Abstract
Full understanding of the functional complexity of the protein interactome requires mapping of biomolecular complexes within the cellular environment over biologically relevant time scales. New approaches to imaging interacting protein partners in vivo will allow the study of functional proteomics of human biology and disease within the context of living animals. Herein, we describe a universal transgenic reporter mouse strain that expresses firefly luciferase (Fluc) under the regulatory control of a concatenated Gal4 promoter (TgG4F(+/−)). Using an adenovirus to deliver a fused binding-domain-activator chimera (Gal4BD-VP16), induction of bioluminescence in TgG4F(+/−) tissues of up to 4 orders of magnitude was observed in fibroblasts, liver, respiratory epithelia, muscle, and brain. The TgG4F(+/−) reporter strain allowed noninvasive detection of viral infectivity, duration of the infection as well as viral clearance in various tissues in vivo. To demonstrate protein–protein interactions in live mice, the well characterized interaction between tumor suppressor p53 (fused to Gal4BD) and large T antigen (TAg) (fused to VP16) was visualized in vivo by using a two-hybrid strategy. Hepatocytes of TgG4F(+/−) mice transfected with p53/TAg demonstrated 48-fold greater induction of Fluc expression in vivo than noninteracting pairs. Furthermore, to demonstrate the feasibility of monitoring experimental therapy with siRNA in vivo, targeted knockdown of p53 resulted in markedly reduced light output, whereas use of a control siRNA had no effect on protein interaction-dependent induction of Fluc. Thus, this highly inducible Gal4→Fluc conditional reporter strain should facilitate imaging studies of protein interactions, signaling cascades, viral dissemination, and therapy within the physiological context of the whole animal.
Keywords: luciferase, molecular imaging, p53, protein–protein interactions, transgenic reporter mouse
Protein–protein interactions are fundamental to living systems, mediating many cellular functions, including signal transduction, cell cycle progression, apoptosis, and metabolic control. On a whole organism scale, protein–protein interactions regulate signals that affect overall homeostasis, patterns of development, normal physiology and disease in living animals (1–3). Protein–protein interactions also have considerable potential as therapeutic targets (4, 5). Traditional techniques in biochemistry for isolation and purification of protein complexes tend to select for only the most robust of interactions, whereas weak and transient protein interactions escape detection (6). Furthermore, many protein interactions arise from host–cell interactions in tissue-specific pathways that cannot be investigated fully with in vitro systems. Thus, there is considerable interest in imaging signal transduction and protein–protein interactions noninvasively in their normal physiological context within living organisms (7). Previous strategies have used transcriptional modification, activated signal transduction, or modification of enzymatic activity to generate readily observable biological or physical reporter readouts (8, 9).
Molecular imaging has become an important technology to noninvasively assess and monitor biological processes in vivo. In recent years, various reporter systems have been developed allowing analysis of dynamic processes of transcriptional regulation, gene expression, protein–protein interactions, signal transduction, and regulatory pathways in living animals in physiological as well as disease contexts (for reviews, see refs. 10–12). Cell trafficking and oncogenic transformation as well as targeted drug action can be followed over time using noninvasive imaging strategies (13–15). Furthermore, use of luciferases to tag genes, cells, and pathogens has proved to be a versatile tool to study infection (16).
Fundamentally, the detection of physical interaction among two or more proteins can be assisted if association between the interactive partners leads to the production of a readily observed biological or physical readout (8). Besides endogenously regulated systems, inducible strategies have been developed that allow regulation of transduced genes in response to exogenous triggers (10). Among the various inducible systems that have been established, one widely known platform is based on an inducible reporter driven by a chimeric transcription factor comprising the DNA-binding domain from the yeast transcription factor Gal4 (Gal4BD) and the activation domain from the VP16 protein (VP16) of herpes simplex virus (HSV-1) (17, 18). When fused to interacting proteins of interest, the separate DNA-binding domain (BD) and activator domain (AD) of the chimeric transcription factor are brought together within the cell nucleus to transactivate a reporter gene. This system has been used to study protein–protein interactions in living animals by injecting cell lines stably expressing components of a two-hybrid system (19–21), but the usefulness can be expanded to various other cellular processes.
To facilitate the study of regulated protein–protein interactions and macromolecular assemblies in cells and living animals in vivo, we herein describe the generation of a transgenic Gal4→Fluc reporter mouse strain that expresses the firefly luciferase gene (Fluc) under the regulatory control of the Gal4 promoter, commonly used in yeast and mammalian two-hybrid systems. This universal reporter mouse enabled the monitoring of protein–protein interactions, viral progression, and experimental therapeutics in vivo.
Results
Expression of Gal4→Fluc Transgene in Cells.
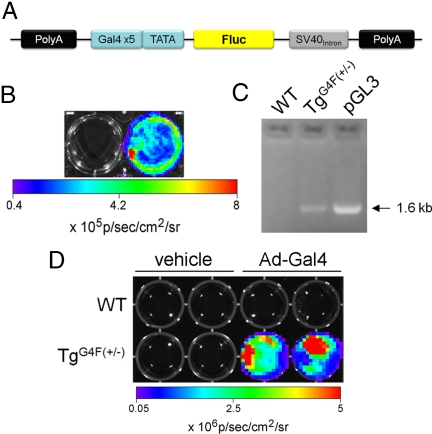
The transgene construct uses a Gal4-TATA promoter to regulate expression of firefly luciferase (Fluc) as depicted in Fig. 1A. As a first test toward confirming induced transactivation in living cells and to confirm the biochemical integrity of the construct, we selected and expanded a stable HeLa cell line expressing the Gal4→Fluc reporter construct (HeLa Fluc #43). After transfection with a positive control vector pM3-VP16 that expresses a fusion of the Gal4BD to the VP16, we imaged cells using a cooled CCD camera system. As shown in Fig. 1B, there was no detectable signal in mock transfected cells compared with a 43-fold induction of signal in cells transfected with the fusion transactivator.
Fig. 1.
Generation and analysis of the reporter transgene. (A) Schematic representation of the transgene vector that was injected into pronuclei of FVB oocytes. (B) HeLa Fluc #43, a cell line stably expressing the Gal4→Fluc reporter construct, was transfected with a positive control vector pM3-VP16 that expresses a fusion of Gal4BD to VP16 (right well). A 43-fold induction was observed when compared with mock-transfected cells (left well). (C) PCR genotyping from a negative mouse (WT), a positive littermate (TgG4F(+/−)), and pGL3 as positive control are shown. (D) Mouse embryonic fibroblasts (MEF) of a negative embryo (WT) and a positive embryo (TgG4F(+/−)) were treated with either vehicle or 4 × 107 pfu of Ad5Gal4BD-VP16 (Ad-Gal4). Cells were imaged 24 h after infection. MEFs from the positive embryo were induced 3-log fold when compared with the uninducible negative MEFs.
Generation of a Gal4→Fluc Transgenic Mouse.
DNA was injected into pronuclei of FVB oocytes, embryos transferred to pseudopregnant FVB mice, and the resulting animals screened for the presence of the transgene. To confirm integration of the luciferase expression cassette into the mouse genome, tail DNA samples were analyzed by PCR, using primers that amplified the entire 1.6-kb coding region of Fluc. Four founder lines were established and bred for 9 generations. In one line, the transgene (Tg) was stably expressed and passed to offspring in near Mendelian ratios. Fig. 1C shows the PCR result of a positive heterozygous offspring (hereafter referred to as TgG4F(+/−)), a negative littermate (WT), and pGL3 as a positive control for Fluc.
We next isolated mouse embryonic fibroblasts (MEFs) on day 13.5 from the selected line of transgenic animals. MEFs from a WT embryo and a positive embryo (TgG4F(+/−)), as verified by PCR genotyping, were treated with either vehicle or 4 × 107 pfu of the adenovirus Ad5Gal4BD-VP16 (Ad-Gal4). The adenovirus was engineered from the same fusion construct (Gal4BD fused to VP16) as the positive control vector used in earlier experiments. Upon bioluminescence imaging 24 h postinfection, photon flux emitted by Ad-Gal4-infected MEFs from the TgG4F(+/−) embryo were induced 1,000-fold compared with the uninducible infected WT MEFs (Fig. 1D).
We also tested primary adult skin fibroblast cells derived from either a WT mouse or a TgG4F(+/−) littermate. To transfect these primary cells with pM3-VP16, we used a nucleofector device and cotransfected with Renilla luciferase as a transfection control. The Fluc/Rluc signal ratio in positive TgG4F(+/−) mouse fibroblasts was 6.5-fold higher than fibroblasts from a negative WT littermate (data not shown).
Expression of Gal4→Fluc Transgene in Living Animals.
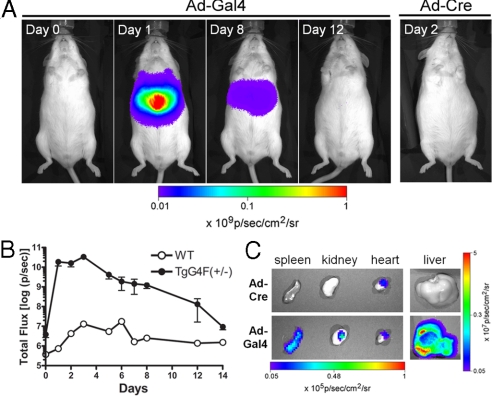
To show that the transactivated reporter gene could be imaged in living animals, we administered Ad-Gal4 (1.4 × 107 pfu per mouse) by tail vein injection into TgG4F(+/−) and negative (WT) mice (n = 3 for TgG4F(+/−), n = 2 for WT). We obtained an image of the mice before treatment (day 0), then injected the virus, and began imaging 24 h after viral administration (Fig. 2A). Signals arising from Fluc expression in the abdomen progressed for 14 days, wherein the signal declined back to almost pretreatment levels (Fig. 2B). A low baseline whole body signal ≈10-fold above instrument background noise could be detected in TgG4F(+/−) mice from this founder line in the pretreatment images. Nonetheless, signal transactivation on day 1 after viral treatment was almost 4 orders of magnitude over baseline (6,000-fold induction), achieving photon flux values of >1010 photons/sec. Because of the highly inducible nature of the photon output, images on day 0 were obtained with different exposure and binning settings compared with the rest of the experiment, otherwise, remarkably, signal output from induced TgG4F(+/−) mice would have saturated the CCD camera. As a control, TgG4F(+/−) mice were also injected with an off-target adenovirus, Ad5CMVCre (Ad-Cre), at the same pfu per mouse and imaged as described. Ad-Cre had no effect on transactivation of the reporter gene, as shown on day 2 (Fig. 2A). Photon flux values were comparable with WT mice.
Fig. 2.
Transactivation of the reporter gene in the abdomen. (A) TgG4F(+/−) and WT mice were injected i.v. with adenovirus Ad-Gal4 expressing the fusion construct (Gal4BD fused to VP16). Mice were imaged before treatment with virus (day 0) and then followed over time as indicated, starting 24 h postinjection (day 1). Images from a representative TgG4F(+/−) mouse are shown. The signal on day 1 was induced over 3 orders of magnitude. As a control, TgG4F(+/−) mice were injected i.v. with an off-target adenovirus, Ad-Cre, and imaged over time. No signal was detected (day 2 is shown). (B) Signal progression of Fluc activity in these mice was followed for 14 days until the signal declined back to almost pretreatment levels. Shown is total photon flux plotted over time. Data are presented as mean ± SEM (n = 3 for TgG4F(+/−) mice) and mean ± range (n = 2 for WT); error bars for WT are smaller than symbol size. (C) Organs from TgG4F(+/−) mice administered Ad-Gal4 or Ad-Cre (as indicated) were also imaged ex vivo (day 2). Signal from the liver of an Ad-Gal4-treated mouse was 3-log greater than the liver of an Ad-Cre-treated mouse. Modest signal was also detectable from the spleen and kidney of the Ad-Gal4-treated mouse, whereas low basal activity was observed in the hearts of both.
For refined imaging on day 2, organs (liver, spleen, kidney, and heart) from several TgG4F(+/−) mice administered Ad-Gal4 or Ad-Cre were imaged ex vivo. Signal detected in the heart of both animals represented a low organ-specific basal activity in this transgenic mouse strain. However, as expected after Ad-Gal4, induced signal was highest from the liver, with photon flux values of >109 photons/sec observed, 3 orders of magnitude above the signal from the liver of an Ad-Cre-treated mouse (Fig. 2C). Signal was also induced in the spleen and kidney in Ad-Gal4-treated mice.
Panexpression of Gal4→Fluc Transgene.
To test for transactivation of the transgene in multiple tissues, we also delivered the transactivating virus Ad-Gal4 through inhalational, intramuscular, and intracranial routes. To control for nonspecific effects, TgG4F(+/−) mice were also treated with an off-target virus (Ad-Cre).
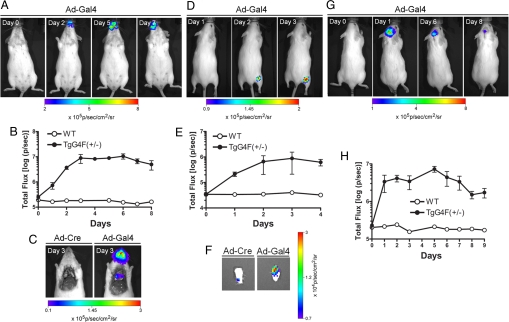
Nasal delivery of Ad-Gal4 (2 × 106 pfu per mouse in 10 μl of PBS) resulted in a steady signal increase when compared with the pretreatment image on day 0 (Fig. 3A). Imaging signals were highest after 3 days, showing a 50-fold increase over background (Fig. 3B), but were detectable only in nasal passages in living animals. To determine whether signal was also induced in the lungs, several mice were killed on day 3 and imaged through an open chest cavity. The reporter transgene was clearly activated in the lungs of Ad-Gal4-treated mice, whereas no signal was detected in the control Ad-Cre-treated animal (Fig. 3C).
Fig. 3.
Transactivation of the reporter gene in multiple tissues. (A) TgG4F(+/−) and WT mice inhaled 2 × 106 pfu per mouse of Ad-Gal4. Mice were imaged before treatment with virus (day 0) and then followed over time, starting with 24 h post inhalation treatment. Images from a representative TgG4F(+/−) mouse are shown. (B) Signal progression of the mice was followed for 9 days until the signal declined back to almost pretreatment levels and is shown as total photon flux plotted over time. Data are presented as mean ± SEM (n = 3 each); error bars for WT are smaller than symbol size. (C) TgG4F(+/−) mice administered Ad-Gal4 or off-target Ad-Cre (as indicated) were imaged through an open chest cavity on day 3. Signal was detectable from the nasal passages and lung of only the Ad-Gal4-treated mouse. (D) TgG4F(+/−) and WT mice were injected with 2.3 × 105 pfu per mouse of Ad-Gal4 (right calf muscle) and with vehicle (left calf muscle). Mice were imaged on day 1 (24 h after virus injection) and followed over time. Images from a representative TgG4F(+/−) mouse are shown. (E) Signal progression of Fluc activity was followed for 4 days and is shown as total photon flux plotted over time. Data are presented as mean ± range (n = 2 each); error bars for WT are smaller than symbol size. (F) TgG4F(+/−) mice were injected identically with Ad-Gal4 (right calf muscle) or with Ad-Cre (left calf muscle). Muscles were excised on day 3 and imaged ex vivo. (G) TgG4F(+/−) and WT mice were injected intracranially with 2 × 106 pfu per mouse of Ad-Gal4 2 mm lateral and posterior of the bregma and 3 mm below the dura. Mice were imaged before treatment with virus (day 0) and then followed over time, starting with 24 h postinjection. Images from a representative TgG4F(+/−) mouse are shown. (H) Signal progression was followed for 9 days and is shown as total photon flux plotted over time. Data are presented as mean ± SEM (n = 3 for TgG4F(+/−) mice) and without error bars for WT (n = 1).
For intramuscular delivery, TgG4F(+/−) mice were injected with Ad-Gal4 (2.3 × 105 pfu in 50 μl of PBS) on one side and injected with vehicle alone or Ad-Cre on the other side (Fig. 3D). The observed signal increase was highest after 3 days with an induction of 22-fold above background (vehicle mock). Because of the low signal induction from this delivery method, the signal was followed for 4 days only (Fig. 3E). TgG4F(+/−) mice administered control Ad-Cre virus did not show any significant signal induction, even after imaging the muscle ex vivo (shown on day 3 in Fig. 3F).
For intracranial delivery, Ad-Gal4 (2 × 106 pfu per mouse in 10 μl of PBS) was injected over 2 minutes into a site 2 mm lateral and posterior of the bregma and 3 mm below the dura. Signal 24 h after injection was 15.6-fold higher in TgG4F(+/−) mice than in a negative WT mouse (Fig. 3G). The signal remained elevated for 7 days and then started to decline (Fig. 3H).
Imaging Protein–Protein Interactions in Vivo.
To demonstrate that protein–protein interactions could be visualized in vivo using this genomic transactivating strategy, we used the well characterized interaction between the tumor suppressor p53 and large T antigen (TAg) from SV40. p53 was fused to the Gal4BD, whereas TAg was fused to the activator domain from VP16. As a control negative, noninteracting partner of p53, we used the polyoma virus coat protein (CP) fused to VP16.
To transfect hepatocytes of the TgG4F(+/−) reporter mice in vivo with various protein pairs (interacting p53/TAg or noninteracting p53/CP) as well as each protein by itself, we used the hydrodynamic somatic gene transfer method. Renilla luciferase (Rluc) driven by the CMV promoter was added to the plasmid mixture and used to control for efficiency of plasmid delivery. Mice were imaged for Renilla expression first, and generally we found that the bioluminescence output was similar in all imaged mice, providing evidence that all plasmids were delivered and processed in equal amounts in these animals. After the signal from Rluc had declined to background levels (≈4 h), mice were imaged for Fluc expression (Fig. 4A).
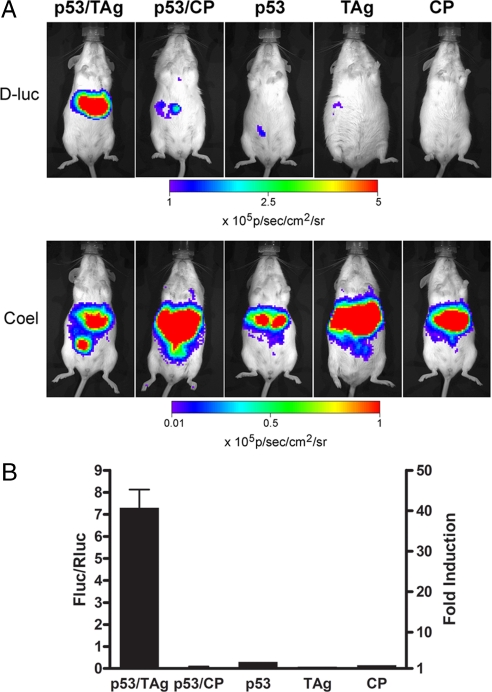
Fig. 4.
Imaging protein–protein interactions in vivo using a Gal4→Fluc reporter mouse. (A) Hepatocytes of TgG4F(+/−) mice were transfected in vivo by hydrodynamic somatic gene transfer with different plasmid combinations of pGal4BD-p53 together with pVP16-TAg or off-target pVP16-CP, or each plasmid separately as indicated. A plasmid for Renilla luciferase was used as transfection control. Mice were imaged before (pretreatment) and 24 h after transfection for expression of firefly luciferase (Fluc with D-luciferin) and Renilla luciferase (Rluc with coelenterazine). Images of representative mice are shown. CP; polyoma virus coat protein. (B) Data are plotted for the different plasmid combinations as controlled for transfection efficiencies (Fluc/Rluc; left axis) and as fold-induction (right axis) 24 h after transfection. Data are presented as mean ± range for plasmid combinations (n = 2), and without error bars for single plasmids (n = 1). Four independent experiments showed similar results.
Only the interacting pair induced Fluc expression in the liver, showing 48-fold induction. Data from all mice are represented as total flux of Fluc over total flux of Rluc (Fig. 4B). The mean interacting protein Fluc/Rluc ratio was 7.3, whereas the noninteracting protein Fluc/Rluc ratio was 0.15.
Use of siRNA to Modulate Protein–Protein Interactions in Vivo.
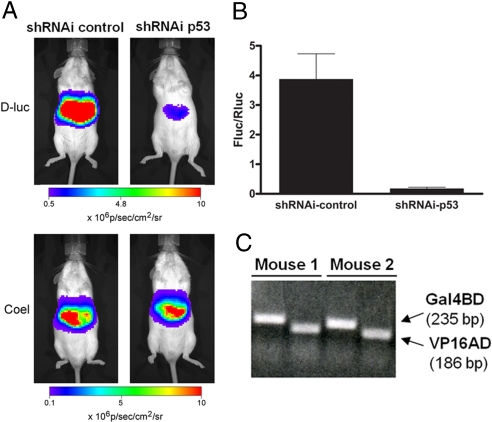
Having shown that protein–protein interactions could be visualized in vivo, we wanted to test whether we could modulate the interaction between p53 and TAg. Therefore, we introduced a shRNAi against p53 into the in vivo experimental protocol. Hepatocytes of the TgG4F(+/−) reporter mice were transfected with the protein pair p53/TAg together with the empty shRNAi vector (shRNAi-control) or the shRNAi vector against p53 (shRNAi-p53). Again, Rluc was added to control for efficiency of plasmid delivery, and imaging was performed as described. As expected, shRNAi-control had no effect on the spontaneous interaction between p53 and TAg to transactivate expression of Fluc. Knockdown of p53, however, was efficient and modulated the interaction-dependent transactivation of Fluc (Fig. 5A). Data from all TgG4F(+/−) mice were represented as normalized Fluc over Rluc activity and showed that knockdown of p53 reduced bioluminescence output to only 4.5% of the shRNAi-control-treated mice (Fig. 5B). To independently document equivalent hepatocellular delivery of the plasmids encoding the interacting proteins (p53 and TAg), DNA from livers was extracted after in vivo imaging. PCR amplification of Gal4BD and VP16 sequences confirmed the equal presence of p53 (fused to Gal4BD) and TAg (fused to VP16) plasmids in both treatment groups (Fig. 5C). Therefore, because plasmid delivery was equal, we concluded that the decrease in bioluminescence signal was attributable to abrogated expression of p53 fusions arising from interference mediated by shRNAi-p53.
Fig. 5.
Imaging shRNAi abrogation of protein–protein interactions in vivo. (A) Hepatocytes of TgG4F(+/−) mice were transfected in vivo by hydrodynamic somatic gene transfer with plasmid combinations of pGal4BD-p53 together with pVP16-TAg plus shRNAi-control or shRNAi-p53. A plasmid for Renilla luciferase was used as transfection control. Mice were imaged 24 h after transfection for expression of firefly luciferase (D-luciferin) or Renilla luciferase (coelenterazine). Images from representative mice are shown. (B) Data are plotted for the shRNAi-control and shRNAi against p53. Total photon flux was normalized for transfection efficiencies (Fluc/Rluc). Data are presented as mean ± SEM for shRNAi-53 (n = 3) and mean ± range for shRNAi-control (n = 2). Three independent experiments showed similar results. (C) PCRs of DNA isolated from livers of TgG4F(+/−) mice that had been injected with plasmid combinations of pGal4BD-p53 together with pVP16-TAg (mouse 1) or pGal4BD-p53 together with pVP16-TAg plus shRNAi-p53 (mouse 2) demonstrate equal delivery of the plasmids encoding the interacting proteins.
Discussion
In this study, we engineered a transgenic reporter mouse (TgG4F(+/−)) that expresses firefly luciferase (Fluc) under the regulatory control of a concatenated (5×) Gal4 promoter. The reporter gene, highly responsive to a two-hybrid transactivation strategy, was inducible in all tissues tested, including liver, spleen, kidney, respiratory epithelia, lungs, muscle, and brain, as well as MEFs and primary adult mouse fibroblasts derived from the mice.
Using in vivo bioluminescence imaging after viral delivery of the binding/activation-domain hybrid, luciferase expression from the TgG4F(+/−) reporter animals was directly visualized and could be used to study viral uptake, localization, and retention over time. As could be clearly seen from the results, different viral delivery methods yielded a variety of responses. Systemic injection (i.v.) of Ad-Gal4 virus resulted in the highest inducibility (4 orders of magnitude) and fastest transactivation of the reporter (<24 h) and was detected primarily within the liver, as expected. After i.v. injection, modest reporter activation in spleen and kidney could also be visualized ex vivo. Adenovirus-mediated signal induction by Ad-Gal4 in muscle, however, was low and corresponded well with the known low expression in muscle of the Coxsackie virus B Ad receptor (CAR) (22), a receptor necessary for cell uptake of the virus. Nonetheless, that a variety of tissues low in CAR levels were readily visualized after adenoviral injections emphasized the sensitivity and high inducibility of this transactivating TgG4F(+/−) reporter strain. Conversely, no reporter activation was detected in mice treated with a control off-target adenovirus (Ad-Cre).
Several other conditional luciferase pan-reporter mice have been described. The Berns group generated a LucRep reporter mouse strain wherein a constitutive β-actin promoter drives a floxed GFP upstream of Fluc (23). Cre-dependent Fluc expression was observed in every organ analyzed with signal increases of 4- to 6-log per mg of tissue ex vivo. Although signals produced ex vivo and in vivo are difficult to compare to each other and across animal models, it may be instructive, nonetheless, to note that delivery of Ad-Cre particles to the lungs of compound LucRep/Cre-dependent Kras2v12 mice produced relative photon fluxes on the order of 106 photons/sec. By comparison, delivery of Ad-Gal4 to respiratory epithelia of TgG4F(+/−) mice produced 107 photons/sec. The same LucRep strain crossed to a compound prostate-restricted (ARR2PB) Cre-mediated PTEN-null mouse produced a 5- to 50-fold increase in pelvic bioluminescence with prostate tumor growth (24). Alternatively, targeted knockin strategies were used to generate a ROSA26 floxed-stop Fluc conditional reporter strain (25). After i.v. and intracranial delivery of Ad-Cre to ROSA26 floxed-stop Fluc mice, maximal bioluminescence signals produced by recombination within liver and brain were on the order of 106 and 105 photons/sec/cm2/steradian, respectively, lower than the bioluminescence signals of 109 and 106 photons/sec/cm2/steradian produced by transactivation in TgG4F(+/−) mice similarly injected with Ad-Gal4. Indeed, the efficiency of Cre-dependent recombination at the ROSA26 locus remains under rigorous investigation (23).
Iyer et al. (26) describe the generation of a transgenic mouse model in which a tissue-restricted Gal4 promoter drives expression of Fluc. In contrast to the TgG4F(+/−) universal reporter strain described herein, the reporter and effector targeting genes were linked on a single construct and thus, tissue-restricted expression of the Gal4→Fluc cassette was transactivated by the weak prostate-specific antigen promoter (PSA) driving a Gal4BD-(VP16)2 fusion construct. Modest hormone-dependent bioluminescence signals were identified, as would be expected, in the dorso-lateral prostate (≈106 photons/sec/cm2/steradian). Using a similar strategy, Wang et al. (27) also linked effector and reporter targeting genes, in this case comprising a VEGF promoter driving a Gal4BD-(VP16)2 fusion to transactivate Gal4→Fluc, revealing an 8-fold increase in bioluminescence upon VEGF activation during wound healing (27). The investigators reported that pairing the Gal4BD-(VP16)2 or the Gal4BD-VP16 transactivators with five concatenated Gal4 sequences (G5) driving the reporter resulted in high absolute reporter signals, but at the cost of high background before induction. Overall, pairing the Gal4BD-(VP16)2 transactivator with two concatenated Gal4 sequences (G2) driving the reporter provided a reasonable compromise between low background and acceptable fold-inducibility. In effect, experiments with our TgG4F(+/−) universal reporter strain comprised transactivating five concatenated Gal4 sequences (G5) with an exogenously delivered Gal4BD-VP16 fusion to drive expression of the Fluc reporter, resulting in a strain with favorably low background and extremely high inducibility. Whether gene locus-specific effects on the Fluc reporter or modes of transactivation underpin differences in reporter magnitude and behavior remain to be determined.
To demonstrate that the TgG4F(+/−) universal reporter strain could also be used to study more complex systems, such as protein–protein interactions in vivo, we transfected mouse hepatocytes with expression cassettes encoding a well known interacting protein pair, p53 (fused to Gal4BD) and large T-antigen (fused to VP16). A 48-fold induction of bioluminescence was observed upon expressing the interacting pair, a level far greater than that detected with noninteracting pairs. We also successfully demonstrated that the p53/TAg protein–protein interaction system could be abrogated in the presence of a siRNA targeted to p53. Binding and dissociation of proteins frequently are dynamic processes, and thus, protein–protein interactions are modified by physiological and pathophysiologic conditions that exist only in intact organisms. Development and application of noninvasive imaging techniques for monitoring protein–protein interactions should enable investigations of mechanisms that regulate the dynamics of protein complexes in vivo, as well as small molecule inhibitors and related therapeutics. However, future studies remain to establish at what level transient and weak protein interactions can be detected in vivo.
In summary, we have generated a Gal4→Fluc transgenic reporter strain with extremely high inducibility, which is amenable to studies on a wide variety of pathways and biological responses. Because the Fluc reporter appears to be widely inducible, readouts are not limited to one tissue, but can be applied to a variety of tissues in vivo and ex vivo. Conversely, breeding strategies incorporating transgenic animals selectively expressing Gal4BD-VP16 fusions may unlock investigation into a variety of tissue-restricted imaging queries. This universal TgG4F(+/−) reporter strain should enhance access to dynamic analysis in living animals of signal transduction, metabolism, tumor progression and metastasis, cell fusion events, gene therapy, mammalian development, infectious disease, and host response.
Materials and Methods
Additional details are available online as supporting information (SI) Text.
Generation of Gal4→Fluc Transgenic (TgG4F(+/−)) Mice.
The plasmid contained a polyadenylation site upstream of five consensus repeats of the upstream activation sequence of Gal4 and an adenovirus E1b minimal TATA box driving the firefly luciferase (P. pyralis) gene (derived from pGL3; Promega). Downstream of the expression cassette, the construct contained a SV40 intron and another polyadenylation site to enhance the stability and expression efficiency of the transgene. The purified linear plasmid was injected into pronuclei of fertilized FVB oocytes, which were transferred into pseudopregnant FVB mice (Washington University Mouse Genetics Core). Pups were weaned at 3 weeks of age and tails sampled for DNA analysis.
DNA Extraction and PCR.
Mouse tail genomic DNA samples were harvested using a standard phenol/chloroform/isoamyl alcohol extraction protocol. To test for the presence of the transgene, a PCR was performed with primers spanning the entire ORF of the luciferase coding sequence. The primers were 5′-ATGGAAGACGCCAAAAACATAAAGAAAGGCCC-3′ at the 5′ end and 5′-CACGGCGATCTTTCCGCCCTTCTTG-3′ at the 3′ end, which amplified a 1.6-kb fragment. PCRs were carried out with each primer, mouse genomic DNA or plasmid DNA of pGL3-control (Promega) using Platinum Pfx Polymerase with Enhancer (Invitrogen).
After hydrodynamic injections, to document the presence of the plasmids encoding the interacting protein pair (pGal4BD-p53 and pVP16-TAg), DNA was extracted from mouse livers using a tissue kit (Qiagen). The primers to amplify a 235-bp fragment of Gal4BD were 5′-CGCTACTCTCCCAAAACCAA-3′ and 5′-CAGTCTCCACTGAAGCCAATC-3′; primers to amplify a 186 bp fragment of VP16 were 5′-GGACGAGCTCCACTTAGACG-3′ and 5′-AGGGCATCGGTAAACATCTG-3′. PCR products were fractionated on 1% or 2% agarose gels and visualized with ethidium bromide.
Generation of Recombinant Viruses.
An Ad5Gal4BD-VP16 adenovirus (Ad-Gal4) that incorporated a fusion construct (Gal4BD fused to VP16) was generated by the Gene Transfer Vector Core at the University of Iowa. Ad5CMVCre (Ad-Cre) was purchased from the same facility.
Cell Culture.
HeLa cells were transfected with pcDNA6/V5-HisA engineered to contain a concatenated Gal4 promoter driving expression of firefly luciferase in place of a mutant thymidine kinase reporter (19). Stable clones were selected in the presence of blasticidin, and clone #43 was used for experiments (HeLa Fluc #43). MEF were isolated on day 13.5, as described (28). Primary mouse fibroblasts were isolated as described in ref. 28.
Bioluminescence Imaging of Cell Lines.
HeLa Fluc #43 cells were transiently transfected with pM3-VP16, a positive control vector that expresses a fusion of Gal4BD and VP16, from the Mammalian Matchmaker Two-Hybrid Assay Kit (Clontech). The next day, cells were imaged in colorless MEBSS containing 150 μg/ml D-luciferin as described in ref. 29 using a cooled CCD camera (IVIS 100; Caliper Life Sciences). MEFs (P1) were incubated with DMEM containing polybrene and 4 × 107 pfu of Ad-Gal4 for 90 min. After that time, complete medium was added. Cells were imaged after 24 h using 150 μg/ml D-luciferin in colorless MEBSS. Primary mouse skin fibroblast cells were cotransfected using a nucleofector device (Amaxa) with pM3-VP16 along with pRluc-N3 (BioSignal Packard) encoding Renilla luciferase as a transfection control. Cells first were imaged for Renilla luciferase activity with MEBSS containing a final concentration of 400 nM coelenterazine. To image firefly luciferase, the media was changed to MEBSS containing 150 μg/ml D-luciferin and images acquired with a >590 nm bandpass filter. Data analysis was performed as described (29, 30).
Animal Studies.
Animal care and euthanasia were approved by the Washington University Medical School Animal Studies Committee. Bioluminescence imaging was performed on the IVIS under 2.5% isoflurane anesthesia at the indicated time points as described (29, 30). After imaging, animals were killed by cervical dislocation. For i.v. injections, mice were anesthetized with isoflurane before tail vein injections of Ad-Gal4 or Ad-Cre [1.4 × 107 pfu per mouse in 100 μl of PBS (pH 7.4)]. Imaging of mice was performed by injection of D-luciferin (150 μg/g of body weight, i.p.), starting 24 h after virus delivery. On day 2, after imaging in vivo, some mice were killed by cervical dislocation, organs quickly harvested, and immediately imaged ex vivo. For inhalational delivery, mice were anesthetized with ketamine/xylazine mixture. To straighten the airway, mice were hung by their front teeth on twisted surgical tape. Ad-Gal4 or Ad-Cre [2 × 106 pfu per mouse in 10 μl of PBS (pH 7.4)] was then added dropwise onto the nose. Imaging was performed before virus administration (day 0) and 24 h after virus delivery (day 1) and followed for 9 days. On day 3, a few mice were killed by cervical dislocation, the chest cavity quickly opened, and mice imaged to detect signal originating from the lungs. For intramuscular injections, mice were anesthetized with isoflurane and injected with Ad-Gal4 (2.3 × 105 pfu per mouse in 50 μl of PBS) into the right calf muscle. The left calf muscle was injected with vehicle alone or Ad-Cre (2.3 × 105 pfu per mouse in 50 μl of PBS). Imaging was started 24 h after viral inoculation and followed for 4 days. On day 3, a few mice were killed by cervical dislocation, calf muscle quickly harvested, and immediately imaged ex vivo. For intracranial injections, mice were anesthetized with ketamine/xylazine mixture and fixed in a stereotactic frame (Stoelting). Ad-Gal4 [2 × 106 pfu per mouse in 10 μl of PBS (pH 7.4)] were injected through a 27-gauge needle over 2 min at 2 mm lateral and posterior to the bregma and 3 mm below the dura. The incision was closed with Vetbond (3M). Imaging was performed prior (day 0) and 24 h after virus injection (day 1) and followed for 9 days.
Hydrodynamic Injections.
In vivo transfection of mouse hepatocytes was performed using the hydrodynamic somatic gene transfer method as described (29, 31). Briefly, different plasmid combinations of pM-53 together with pVP16-T or pVP16-CP (Mammalian Matchmaker Kit Two-Hybrid Assay Kit; Clontech) (15 μg) and pRluc-N3 (2 μg) were combined as indicated. For the shRNAi experiments, different plasmid combinations of pM-53 together with pVP16-T (5 μg) combined with pSuper (shRNAi-control) or pRetrosuper-p53 (shRNAi-p53) (50 μg) (29, 32) and pRluc-N3 (1 μg) as injection control were combined as indicated. Plasmids were diluted in PBS (pH 7.4) in a volume of 1 ml of per 10 g of body weight and rapidly injected into tail veins of mice (29). Imaging of mice for Rluc and Fluc activity, respectively, was performed before and 24 h after somatic gene transfer by injection of coelenterazine (1 μg/g of body weight, i.v.), followed 4 h later (generally no residual signal) by injection of D-luciferin (150 μg/g of body weight, i.p.).
Analysis and Statistics.
Corresponding grayscale photographs and color luciferase images were superimposed and analyzed with LivingImage 2.50 (Xenogen) and Igor (Wavemetrics) image analysis software. Signal intensities for luciferase were obtained from regions of interest defined either with a grid template for cells grown in tissue culture plates or manually for the viral induction sites as well as the hepatocyte transfections. Data were expressed as total photon flux (photons/sec) or as normalized total flux (Fluc photons/sec over Rluc photons/sec). Fold-induction was expressed as normalized posttreatment/pretreatment images. Values (n = indicated animals within each group) were reported as mean ± SEM or mean ± range.
Supplementary Material
Acknowledgments.
We thank Lynne Collins for technical assistance and the Washington University Mouse Genetics Core for oocyte injections. This work was supported by National Institutes of Health Grant P50 CA94056.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801075105/DCSupplemental.
References
- 1.Zhang H, et al. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark G, Kerr I, Williams B, Silverman R, Schreiber R. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Ogawa H, Ishiguro S, Gaubatz S, Livingston D, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 4.Heldin C. Signal transduction: Multiple pathways, multiple options for therapy. Stem Cells. 2001;19:295–303. doi: 10.1634/stemcells.19-4-295. [DOI] [PubMed] [Google Scholar]
- 5.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 6.Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- 7.Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein–protein interactions in vivo with genetically encoded reporters. Annu Rev Biomed Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- 8.Toby G, Golemis E. Using the yeast interaction trap and other two-hybrid-based approaches to study protein–protein interactions. Methods. 2001;24:201–217. doi: 10.1006/meth.2001.1182. [DOI] [PubMed] [Google Scholar]
- 9.Luker GD, Sharma V, Piwnica-Worms D. Visualizing protein–protein interactions in living animals. Methods. 2003;29:110–122. doi: 10.1016/s1046-2023(02)00285-2. [DOI] [PubMed] [Google Scholar]
- 10.Gross S, Piwnica-Worms D. Spying on cancer: Molecular imaging in vivo with genetically encoded reporters. Cancer Cell. 2005;7:5–15. doi: 10.1016/j.ccr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 12.Tsien RY. Imagining imaging's future. Nat Rev Mol Cell Biol. 2003;(Suppl):SS16–SS21. [PubMed] [Google Scholar]
- 13.Negrin RS, Contag CH. In vivo imaging using bioluminescence: a tool for probing graft-versus-host disease. Nat Rev Immunol. 2006;6:484–490. doi: 10.1038/nri1879. [DOI] [PubMed] [Google Scholar]
- 14.El-Deiry WS, Sigman CC, Kelloff GJ. Imaging and oncologic drug development. J Clin Oncol. 2006;24:3261–3273. doi: 10.1200/JCO.2006.06.5623. [DOI] [PubMed] [Google Scholar]
- 15.Gross S, Piwnica-Worms D. Molecular imaging strategies for drug discovery and development. Curr Opin Chem Biol. 2006;10:334–342. doi: 10.1016/j.cbpa.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Luker G, Luker K. Optical imaging: Current applications and future directions. J Nucl Med. 2008;49:1–4. doi: 10.2967/jnumed.107.045799. [DOI] [PubMed] [Google Scholar]
- 17.Fields S, Song O. A novel genetic system to detect protein–protein interaction. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 18.Fields S, Sternglanz R. The two-hybrid system: An assay for protein–protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 19.Luker G, et al. Noninvasive imaging of protein–protein interactions in living animals. Proc Natl Acad Sci USA. 2002;99:6961–6966. doi: 10.1073/pnas.092022399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luker G, et al. Molecular imaging of protein–protein interactions: Controlled expression of p53 and large T antigen fusion proteins in vivo. Cancer Res. 2003;63:1780–1788. [PubMed] [Google Scholar]
- 21.Ray P, et al. Noninvasive quantitative imaging of protein–protein interactions in living subjects. Proc Natl Acad Sci USA. 2002;99:2105–3110. doi: 10.1073/pnas.052710999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2003;5:451–462. doi: 10.1002/jgm.409. [DOI] [PubMed] [Google Scholar]
- 23.Lyons SK, Meuwissen R, Krimpenfort P, Berns A. The generation of a conditional reporter that enables bioluminescence imaging of Cre/loxP-dependent tumorigenesis in mice. Cancer Res. 2003;63:7042–7046. [PubMed] [Google Scholar]
- 24.Liao C-P, et al. Mouse models of prostate adenocarcinoma with the capacity to monitor spontaneous carcinogenesis by bioluminescence or fluorescence. Cancer Res. 2007;67:7525–7533. doi: 10.1158/0008-5472.CAN-07-0668. [DOI] [PubMed] [Google Scholar]
- 25.Safran M, et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- 26.Iyer M, et al. Non-invasive imaging of a transgenic mouse model using a prostate-specific two-step transcriptional amplification strategy. Transgenic Res. 2005;14:47–55. doi: 10.1007/s11248-004-2836-1. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, et al. Noninvasive indirect imaging of vascular endothelial growth factor gene expression using bioluminescence imaging in living transgenic mice. Physiol Genomics. 2006;24:173–180. doi: 10.1152/physiolgenomics.00308.2004. [DOI] [PubMed] [Google Scholar]
- 28.Bonifacino JS, et al. Current Protocols in Cell Biology. New York: Wiley; 1998. [Google Scholar]
- 29.Pichler A, Zelcer N, Prior JL, Kuil AJ, Piwnica-Worms D. In vivo RNA interference-mediated ablation of MDR1 P-glycoprotein. Clin. Cancer Res. 2005;11:4487–4494. doi: 10.1158/1078-0432.CCR-05-0038. [DOI] [PubMed] [Google Scholar]
- 30.Gross S, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods. 2005;2:607–614. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 32.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.