Abstract
High-level production of membrane proteins, particularly of G protein-coupled receptors (GPCRs) in heterologous cell systems encounters a number of difficulties from their inherent hydrophobicity in their transmembrane domains, which frequently cause protein aggregation and cytotoxicity and thus reduce the protein yield. Recent advances in cell-free protein synthesis circumvent those problems to produce membrane proteins with a yield sometimes exceeding the cell-based approach. Here, we report cell-free production of a human olfactory receptor 17-4 (hOR17-4) using the wheat germ extract. Using the simple method, we also successful produced two additional olfactory receptors. To obtain soluble olfactory receptors and to increase yield, we directly added different detergents in varying concentrations to the cell-free reaction. To identify a purification buffer system that maintained the receptor in a nonaggregated form, we developed a method that uses small-volume size-exclusion column chromatography combined with rapid and sensitive dot-blot detection. Different buffer components including salt concentration, various detergents and detergent concentration, and reducing agent and its concentrations were evaluated for their ability to maintain the cell-free produced protein stable and nonaggregated. The purified olfactory receptor displays a typical a α-helical CD spectrum. Surface plasmon resonance measurements were used to show binding of a known ligand undecanal to hOR17-4. Our approach to produce a high yield of purified olfactory receptor is a milestone toward obtaining a large quantity of olfactory receptors for designing bionic sensors. Furthermore, this simple approach may be broadly useful not only for other classes of GPCRs but also for other membrane proteins.
Keywords: detergent screen, G protein-coupled receptor purification, membrane protein, odorant interaction, surface plasmon resonance
Membrane proteins play vital roles in all living systems. Approximately 30% of all genes in almost all sequenced genomes code for membrane protein (1–3). However, our understanding of their structures and function falls far behind that of soluble proteins. As of August 2008, there are only 167 unique membrane protein structures of total 386 variations known (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html) among >53,000 structures in the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The reason is that there are several notoriously difficult steps to obtain membrane proteins that include: (i) production of large quantities, (ii) purification of stable and functional membrane proteins, and (iii) long-term stabilization of nonaggregated membrane proteins. Membrane proteins are exquisitely fine nature-made molecular devices that will be very useful for a wide range of applications including solar energy harvesting and ultrasensitive sensing. To accelerate membrane protein structural studies and use them for design and fabrication of nanobiodevices, new and simple methods are crucial.
Among membrane proteins, G protein-coupled receptors (GPCRs) represent the largest family (4, 5). The olfactory receptors are the most abundant GPCRs (6, 7) and perhaps are one of the oldest sensory GPCRs. Although olfaction is an important part of our perception, the olfactory receptor molecular structure currently remains unknown. The question of how the finite numbers of olfactory receptors recognize seemingly infinite odorants remains a tantalizing enigma. This lack of understanding is mainly due to the difficulty of obtaining large quantities of olfactory receptors.
Heterologous expression of olfactory receptors is extremely difficult, with only a few examples found in the literature (8–10). The heterologous expression system is often performed in Escherichia coli, yeast, or a mammalian cell line. Olfactory receptors expressed in such systems are not only partly located in the membrane but are also found in cell organelles or in inclusion bodies, probably because of inappropriate processing, and the expression levels are often low (11). In addition, expression in mammalian cell lines is time consuming and expensive (12). For structural, biochemical, and biophysical studies as well as nanobiodevice design, high-yield production is a prerequisite. Therefore, the expression of the gene of interest is often driven by a strong promoter, resulting in overloading of the different translocation machineries in the cell (13, 14). When production levels increase, so does the burden on the translocation machinery resulting in cellular toxicity and eventually cell death. High-level production of membrane proteins in cells as bacteria, yeast, and mammalian cells, therefore, has an inherent fundamental problem.
Cell-free protein production employing extract from various sources such as rabbit, insect, wheat germ, and E. coli provide an attractive alternative because the extract contains all of the necessary components for transcription and translation but without membranes. To compensate for the lack of a natural membrane, addition of suitable detergent is crucial for the solubilization and conservation of the freshly produced membrane protein. In addition to detergents, other additives, such as GroE and DnaK, can be supplemented to the reaction to enhance folding (15).
Recent advances in cell-free protein production technology have improved the protein yields, with the single most important development being continuous-exchange feeding systems developed by Spirin and colleagues (16) in which up to ≈9.7 mg of protein per milliliter of reaction solution have been produced (17). Cell-free production is also fast and convenient in a high-throughput setting and is currently routinely used at RIKEN Japan for protein synthesis for their structural genomics projects (18). Currently, >1,000 soluble protein structures have been determined from proteins synthesized by the cell-free method (19). However, not a single membrane protein structure has been determined from proteins by using the cell-free method.
Klammt et al. (20) investigated the effects of 24 different detergents on the cell-free production of membrane proteins including the GPCR V2R in E. coli extracts. Most detergents did not affect the yield of either total or soluble membrane proteins. However, detergents such as phosphocholines inhibited the transcription and translation machinery, despite their similarity to natural lipids in the cell membrane. Glycosidases and CHAPS, although both are known as mild detergents, also inhibited protein production. The optimal detergent was found to be protein-specific. In the case of a GPCR vasopressin receptor, digitonin and Brij generated the highest yields of soluble receptor. Similar results have also been reported for 10 other GPCRs produced in E. coli cell-free-based extracts (20, 21). Although the selection of the optimal detergent is crucial to achieve the highest possible yield of soluble membrane protein, the composition of the buffer for subsequent purification and storage is equally important.
The ultimate technique for screening buffers' composition for optimal membrane protein stability is a protein activity assay. However, these assays are difficult to develop for many membrane proteins (22, 23) and particularly for olfactory receptors. A more feasible approach is to study the protein homogeneity by using size-exclusion chromatography (SEC). SEC provides information about the protein size and monodispersity because a single symmetrical protein peak may suggest that the protein is correctly folded and stable, making it a promising candidate for functional and structural studies (24, 25). By using a short SEC column with a small bed volume, the protein quantity needed per run is small, the short running time allowing for multiple runs per day and efficient buffer scouting. Additionally, a small bed volume reduces the required amount of buffer containing costly detergent.
Here, we report high-level protein production of three olfactory receptors, hOR17-4, mOR23, and mS51, using cell-free protein production technology as well as a time-efficient downstream buffer optimization and stabilization method. The production yields ≈1 mg protein per 3 ml of cell-free reaction solution. This quantity is sufficient to carry out secondary structural and odorant binding analyses as well as initial crystallization screening trials.
Results and Discussion
General Considerations.
Following the reports in the literature of producing GPCRs by using E. coli extract and also for the lower-price consideration of E. coli extract as compared with wheat germ-based extract; we first used E. coli cell-free extracts from both Roche Diagnostics and Qiagen. However, we found both produced hOR17-4 in soluble and insoluble proteins at very low or nondetectable levels (results not shown).
Because E. coli thioredoxin has been shown to greatly enhance the levels of GPCRs in E. coli-based extracts, hOR17-4 was cloned into a plasmid including a thioredoxin N-terminally of hOR17-4. The addition of thioredoxin did, however, not affect the production yield of hOR17-4. The production is driven by a T7-promotor, and any vector carrying a T7 promotor and termination sequence and ribosomal binding site should suffice for hOR17-4. Roche Diagnostics provides optimized vectors and linear generation template kits for high-yield production. When production was performed by using a linear template, the production levels were not improved.
For production in wheat germ extract, the hOR17-4 gene was cloned into pVEX1.3 and pVEX1.4 including a six-residue histidine tag at either C or N terminus, respectively. Compared with production in E. coli, wheat germ lysate produced the olfactory receptor in detectable amounts. In addition, pilot studies showed that production from pVEX1.3 was far superior to pVEX1.4 (results not shown).
Effect of Detergents for Olfactory Receptor Yield and Solubility.
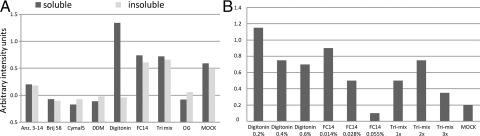
Detergents are a prerequisite when working with membrane proteins in solution, and this also holds true for membrane proteins produced in cell-free extracts because both E. coli and wheat germ extracts are devoid of lipids. We therefore carefully studied the optimal types and concentrations of various detergents in the cell-free system. A panel of eight detergents was chosen based on their efficacy in previous studies of GPCR production in cell-free extracts or on their ability to solubilize GPCRs expressed in vivo (Table S1). The detergents were all tested at concentrations above their respective critical micelle concentration (CMC) value measured by the suppliers. The detergents' effect on the amount of soluble and insoluble produced hOR17-4 was quantitatively measured by using the dot-blot method (Fig. 1A). OG, DDM, Cymal5, and Brij58 all reduced the production level, and Anzergent3-14, FC14, and the Tri-mix either did not or only marginally affect the production level. Digitonin was found to be very effective in maintaining the solubility of the protein, with no insoluble protein detected. This is in agreement with previous studies of other GPCRs produced in E. coli-based systems (20).
Fig. 1.
The effects of first screen of different detergents on olfactory receptor production levels in wheat germ cell-free production system. (A) The amount of soluble (black bars) and insoluble (gray bars) produced hOR17-4 in different detergents. (B) Titration of detergent concentration for production of soluble hOR17-4.
To avoid large protein–detergent micelles and to avoid inhibiting the production system at high concentrations of detergents, we systematically optimized detergent concentration by either decreasing or increasing its concentrations. Increasing the digitonin concentration resulted in very low levels of insoluble protein. In addition to digitonin, the optimal concentration of FC14 and the Tri-mix was also evaluated by increasing the concentration, because the production levels were reasonable, but the protein was not completely soluble. The yield of soluble receptor could not be increased by increasing concentrations of FC14 or Tri-mix. This is most likely because of an inhibitory effect of these detergents on the cell-free production system (Fig. 1B). The most favorable digitonin concentration was found to be 0.2% for hOR17-4 and was consequently chosen as the optimal concentration for large-scale hOR17-4 production (Fig. 1B).
To test whether the identified production conditions would also be suitable for other olfactory receptors, mouse receptors mOR23 and mS51 were also produced. The initial detergent screen was reduced to three detergents, namely FC14, Brij58, and the Tri-mix, mainly chosen for their efficacy in producing soluble hOR17-4. Production of soluble mOR23 was similar to the production of hOR17-4, with the highest yield in digitonin, but at a slightly higher digitonin concentration: 0.6% (Fig. 2). The mS51, on the other hand, was not produced at all in digitonin, but Brij58 at 0.1% and the Tri-mix at 1× produced high levels of soluble protein (Fig. 2). Once the detergent composition was optimized, production in cell-free extract was proven to be very efficient, as well as time-saving, in producing soluble olfactory receptors compared with other production systems.
Fig. 2.
The effects of additional screen of different detergents on olfactory receptor production levels in wheat germ cell-free production system. The amount of soluble (black bars) and insoluble (gray bars) produced mOR23 (A) and mS51 olfactory receptor production (C) with Brij58, digitonin, and Tri-mix. Titration of detergent concentration for production of soluble mOR23 (B) and mS51 (D and E).
Optimizing Buffer Conditions.
The use of GFP as a reporter in SEC-based buffer optimization is straightforward and time-saving. Protein purification is not necessary, and detection can be carried out on-line by using a chromatography system (25). However, one drawback of the use of GFP fusions is that GFP has to be cleaved off for down-stream applications such as crystallization. This process has to be optimized so as to avoid loss of protein activity and protein yield. We thus used an alternative approach where a sample from each SEC fraction was dot-blotted on a membrane and developed by using an antibody directed against a short C-terminal tag. By using a short SEC column with only a 3-ml bed volume, up to 12 runs with different buffers could be performed in a single day. This time-saving step allowed us to investigate the effect of 12 different detergents, six different concentrations of NaCl, two pH points, and the presence of a reducing agent at three different concentrations.
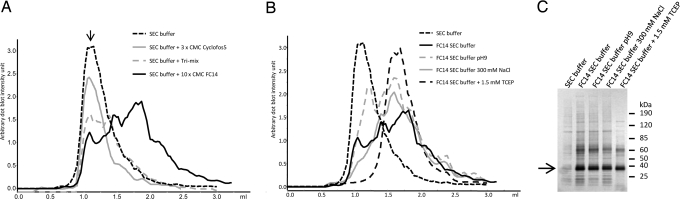
Initially, the presence of 10 different detergents (Table S2) in the run was evaluated. Immediately after cell-free production, the protein was captured on anti-bovine rhodopsin monoclonal antibody 1D4-coated beads, and the new buffer was introduced during the washing step. The beads were washed with 100-bed volumes for complete detergent exchange. Exchange to buffers containing C8E4, DDOMG, LDAO, or no detergent resulted in very low or nondetectable levels of hOR17-4 in the eluate from the beads, possibly because of receptor aggregation on the beads. The Tri-mix, Cyclophos5, and DDM, on the other hand, resulted in high yields, but the receptor eluted from the SEC with the void, indicating that the protein had aggregated to complexes >600 kDa, which is the exclusion volume of the SEC column (Fig. 3). Three of the detergents, Anzergent3–14, Brij58, and FC14, resulted in protein eluting as monomers or as higher-molecular-mass oligomers (Fig. 3). The receptor in FC14 buffer eluted at 1.71 ml, corresponding to 114 kDa. The amount of FC14 that binds to the hOR17-4 molecule has been calculated to be 76 kDa, and, when added to the molecular mass of the receptor (36 kDa), it is very close to the observed molecular mass of 112 kDa. The buffers including the three most promising detergents were further optimized by varying the pH (pH 7.5 and 9.5), NaCl concentration (150 and 300 mM), and the presence of a reducing agent TCEP (0 and 1.5 mM).
Fig. 3.
Size-exclusion chromatography analysis of hOR17-4 aggregation states. (A) We used Superdex 200 GL 5/150. SEC analysis in different detergents, running buffer: SEC buffer [25 mM Tris·HCl (pH 7), 150 mM NaCl, 10% glycerol) supplemented with 3× CMC cyclofos 5 (gray line), Tri-mix (gray dashed line), 10× CMC FC14 (black line), and SEC buffer only (black dotted line). The arrow indicates the void volume. (B) Monitoring the effect of hOR17-4 aggregation states in different buffers with different pH, NaCl concentration, and reducing agent. (C) SDS/PAGE of the elution fractions from affinity purification with the same buffers as in B.
The elution profiles from SEC in the buffers containing Anzergent3-14 and Brij58 were not affected by the different salt concentrations, pH, and reducing agent; the receptor was eluted as larger aggregates/oligomeric forms. In the case of FC14 detergent-containing buffers, there was a marked difference between the buffers (Fig. 3). Higher pH decreased the monomer: aggregate ratio, and higher salt concentration increased the same ratio. TCEP fully prevented aggregation and, as indicated by the Gaussian-shaped peak from SEC, the eluted protein monodispersity. Addition of TCEP also resulted in a purer sample from affinity purification (Fig. 3C). As a consequence, FC14 was chosen as detergent for further studies.
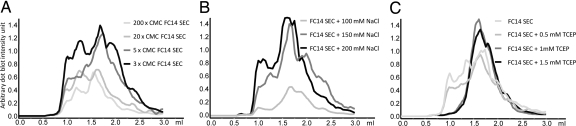
Because high salt and detergent concentration could be detrimental for crystallization, and because high concentration of reducing agent could hamper protein activity, a careful screening of those variables was performed to find the lowest concentration that still kept the protein in a monodispersed form. Increasing detergent concentration, up to 200× CMC, resulted in faster elution; 1.56 ml compared with 1.71 ml. This corresponds to a difference in calculated molecular mass of 100 kDa, indicating a lower protein/detergent ratio at 200× CMC compared with 3× CMC (Fig. 4A). A low protein/detergent ratio has been reported to impede crystallization and protein activity. The 3× CMC resulted in the most well defined peaks. The salt concentration was screened between 50 and 300 mM NaCl, and concentrations <150 mM were found with lower yields of receptor (Fig. 4B). Interestingly, varying salt concentration did not affect the receptor retention volume. Addition of reducing agent prevented aggregation at concentrations as low as 1 mM (Fig. 4C). The optimal buffer conditions were finally identified after testing different combinations of the salt, detergent, and TCEP concentrations identified above. Two buffers, one with TCEP and one without, were chosen for further studies. Buffer 1: 25 mM Tris (pH 7), 10% glycerol, 3× CMC FC14, and 200 mM NaCl and buffer 2: 25 mM Tris (pH 7), 10% glycerol, 3× CMC FC14, 150 mM NaCl, and 1 mM TCEP. SEC in a TCEP-containing buffer resulted in a single Gaussian-shaped peak, indicative of a monodispersed protein sample. SEC without TCEP resulted in monomer and dimer as well as aggregation peaks, but monomers could be separated from the other forms. Models of hOR17-4 predict the presence of disulfide bonds, and their reduction by TCEP could hinder protein folding and function. TCEP, however, breaks down over time, and disulfide bonds will form again.
Fig. 4.
Fine coarse size-exclusion chromatography analysis of hOR17-4 aggregation states. Different concentrations of FC14 (A), NaCl (B), and TCEP (C) using a Superdex 200 GL 5/150. Running buffer is SEC buffer supplemented with 10× CMC FC14 if not stated otherwise.
Large-Scale Purification.
For further studies of secondary and tertiary structure as well as crystallization studies, pure receptor at high concentration is needed. Crystallization is usually required at concentration >5 mg/ml, and CD measurements require concentrations ≈0.2 mg/ml. Large-scale purification of the hOR17-4 receptor from up to 6 ml of reaction solution was carried out by increasing the bed volume of 1D4 antibody-coated beads. The same pattern as in the buffer optimization experiments was observed, with TCEP resulting in a purer sample. The 1D4 monoclonal antibody is highly specific, and compared with a Ni2+-chelate-based affinity purification, the 1D4 was able to purify the receptor from very low levels in the reaction solution to 70% purity in one step (results not shown). For further purification, the eluted fractions were concentrated and applied to a 24-ml SEC column. The peaks containing hOR17-4 were pooled and concentrated again. The yield was determined to be ≈0.3 mg of pure hOR17-4 per milliliter of cell-free reaction solution. Eleven GPCRs have previously been produced in cell-free lysates at high levels. The yield of unpurified receptors ranged between 0.15 and 6 mg per milliliter of reaction lysate (20, 21). A few of the receptors have been purified, but no yields have been reported. Each purification step always results in protein loss; for example, a 50% yield has been reported for a two-step purification scheme of the GPCR neurotensin (26). The yield of pure hOR17-4 is well in agreement with what could be expected from the literature describing unpurified GPCRs produced in cell-free systems.
Secondary Structure Analysis of Purified Receptors.
Correct protein structural folding largely depends on the mode of production and on the properties of the buffer milieu in which the protein is stored. Olfactory receptors are one member of the GPCRs, which are predicted to have mostly α-helical structure with seven transmembrane helices. The GPCR α-helical feature has been verified by three high-resolution structures that have been recently solved (27–29). Both CD spectra from purified hOR17-4 in buffer with and without reducing agent (buffer 1 and buffer 2) display the typical α-helical features (Fig. 5). The mean residue ellipticity was more distinct from hOR17-4 purified in buffer 2 that contains the reducing agent TCEP.
Fig. 5.
CD spectra of hOR17-4. Purified hOR17-4 diluted to 0.3 mg/ml in Buffer 1 (black line) 25 mM Tris (pH 7), 10% glycerol, 3× CMC FC14 and 200 mM NaCl; and buffer 2 (gray line) 25 mM Tris (pH 7), 10% glycerol, 3× CMC FC14, 150 mM NaCl, and 1 mM TCEP. It is noteworthy that the identical amount of protein has different helical content in two different buffers.
Surface Plasmon Resonance (Biacore T100) Detection of Odorant Interaction with hOR17-4.
The CD measurements suggest that hOR17-4 is correctly folded. We ask whether the receptor is able to bind to the odorants. The activity of a solubilized olfactory receptor is very difficult to assess because its ligands are mostly <300 Da, the receptor itself is >100 times larger, ≈36,000 Da. In addition, the ligand-binding pocket is predicted to be buried within the protein, which can make it difficult to measure odorant binding through traditional methods. Biacore is an SPR-based label-free technology that is sensitive enough to enable detection of extremely small changes in mass when an odorant binds to the receptor captured on the sensor chip surface. Dose-dependent specific binding of undecanal, a known ligand, to the hOR17-4 receptor captured on the Biacore sensor surface was observed in the present study. (Fig. 6A). Furthermore, the data generated could be used to derive an affinity constant, KD, of ≈22 μM (Fig. 6B), in agreement with other in vitro experiments that have shown that odorants bind to hOR17-4 with EC50s in the micromolar range (30).
Fig. 6.
Surface plasmon resonance detection of the interaction between hOR17-4 with the known hOR17-4-binding odorant undecanal. (A) Responses from injections at 1.2, 3.7, 11, 33, and 100 μM. Sensorgram are double-referenced and solvent-corrected. Experiments with an odorant that is a known nonbinder for hOR17-4 did not show any interaction (results not shown). (B) Equilibrium binding responses plotted versus undecanal concentration and fitted to a simple binding isotherm to yield an affinity of ≈22 μM for hOR17-4.
Whereas these experiments represent a preliminary investigation into understanding such intriguing interactions, future experiments where odorants or ligands having more homogenous solution behavior are used will likely yield more detailed mechanistic and functional information.
Conclusion
Our study of producing three olfactory receptors by using wheat germ cell-free extracts proved the simple technology to be very useful in obtaining correctly folded and active GPCR membrane proteins. Together with efficient buffer scouting using small-volume size-exclusion chromatography, appropriate detergent and other buffer components could be identified. In addition, the high-yield production provided sufficient olfactory receptor to initiate detailed structural analysis.
Because most membrane proteins are natural molecular devices, our work will likely facilitate the design of membrane protein based nano–bio devices for a wide range of applications, from detection of infinitesimal amounts of odorants, emitted from diverse diseases and environment to direct harvest of solar energy.
Materials and Methods
Reagents.
All detergents where purchased from Anatrace except Digitonin EMD (Merck). The cell-free protein production kits (both E. coli and wheat germ systems) were purchased from Roche Diagnostics. Protein purification materials are purchased from GE Healthcare Life Science. Others are described below.
Cell-Free Production.
Generation of DNA template.
The ORF for the human olfactory receptor 17-4 (hOR17-4) (UniProt accession number P34982) was generated by using PCR-based gene synthesis. By using the free program DNAWorks (http://helixweb.nih.gov/dnaworks), oligonucleotides were designed to build the ORFs of the olfactory receptors for PCR-based gene synthesis. The ORF of hOR17-4 was optimized for E. coli class II codon usage with the addition of a six-residue-long C-terminal histidine tag, followed by a stop codon and N- and C-terminal att-sites. The following parameters were used for automatic design of the oligos: oligo size 45 nucleotides, annealing temperature 58°C, 25 nM oligonucleotide, 10 mM sodium, and 2.0 mM Mg2+, and the codon frequency threshold was set at 100%. The PCR product was cloned into pDEST42 and pBAD-DEST49 (Invitrogen) for nonfused and thioredoxin-fused protein production, respectively, according to manufacturer's instructions. Linear templates for production in RTS 100 HY E. coli kit was generated by using RTS E. coli Linear Template Generation Set (Roche) according to the manufacturer's instructions with a six-residue-long C-terminal histidine tag. For production of hOR17-4, OR23, and S51 in wheat germ extract, a human codon-optimized version was produced in a similar manner but with NcoI and SmaI restriction sites for cloning into pIVEX1.3 WG and pVEX1.4 WG, which includes a six-residue-long C- or N-terminal histidine tag, respectively, according to the manufacturer's instructions. A third construct for production in wheat germ extract was generated with the nine-residue-long Rho-tag (TETSQVAPA) instead of the C-terminal histidine tag. The constructs encoding the olfactory receptors were verified by DNA sequencing. Plasmid DNA template for cell-free production was obtained from the Genopure Maxi kit (Roche) with an OD260/280 < 1.7. The plasmid was aliquoted and stored at −20°C, and the same batch was used throughout the study.
Cell-free production in E. coli extracts.
Production of hOR17-4 in E. coli extracts was performed by using the RTS 100 HY E. coli kit, RTS E. coli Disulphide kit, and EasyXpress Protein Synthesis Mini kit (Qiagen) according to the manufacturer's instructions in pDEST42 and pBAD-DEST49 production plasmids. A linear template was also used in the case of the RTS 100 HY E. coli kit.
Cell-free production in wheat germ extracts.
Small-scale production in wheat germ lysate was performed in 50-μl reaction chambers by using a RTS 100 Wheat Germ CECF kit. Large-scale 1-ml reactions were carried out by using a RTS 500 Wheat Germ CECF kit.
An initial screen was set up to test the effect of detergents on the yield of soluble receptor hOR17-4 by adding different detergents to the reaction chambers, see Table S1. The concentrations tested were all above the CMC of the respective detergent. The concentrations of respectively Digitonin, Brij58, DDM, OG, and DDM were based on previous results of production of GPCRs in E. coli cell-free systems (20). The optimal concentration of FC14, Digitonin, and the Tri-mix to keep the freshly produced receptor soluble was further tested at concentrations between 0.014% and 0.055%, 0.2% and 0.6%, and 1–3×, respectively. Initial detergent screening for soluble production of OR23 and S51 was performed in FC14, Digitonin, and the Tri-mix, and the optimal concentration was identified by titration of the detergent that resulted in the highest production in the initial screen with hOR17-4. When the production cycle had terminated, the reaction solution was centrifuged for 10 minutes at 16,000 × g at 4°C to separate soluble and insoluble proteins.
To analyze the effect of the different detergents, 1.5 μl of the soluble and insoluble fraction was dotted on a Protran BA85 nitrocellulose membrane (Whatman). The membrane was blocked, washed, and probed with either a 1D4 antibody directed against the Rho-tag or an anti-His antibody (Novagen/Merck), and developed as previously described (31). The intensity of the spot was recorded using an AlphaImager (Alpha Innotech).
Olfactory Receptor Purification.
Purification of the receptor was carried out by using Sepharose-4B beads (GE Healthcare) with covalently linked 1D4 antibodies specific for the Rho tag. The antibody-coated beads were prepared as described (32). For small-scale purification from 75 μl of production reaction solution, 50 μl of beads washed in PBS was used. The reaction solution–bead mix was incubated end-over-end for 4 hours at 4°C. Purification was thereafter performed in empty spin columns, and unbound material was removed by gravity flow. The beads were washed five times by using, in total, 5 ml of purification buffer. Finally, hOR17-4 was eluted by adding 50 μl of purification buffer containing 200 μM peptide TETSQVAPA to the beads, and the column was capped and incubated for 1 hour at room temperature with shaking. The protein was collected by centrifugation at 800 × g for 10 s.
Large-scale purification was performed as described above with the following exceptions. The amount of 1D4 antibody-coated beads was increased to a final 3:2 ratio of cell-free reaction solution to beads. The receptor was eluted in 5–7 bed volumes of buffer supplemented with the elution peptide. The eluted protein was concentrated to one-third of the volume by using a 10-NMWL Microcon spin filter (Millipore) and injected on a Superdex 200 10/300 24-ml SEC column (GE Healthcare Life Science) for further purification and to remove elution peptide from the sample. For stabilization studies and crystallization, the eluted receptor was concentrated by using a 10-NMWL Amicon Ultra-4 Centrifugal Filter (Millipore). Protein concentration was measured throughout the study by a reducing agent-compatible microplate BCA assay (Pierce).
Optimization of Buffer Conditions.
After small-scale affinity purification using monoclonal antibody of anti-bovine rhodopsin 9-residue TETSQVAPA C-terminal tag 1D4-coupled beads, the oligomeric state of the protein eluted in different buffers was assayed by using a short 3-ml size-exclusion column, Superdex 200 5/150GL (GE Healthcare Life Science). Fifty-microliter fractions were collected in microwell plates. and 1.5 μl of each fraction was dotted on a cellulose membrane (Protran BA85 nitrocellulose membrane) to assay protein amount. The intensity recorded from each spot was inserted in an activity histogram in the ÄKTA software Unicorn version 5.11 (GE Healthcare Life Science) and smoothed over two fraction volumes. For buffer optimization, a 1-ml wheat germ reaction was used for scouting up to 12 buffers.
Secondary Structural Analysis Using CD.
CD measurements of the hOR17-4 were performed with protein at 0.3 mg/ml. The investigations were carried out on an Aviv 202 Spectropolarimeter (Aviv Biomedical) by using a 1-mm path-length cell, equilibrated at 25°C. Spectra were recorded between 200 and 250 nm with 1-nm resolution with a 2-s averaging time. The final spectra were baseline-corrected by subtracting the corresponding buffer spectra obtained under identical conditions. Results were expressed as the molar mean residue ellipticity (θ) at a given wavelength.
Surface Plasmon Resonance (Biacore T100) Odorant Binding Assay.
SPR analyses were conducted by using a Biacore T100 (GE Healthcare). Sensor surface preparation and interaction analyses were performed at 25°C in a PBS [10 mM sodium phosphate, 137 mM sodium chloride, 2.7 mM potassium chloride (pH7.4)] running buffer containing 3% DMSO. Sensor chip CM4, amine coupling reagents [N-ethyl-N′-dimethylaminopropylcarbodiimide, EDC; N-hydroxysuccinimide, NHS; and 1 M ethanolamine HCl (pH 8.5)] and PBS, were obtained from GE Healthcare.
Monoclonal anti-polyhistidine antibody (R & D Systems) was diluted to 5 μg/ml in 10 mM sodium acetate (pH 4.75) and immobilized onto series S sensor chip CM4 via standard amine-coupling procedures (33). Typical immobilization levels were ≈12,000 resonance units (RU). Control surfaces were prepared similarly and were used as reference surfaces for odorant binding experiments.
Cell-free lysate containing the expressed olfactory receptor was centrifuged for 10 min at 14,000 × g at 4°C to remove larger particles. The neat supernatant, containing ≈0.6 mg/ml olfactory receptor, was immediately captured on the antipolyhistidine-derivatized CM4 surface by using a 4-min injection at 10 μl/min. Resultant receptor surface densities were ≈4,000 RU.
Fresh odorant undecanal samples were prepared by dilution in running buffer containing 3% (vol/vol) DMSO to obtain a concentration series of 1.2, 3.7, 11, 33, and 100 μM of odorant undecanal. For odorant interaction analyses, odorant samples were flowed over control and receptor-derivatized surfaces for 60 s at a flow rate of 60 μl/min. Zero-concentration blank buffer cycles were included as negative control samples. Solvent-correction procedures were included to compensate for any DMSO-related bulk refractive index variations and performed as described (34). Nonspecific odorant binding to antipolyhistidine-derivatized sensor surfaces was absent for all analyses reported.
Data analysis was carried out by using Biacore T100 evaluation software (v1.1.1; GE Healthcare). Data were prepared by subtraction of reference surface data and blank buffer sample data, a procedure commonly referred to as “double referencing” (35). Solvent correction was applied as described (34). A plot of the corrected equilibrium odorant binding responses versus odorant concentration was fitted by using the following equation to yield the KD value: Req = KA C Rmax/KA C + 1, where Req is the odorant binding response at equilibrium, C is the odorant concentration, Rmax is the maximum binding capacity of the captured receptor, and KA is the association equilibrium constant. KD, the dissociation equilibrium constant, is calculated as the inverse of KA (KD = 1/KA).
Supplementary Material
Acknowledgments.
L.K. gratefully acknowledges a generous fellowship from the Knut and Alice Wallenberg Foundation, Sweden; J.G-B gratefully acknowledges a generous fellowship from the Netherlands Organization for Scientific Research; and S.Z. gratefully acknowledges a John Simon Guggenheim Fellowship to provide freedom to pursue new frontiers of research. This work was also supported in part by a generous grant from ROHM Corporation, Kyoto, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804766105/DCSupplemental.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loll PJ. Membrane protein structural biology: The high throughput challenge. J Struct Biol. 2003;142:144–153. doi: 10.1016/s1047-8477(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Nilsson J, Persson B, von Heijne G. Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins. 2005;60:606–616. doi: 10.1002/prot.20583. [DOI] [PubMed] [Google Scholar]
- 4.Strader CD, Fong TM, Tota MR, Underwood D, Dixon RA. Structure and function of G protein-coupled receptors. Annu Rev Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- 5.Burne H, Horuk R, Kuhnke J, Michel H. GPCRs: From Deorphanization to Lead Structure Identification. Berlin: Spriner; 2007. [Google Scholar]
- 6.Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 7.Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 8.Breer H, Krieger J, Meinken C, Kiefer H, Strotmann J. Expression and functional analysis of olfactory receptors. Ann NY Acad Sci. 1998;855:175–181. doi: 10.1111/j.1749-6632.1998.tb10563.x. [DOI] [PubMed] [Google Scholar]
- 9.Kiefer H, et al. Expression of an olfactory receptor in Escherichia coli: Purification, reconstitution, and ligand binding. Biochemistry. 1996;35:16077–16084. doi: 10.1021/bi9612069. [DOI] [PubMed] [Google Scholar]
- 10.Vidic JM, Grosclaude J, Persuy MA, Aioun J, Salesse R, et al. Quantitative assessment of olfactory receptors activity in immobilized nanosomes: A novel concept for bioelectronic nose. Lab Chip. 2006;6:1026–1032. doi: 10.1039/b603189g. [DOI] [PubMed] [Google Scholar]
- 11.Ivic L, Zhang C, Zhang X, Yoon SO, Firestein S. Intracellular trafficking of a tagged and functional mammalian olfactory receptor. J Neurobiol. 2002;50:56–68. doi: 10.1002/neu.10016. [DOI] [PubMed] [Google Scholar]
- 12.Lundstrom K. Structural genomics on membrane proteins: Mini review. Comb Chem High Throughput Screen. 2004;7:431–439. doi: 10.2174/1386207043328634. [DOI] [PubMed] [Google Scholar]
- 13.Tate CG. Overexpression of mammalian integral membrane proteins for structural studies. FEBS Lett. 2001;504:94–98. doi: 10.1016/s0014-5793(01)02711-9. [DOI] [PubMed] [Google Scholar]
- 14.Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Buchner J, Pastan I, Brinkmann U. A method for increasing the yield of properly folded recombinant fusion proteins: Single-chain immunotoxins from renaturation of bacterial inclusion bodies. Anal Biochem. 1992;205:263–270. doi: 10.1016/0003-2697(92)90433-8. [DOI] [PubMed] [Google Scholar]
- 16.Spirin AS, Baranov VI, Ryabova LA, Ovodov SY, Alakhov YB. A continuous cell-free translation system capable of producing polypeptides in high yield. Science. 1988;242:1162–1164. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- 17.Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol. 2006;17:373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama S. Protein expression systems for structural genomics and proteomics. Curr Opin Chem Biol. 2003;7:39–43. doi: 10.1016/s1367-5931(02)00019-4. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama S, Terwilliger TC, Kuramitsu S, Moras D, Sussman JL. RIKEN aids international structural genomics efforts. Nature. 2007;445:21. doi: 10.1038/445021a. [DOI] [PubMed] [Google Scholar]
- 20.Klammt C, et al. Evaluation of detergents for the soluble expression of alpha-helical and beta-barrel-type integral membrane proteins by a preparative scale individual cell-free expression system. FEBS J. 2005;272:6024–6038. doi: 10.1111/j.1742-4658.2005.05002.x. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara G, et al. Expression of G protein coupled receptors in a cell-free translational system using detergents and thioredoxin-fusion vectors. Protein Expr Purif. 2005;41:27–37. doi: 10.1016/j.pep.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Columbus L, et al. Expression, purification, and characterization of Thermotoga maritima membrane proteins for structure determination. Protein Sci. 2006;15:961–975. doi: 10.1110/ps.051874706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savage DF, Anderson CL, Robles-Colmenares Y, Newby ZE, Stroud RM. Cell-free complements in vivo expression of the E. coli membrane proteome. Protein Sci. 2007;16:966–976. doi: 10.1110/ps.062696307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 25.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure (London) 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Grisshammer R, White JF, Trinh LB, Shiloach J. Large-scale expression and purification of a G-protein-coupled receptor for structure determination —An overview. J Struct Funct Genomics. 2005;6:159–163. doi: 10.1007/s10969-005-1917-6. [DOI] [PubMed] [Google Scholar]
- 27.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 29.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spehr M, et al. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 31.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biacore. Biacore Sensor Surface Handbook. Sweden: Biacore Uppsala; 2003. [Google Scholar]
- 34.Karlsson R, et al. Biosensor analysis of drug–target interactions: direct and competitive binding assays for investigation of interactions between thrombin and thrombin inhibitors. Anal Biochem. 2000;278:1–13. doi: 10.1006/abio.1999.4406. [DOI] [PubMed] [Google Scholar]
- 35.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.