuclear gene expression values (right column) are presented, left to right, as COVa, CytC, and ATPc. AB = Abalone Cove; SCN = Santa Cruz; SD = San Diego.
Table 1.
Significant differences between genotypic classes under hypoosmotic stress for mitochondrial, but not nuclear, gene expression
| ABxSD | ABxSCN | SCNxSD | SCNxAB | SDxAB | SDxSCN | ||
|---|---|---|---|---|---|---|---|
| COI | n.d. | n.d. | 0.0248* | 0.2523 | n.d. | 0.0273* | |
| mtDNA | CytB | 0.0051* | 0.0133* | 0.0487* | 0.1124 | 0.0425* | 0.0433* |
| ATP6 | 0.0366* | 0.0311* | 0.0071* | 0.0978 | 0.0215* | 0.0118* | |
| COVa | ND | ND | 0.8163 | 0.6995 | ND | 0.5231 | |
| gDNA | CytC | 0.8575 | 0.8346 | 0.4598 | 0.7316 | 0.4330 | 0.6404 |
| ATPc | 0.7507 | 0.7638 | 0.8817 | 0.4763 | 0.9340 | 0.9450 |
ANOVA of gene expression data partitioned by cross and gene for hypoosmotic stress treatment only. Crosses shown as (female parent population) × (male parent population). ANOVA testing three-way interaction with (i) maternal parent, (ii) matched hybrids, and (iii) matched hybrids for each gene, partitioned by cross (* denotes significant ANOVA result at α=0.05). AB = Abalone Cove; SCN = Santa Cruz; SD = San Diego. ND, not determined.
In hybrids bearing either AB or SCN mtDNA, expression of mitochondrial genes under hypoosmotic stress in mismatched mtRPOL-mtDNA hybrids was lower than that of either parental controls or matched mtRPOL-mtDNA hybrids. This effect was significant in three of the four crosses; the cross of SCN female × AB male exhibited the same pattern, although the effect was not significant (Fig. 2; Table 1; Table S3). No parallel effect was observed for nuclear gene expression. Crosses with SD mtDNA followed a different pattern in which matched mtRPOL-mtDNA hybrids had significantly lower mitochondrial gene expression than either parental or mismatched mtRPOL-mtDNA hybrids. No effect of genotype on expression was observed for nuclear-encoded genes (Fig. 2; Table 1; Table S3). These results indicate that mismatched mtRPOL-mtDNA combinations in hybrids with either AB or SCN mtDNA, and matched mtRPOL-mtDNA combinations for hybrids with SD mtDNA, have a diminished mitochondrial regulatory stress response relative to parental animals.
Viability Effects of mtRPOL Genotype.
In the course of establishing inbred hybrid lines of known mtRPOL genotype, a total of 1,104 F4 T. californicus hybrid adults reared under control conditions were genotyped for mtRPOL. Because lines were initiated as interpopulation hybrids, all F1 individuals were heterozygotes and the neutral expected frequencies for each of the two mtRPOL alleles in each cross would remain at 50%. Changes in allelic frequencies in generations 2–4 and deviations from Hardy–Weinberg expected genotypic proportions provide tests for viability effects. Results are presented in Table 2.
Table 2.
Viability effects of mtRPOL genotype in interpopulation hybrids
| mtRPOL genotypes |
Hardy–Weinberg | Allele frequencies | ||||
|---|---|---|---|---|---|---|
| Match | Heterozygote | Mismatch | ||||
| ABxSCN | ||||||
| Observed | 69 | 94 | 50 | χ2 = 2.5909 | pmatch = 0.54 | χ2 = 3.206 |
| Expected | 63 | 106 | 44 | P = 0.1075 | pmismatch = 0.46 | P = 0.073 |
| ABxSD | ||||||
| Observed | 77 | 79 | 32 | χ2 = 1.2549 | pmatch = 0.62 | χ2 = 21.543 |
| Expected | 72 | 89 | 27 | P = 0.2626 | pmismatch = 0.38 | P < 0.0001 |
| SCNxAB | ||||||
| Observed | 67 | 115 | 35 | χ2 = 1.5118 | pmatch = 0.57 | χ2 = 9.428 |
| Expected | 71 | 106 | 39 | P = 0.2188 | pmismatch = 0.43 | P = 0.002 |
| SCNxSD | ||||||
| Observed | 62 | 71 | 17 | χ2 = 0.2435 | pmatch = 0.65 | χ2 = 26.316 |
| Expected | 63 | 68 | 18 | P = 0.6217 | pmismatch = 0.35 | P < 0.0001 |
| SDxAB | ||||||
| Observed | 22 | 61 | 59 | χ2 = 0.8698 | pmatch = 0.37 | χ2 = 19.282 |
| Expected | 19 | 66 | 56 | P = 0.3510 | pmismatch = 0.63 | P < 0.0001 |
| SDxSCN | ||||||
| Observed | 30 | 89 | 75 | χ2 = 0.1781 | pmatch = 0.38 | χ2 = 20.876 |
| Expected | 29 | 92 | 74 | P = 0.6730 | pmismatch = 0.62 | P < 0.0001 |
Genotypic numbers and χ2 test to Hardy–Weinberg expectations for mtRPOL genotypes in F4 hybrid adults reared under control conditions. Also shown are the mtRPOL allelic frequencies (matched = same source population as for the mtDNA) and chi-square test for deviation from the neutral expected frequency of 0.5. All crosses are listed as (female parent population) × (male parent population). AB = Abalone Cove; SCN = Santa Cruz; SD = San Diego.
Viability effects showed two general patterns: in hybrids with either the AB or the SCN mtDNA genotype, matched mtRPOL-mtDNA was favored, whereas the SD mtRPOL was never favored in hybrids, even with the matched SD mtDNA genotype. The effect was significant in all crosses except for AB female × SCN male. In this case, the direction of the viability effect was the same (favoring the matched genotype), but was not significant. In each cross, animals assayed for mtRPOL genotype did not significantly deviate from Hardy–Weinberg equilibrium.
Discussion
Enzyme complexes participating in the OXPHOS pathway consist of subunits encoded in both nuclear and mitochondrial genomes. Coordinated gene expression is therefore integral to efficient regulation of the OXPHOS pathway and is complicated by the existence of unique nuclear and mitochondrial transcriptional systems (13). Although regulatory networks of nuclear and mitochondrial OXPHOS genes may not be fully independent, mtRPOL is involved only in the transcription of mitochondrial genes and therefore is not expected to directly impact the expression of nuclear genes. Here, we have found that in interpopulation hybrids of T. californicus, mtRPOL genotype has a profound impact on the expression of mitochondrial genes, but no concomitant effect on associated nuclear genes.
The transcriptional profile of OXPHOS genes in interpopulation hybrids of T. californicus depends on mtRPOL genotype. Hybrids generated from the AB and SCN populations with matched mtRPOL-mtDNA genotypes have transcriptional profiles similar to parental controls when under hypoosmotic stress, whereas hybrids with mismatched mtRPOL-mtDNA combinations are significantly different. This observation is primarily driven by reduced transcript levels of mitochondrial genes in mismatched hybrids; expression of nuclear genes was, as expected, largely unaffected by mtRPOL genotype. The transcriptional profiles of hybrids with mismatched genotypes under stress were further found to be more similar to the control treatment groups than the hypoosmotic stress treatment parental controls, suggesting that mismatched hybrids have a reduced capacity to respond to salinity stress. In sum, these data suggest that coadaptation of mitochondrial transcriptional network components within populations of T. californicus may be disrupted by hybridization, thereby impairing the regulatory stress response.
In contrast to the above results, hybrids with the SD mtRPOL homozygous genotype, regardless of mtDNA haplotype, were found to have significantly different transcriptional profiles than parental genotypes when under hypoosmotic stress. Hybrids bearing the SD mtRPOL genotype had transcriptional profiles more similar to the unstressed control treatment. Again, this change in transcriptional profile was a result of altered regulation of mitochondrial genes and little change was observed in nuclear gene expression. It therefore appears that the SD mtRPOL genotype has a diminished salinity stress response compared with either the AB or SCN mtRPOL genotypes.
A priori, it is unclear what scale of change in mitochondrial transcriptional stress response capacity might be required for any deleterious effects on fitness to be visible to selection. In this study, the hybrid mtRPOL genotype associated with lower mitochondrial transcript levels corresponded exactly to the underrepresented mtRPOL allele in F4 in all six of the interpopulation crosses studied. Further, the only cross in which no significant mtRPOL viability effect was observed, SCN females crossed with AB males, was the same cross that did not show a significant effect of mtRPOL genotype on transcriptional profile (Table 2; Fig. 2). This suggests both that increased mitochondrial transcriptional stress response is favored in hybrids and that the effect is visible to viability selection.
Role of Transcription Factors.
The mitochondrial transcription machinery is highly simplified, consisting of only a single-subunit RNA polymerase (mtRPOL) and two transcription factors (15, 16). The simplicity of this system suggests that coadaptation of mtRPOL and the mitochondrial promoter may evolve within isolated lineages. In crosses between the AB and SCN populations of T. californicus, matched mtRPOL-mtDNA combinations are favored over mismatched combinations in hybrids. This may reflect coadaptation between mtRPOL and the mitochondrial promoter evolving within these populations to improve binding specificity and strength independent of transcription factors.
Although only two transcription factors, TFAM and either TFB1M or TFB2M, are generally involved in mitochondrial transcription (15, 16), a number of accessory proteins also appear to be involved in mitochondrial regulatory processes (33, 34). Crosses involving the SD population repeatedly revealed poor function of SD-derived mtRPOL. Remarkably, hybrids with a matched SD mtRPOL-mtDNA genotype were unable to mirror the SD parental transcriptional profile, clearly indicating that genes other than mtRPOL are involved in maintaining the efficacy of the SD mitochondrial transcriptional system in parental lineages. TFB1M has been shown to be in tight linkage with mtRPOL in T. californicus (31) and is therefore unlikely to contribute to the SD mtRPOL dysfunction in hybrids described herein. The role of TFAM and other regulatory accessory proteins in maintaining this regulatory network, however, remains open to investigation.
Evolution of the Mitochondrial Transcriptional Network in T. californicus.
The mechanisms by which hybrid incompatibilities arise are frequently understood in terms of the Dobzhansky-Muller model (2, 3). This model suggests that neutral or nearly neutral mutations accumulating within lineages become deleterious only when placed on a different genetic background, thus permitting the evolution of deleterious incompatibility loci between lineages without strong selection acting against any combination of the same loci within lineages. Landry et al. (11) argue that the complexity of regulatory networks creates “degeneracy” analogous to the redundant nature of the genetic code. In this way, neutral variation may segregate within lineages and eventually diverge from sister groups. However, the reduced complexity of the mitochondrial transcriptional regulatory network compared with nuclear regulatory networks may reduce the applicability of such a model with regard to mitochondrial transcription (8). An alternative model describing the generation of hybrid incompatibilities was proposed by Kimura (35). Under this model of compensatory evolution, deleterious mutations are balanced within lineages by secondary mutations that yield the epistatic network neutral or nearly neutral; dissolution of these epistatic interactions and their compensatory mechanisms in hybrids subsequently exposes the deleterious mutations to selection.
A Dobzhansky-Muller process may explain the pattern of mitochondrial regulatory dysfunction in crosses between the AB and SCN populations of T. californicus, wherein each population appears to have coadapted mtRPOL-mtDNA genotypes. However, this same model is unlikely to explain the pattern observed in crosses involving the SD population. It is clear that the SD mtRPOL genotype is deleterious when placed in a hybrid genomic background. A combination of SD mtRPOL and SD mtDNA in hybrids is not sufficient to reconstitute the SD parental mitochondrial transcriptional profile, therefore one or more deleterious mutations in the SD mitochondrial transcription machinery must be maintained by an unlinked compensatory locus. This could involve a mitochondrial transcription factor (such as TFAM) or accessory protein, although this study lacks the resolution to specifically identify the relevant compensatory locus or loci.
The evolution of cis–trans regulatory interactions, and their modification in hybrids, has been surveyed extensively (36–38), but little is known about individual cis-–trans interactions between promoters and RNA polymerases, transcription factors, or accessory proteins in these cases. Here, we evaluate the effect of hybridization specifically on the mitochondrial regulatory system and find evidence (1) that mtRPOL genotype has a large impact on mitochondrial transcription in hybrids and (2) that mitochondrial regulatory dysfunction may result in decreased responsiveness to stress conditions. Such an effect may have broadly pleiotropic effects on hybrid fitness, including delayed development and reduced fecundity and survivorship. We conclude that compensatory epistatic mutations in the mitochondrial transcriptional regulatory system within populations of T. californicus are disrupted by interpopulation hybridization. This may contribute to a reduced stress response capacity and, consequently, reduced fitness in hybrids.
Materials and Methods
Generation of Recombinant Hybrid Inbred Lines.
Interpopulation hybrids were generated for six pairwise crosses beginning with three populations of T. californicus: Santa Cruz, CA (SCN: 36°57′N, 123°03′W, collected April 2006), Abalone Cove, Palos Verdes, CA (AB: 33°44′N, 118°22′W, collected May 2006), and San Diego, CA (SD: 32°45′N, 117°15′N collected June 2006). Stock cultures of each population were kept in beakers containing 200 ml of seawater at 20°C and fed dried Spirulina algae. All experimental crosses were completed in 100-mm diameter Petri dishes containing 0.1 mg of ground Spirulina per liter filtered seawater. Animals were transferred to fresh dishes with each generation.
T. californicus females mate only once and are typically clasped by mature males until reproductively mature. Clasped males and virgin females were dissected apart and transferred to appropriate culture dishes to allow repairing and mating with individuals from different populations. Beginning with the three populations listed above (AB, SCN, and SD), all six possible pairwise crosses were generated. For the F1, F2, and F3 hybrid generations, clasped hybrid pairs were removed from culture, separated, and crossed with individuals from a replicate hybrid culture to eliminate the possibility of inbreeding before the F4 generation.
Clasped pairs of F4 adults were isolated in culture to initiate recombinant hybrid inbred lines. After the appearance of F5 juveniles (copepodids), F4 parental animals were removed from culture for subsequent genotyping of mtRPOL. Only lines determined to be homozygous for mtRPOL were maintained as recombinant hybrid inbred lines. Lines were inbred by full-sib mating for three fully discrete generations. Offspring of the fourth inbred generation were used to measure OXPHOS transcriptional profiles. Inbred parental lines were generated in parallel as controls from single pairs of population stocks and were inbred for an identical number of generations.
Genotyping of mtRPOL.
F4 hybrid adults used to initiate recombinant hybrid inbred lines were removed from culture and genotyped for mtRPOL using the protocol of Ellison and Burton (26). Briefly, DNA was prepared by digesting single copepods with 25 μl of proteinase-K cell-lysis buffer at 65°C for 1 h followed by 85°C for 15 min. PCR amplification was performed with a combination of either MTRP-gtype.1F and MTRP-gtype.1R primers (for SD × AB and SD × SCN crosses) or MTRP-MspI.F and MTRP-MspI.R primers (for AB × SCN crosses). Primer sequences are in Table S4. Amplification products were digested overnight with Hinf1 or MspI restriction enzymes, respectively (New England Biolabs) and scored on 2% agarose gels.
Isolation of RNA and Generation of cDNA.
Animals were subjected to one of two treatments before isolation of nucleic acids. A control treatment consisted of transferring animals to 100% seawater at 20°C for 30 min before initiation of RNA and DNA isolation procedure. In a stress treatment, animals were transferred to a hypoosmotic environment (50% seawater) at 20°C for 30 min before the isolation of RNA. The nature and duration of this stress treatment were determined empirically with pure strain individuals.
Animals were pooled in groups of five individuals, with all animals within a sample being drawn from a single clutch within a recombinant hybrid inbred line. RNA was isolated by using TRI Reagent (Sigma) according to manufacturer's specifications. Tissues were disrupted by bead-beating on a MiniBeadbeater (BioSpec Products) with zirconia/silica beads for 20 seconds at 4.5 m · sec−1. cDNA synthesis was completed by using gene-specific primers (Table S4) and the Stratascript Reverse Transcription kit (Stratagene), according to the manufacturer's specifications, using 30-μl reaction volumes. For each sample, cDNA was generated for α-tubulin (ATU), and either cytochrome oxidase I (COI) and cytochrome oxidase Va (COVa), cytochrome B (CytB) and cytochrome C (CytC), or ATPase 6 (ATP6) and ATPase c (ATPc). Once first-strand cDNA synthesis was complete, samples were stored at −20°C until subsequent quantification assays.
qPCR Assays.
All qPCR assays were completed on a Stratagene Mx3000P (Stratagene) using the Power SYBR Green PCR Master Mix (Applied Biosystems). Standards were generated by using purified PCR products from parental cDNA samples quantified by using a Lambda 35 UV/VIS spectrophotometer (Perkin–Elmer). Approximate copy number was inferred by using OligoCalc (39) and standards for each target gene were diluted to include concentrations of 1011 copies · μl−1, 1010 copies · μl−1, 109 copies · μl−1, 108 copies · μl−1, and 107 copies · μl−1.
qPCR was performed in 20-μl reaction volumes containing 8 μl of cDNA, 0.25 μg/ml each forward and reverse primer, and 10 μl of Power SYBR Green PCR Master Mix (Applied Biosystems). Reaction conditions included a 2-min denaturing step at 95°C, followed by 40 cycles of 30 seconds at 95°C, 30 seconds at 52°C, and 60 seconds at 72°C. Denaturing curves were generated at the end of each series of assays to verify the specificity of the reaction.
Statistical Analysis.
Quantitative PCR data were normalized to a standard curve. Data for target genes were then normalized to the expression levels of α-tubulin (housekeeping gene). Statistical analysis was performed by using SPSS Graduate Pack 11.0 Software (SPSS). For crosses involving the AB population (ABxSD, ABxSCN, and the respective reciprocal crosses), data on CytB, CytC, ATPc, and ATP6 were included in the analysis. For the crosses involving the SD and SCN populations, more animals were available for analysis and data on these four loci plus COI and COVa were included.
Data were initially classified by using a multivariate discriminant function analysis, including each cross, all six genotypic classes (maternal parent, paternal parent, matched and mismatched mtRPOL-mtDNA genotypes for each cross) for both the control and salinity stress treatments for a total of 72 data categories. Animals from the same hybrid inbred line were treated as identical source material for all analyses. Functions with eigenvalues greater than one were extracted from the data and Wilk's Lambda was used to test (1) equality of group means and (2) power of the primary functions. A structure matrix was calculated for each of the significant discriminant functions to weight the correlation of each of the variables (the genes assayed) with the principal discriminant functions. One-way ANOVA with a Bonferroni posthoc multiple comparisons correction (α = 0.05) was applied to the significant functions to distinguish the classes of data. Identical one-way ANOVA analyses were used to evaluate stress-treatment data independently of the discriminant function analysis. Sample sizes for each class of data are listed in Table S5.
Supplementary Material
Acknowledgments.
We thank three anonymous reviewers for insightful comments. A. Ho and S. Hsu assisted with copepod culture. Funding was provided by National Science Foundation grants (R.S.B.) and a National Institutes of Health Graduate Training Grant (C.K.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804253105/DCSupplemental.
References
- 1.Harrison RG. Hybrid zones: Windows on evolutionary process. Oxford Surveys Evol Biol. 1990;7:69–128. [Google Scholar]
- 2.Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- 4.Michalak P, Noor MAF. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol Biol Evol. 2003;20:1070–1076. doi: 10.1093/molbev/msg119. [DOI] [PubMed] [Google Scholar]
- 5.Ranz JM, Namgyal K, Gibson G, Hartl DL. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 2004;14:373–379. doi: 10.1101/gr.2019804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haerty W, Singh RS. Gene regulation divergence is a major contributor to the evolution of Dobzhansky-Muller incompatibilities between species of Drosophila. Mol Biol Evol. 2006;23:1707–1714. doi: 10.1093/molbev/msl033. [DOI] [PubMed] [Google Scholar]
- 7.Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson NA, Porter AH. Rapid speciation via parallel, directional selection on regulatory genetic pathways. J Theor Biol. 2000;205:527–542. doi: 10.1006/jtbi.2000.2070. [DOI] [PubMed] [Google Scholar]
- 9.Porter AH, Johnson NA. Speciation despite gene flow when developmental pathways evolve. Evolution. 2002;56:2103–2111. doi: 10.1111/j.0014-3820.2002.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 10.Ortíz-Barrientos D, Counterman BA, Noor MAF. Gene expression divergence and the origin of hybrid dysfunctions. Genetica. 2007;129:71–81. doi: 10.1007/s10709-006-0034-1. [DOI] [PubMed] [Google Scholar]
- 11.Landry CR, Hartl DL, Ranz JM. Genome clashes in hybrids: Insights from gene expression. Heredity. 2007;99:483–493. doi: 10.1038/sj.hdy.6801045. [DOI] [PubMed] [Google Scholar]
- 12.Scheffler IE. Mitochondria. New York: Wiley; 1999. [Google Scholar]
- 13.Taylor RW, Turnbull DM. Mitochondrial DNA transcription: Regulating power supply. Cell. 2007;130:211–213. doi: 10.1016/j.cell.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 16.Tiranti V, et al. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum Mol Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 17.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsunaga M, Jaehning JA. Intrinsic promoter recognition by a “core” RNA polymerase. J Biol Chem. 2004;279:44239–44242. doi: 10.1074/jbc.C400384200. [DOI] [PubMed] [Google Scholar]
- 19.Burton RS, Lee BN. Nuclear and mitochondrial gene genealogies and allozyme polymorphism across a major phylogeographic break in the copepod Tigriopus californicus. Proc Natl Acad Sci USA. 1998;91:5197–5201. doi: 10.1073/pnas.91.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burton RS. Intraspecific phylogeography across the Point Conception biogeographic boundary. Evolution. 1998;52:734–745. doi: 10.1111/j.1558-5646.1998.tb03698.x. [DOI] [PubMed] [Google Scholar]
- 21.Burton RS. Evolutionary consequences of restricted gene flow in the intertidal copepod Tigriopus californicus. Bull Mar Sci. 1986;39:526–535. [Google Scholar]
- 22.Burton RS. Hybrid breakdown in developmental time in the copepod Tigriopus californicus. Evolution. 1990;44:1814–1822. doi: 10.1111/j.1558-5646.1990.tb05252.x. [DOI] [PubMed] [Google Scholar]
- 23.Edmands S. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution. 1999;53:1757–1765. doi: 10.1111/j.1558-5646.1999.tb04560.x. [DOI] [PubMed] [Google Scholar]
- 24.Edmands S, Burton RS. Cytochrome c oxidase activity in interpopulation hybrids of a marine copepod: A test for nuclear-nuclear or nuclear-cytoplasmic coadaptation. Evolution. 1999;53:1972–1978. doi: 10.1111/j.1558-5646.1999.tb04578.x. [DOI] [PubMed] [Google Scholar]
- 25.Ellison CK, Burton RS. Interpopulation hybrid breakdown maps to the cytoplasm. Evolution. 2008;62:631–638. doi: 10.1111/j.1558-5646.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 26.Ellison CK, Burton RS. Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution. 2006;60:1382–1302. [PubMed] [Google Scholar]
- 27.Rawson PA, Burton RS. Functional coadaptation between cytochrome c and cytochrome c oxidase within allopatric populations of a marine copepod. Proc Natl Acad Sci USA. 2002;99:12955–12958. doi: 10.1073/pnas.202335899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison JS, Burton RS. Tracing hybrid incompatibilities to single amino acid substitutions. Mol Biol Evol. 2006;23:559–564. doi: 10.1093/molbev/msj058. [DOI] [PubMed] [Google Scholar]
- 29.Willett CS. Deleterious epistatic interactions between electron transport system protein-coding loci in the copepod Tigriopus californicus. Genetics. 2006;173:1465–1477. doi: 10.1534/genetics.105.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton RS, Byrne RJ, Rawson PD. Three divergent mitochondrial genomes from California populations of the copepod. Tigriopus californicus. Gene. 2007;403:53–59. doi: 10.1016/j.gene.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 31.Flowers JM. La Jolla, CA: University of California at San Diego; 2005. PhD dissertation. [Google Scholar]
- 32.Goolish E, Burton RS. Energetics of osmoregulation in the intertidal copepod Tigriopus californicus. Func Ecol. 1989;3:81–89. [Google Scholar]
- 33.Park C, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 34.Falkenberg M, Larsson N-G, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 35.Kimura M. The role of compensatory mutation in molecular evolution. J Genet. 1985;64:7–19. [Google Scholar]
- 36.Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 37.Landry CR, et al. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- 39.Kibbe WA. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acid Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.