Abstract
Similar to many genes involved in programmed cell death (PCD), the caspase 2 (casp-2) gene generates both proapoptotic and antiapoptotic isoforms by alternative splicing. Using a yeast RNA–protein interaction assay, we identified RBM5 (also known as LUCA-15) as a protein that binds to casp-2 pre-mRNA. In both transfected cells and in vitro splicing assay, RBM5 enhances the formation of proapoptotic Casp-2L. RBM5 binds to a U/C-rich sequence immediately upstream of the previously identified In100 splicing repressor element. Our mutagenesis experiments demonstrate that RBM5 binding to this intronic sequence regulates the ratio of proapoptotic/antiapoptotic casp-2 splicing isoforms, suggesting that casp-2 splicing regulation by RBM5 may contribute to its tumor suppressor activity. Our work has uncovered a player in casp-2 alternative splicing regulation and revealed a link between the alternative splicing regulator and the candidate tumor suppressor gene. Together with previous studies, our work suggests that splicing control of cell death genes may be an important aspect in tumorigenesis. Enhancing the expression or activities of splicing regulators that promote the production of proapoptotic splicing isoforms might provide a therapeutic approach to cancer.
Keywords: alternative splicing regulation, cancer, cell death, RNA binding protein
Apoptosis, a type of programmed cell death, is critical not only for embryonic development but also for adult tissue homeostasis. Accumulating evidence supports that insufficient cell death plays a role in tumorigenesis and contributes to the development of resistance to cancer therapies. Apoptosis involves a cascade of activation of cysteine–aspartate proteases, beginning with initiator caspases followed by executor caspases (for recent reviews, see refs. 1–3). Caspase 2 is one of the initiator caspases activated by various stimuli, including chemotherapeutic agents, reactive oxygen species, death receptor ligands, and heat shock treatment (4–13). The caspase 2 gene produces several alternative splicing isoforms. Exclusion or inclusion of exon 9 results in the formation of the major splicing isoforms, Casp-2L and Casp-2S, with antagonistic activities in cell death. Exon 9 inclusion leads to the inclusion of an in-frame stop codon, thus producing a truncated protein, Casp-2S. The Casp-2L mRNA transcript encodes a full-length procaspase, whereas Casp-2S lacks the enzyme active domain and inhibits apoptosis. Casp-2L is expressed in a range of tissues, and Casp-2S is expressed predominantly in the heart, brain, and skeletal muscle (14, 15). Casp-2L promotes apoptosis, and Casp-2S overexpression protects cells against apoptosis induced by chemotherapeutic drugs such as etoposide (16–18). Motor neuron death is accelerated in caspase 2-deficient mice during development (19), suggesting that splicing regulation of Casp-2L/Casp-2S may play an important role in neural development.
To understand molecular mechanisms regulating Casp-2 alternative splicing, we have begun to dissect cis-regulatory elements controlling Casp-2 alternative splicing and identified an intronic element, In100, that suppresses exon 9 inclusion (20, 21). Our recent work suggests that such an intronic element may be a general motif for regulating caspase alternative splicing (22). To search for transacting factors regulating Casp-2 alternative splicing, we used a yeast RNA–protein interaction cloning strategy, with the intronic sequence containing In100 and flanking sequence as bait to identify proteins interacting with this region of Casp-2 pre-mRNA. This led to the identification of RBM5 as a protein interacting with Casp-2 pre-mRNA. Using transfected cells and in vitro biochemical assays, we show that RBM5 regulates Casp-2 alternative splicing. RBM5 overexpression increases the production of Casp-2L isoform and induces apoptosis. By deletion and site-specific mutagenesis, we have mapped the RBM5-binding site to a U/C-rich sequence upstream of the In100 element. Deleting or mutating this RBM5-binding site in the Casp-2 pre-mRNA decreases Casp-2L and increases Casp-2S, demonstrating the functional importance of RBM5 in regulating Casp-2 alternative splicing.
Results
Identification of RBM5 as a Candidate Splicing Regulator for Casp-2.
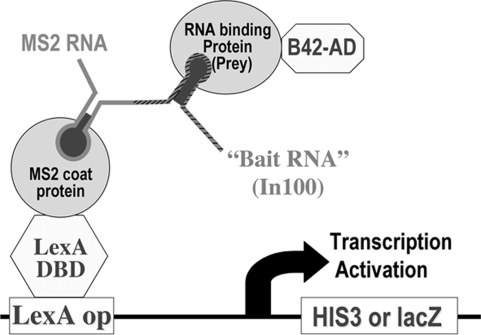
Our previous work revealed that a 100-nucleotide element inside intron 9, named In100, acts as an important sequence regulating casp-2 alternative splicing. To understand molecular mechanisms controlling casp-2 alternative splicing, we used the yeast RNA–protein interaction approach to screen for cDNAs encoding factors interacting with this sequence element. As illustrated in Fig. 1 and described in Materials and Methods, the bait construct contained In100 with the flanking sequence, including 45 nucleotides upstream of and 9 nucleotides downstream of In100, fused to an MS2-binding site in plasmid pRH5′. This bait plasmid was expressed in the yeast cells containing reporter genes and then used to screen a human fetal brain cDNA library. After screening ≈2 × 106 independent colonies, eight cDNA clones were identified, including the cDNA encoding RBM5. Detailed analyses of other cDNA clones are still in progress and will be reported elsewhere. We focus on characterization of RBM5 in this work.
Fig. 1.
Diagram illustrating the yeast RNA–protein interaction expression cloning strategy. As described in Materials and Methods, Casp-2 intronic splicing regulatory element (In100 together with the flanking sequence, depicted by the hatched gray line) is expressed as a RNA fused to MS2 RNA (depicted as the gray line) in yeast expressing reporter genes (LacZ and HIS3) under the control of the promoter containing LexA-binding sites. The yeast also expresses LexA DNA-binding protein fused to MS2 coat protein. The cDNA library encodes proteins fused to B42 transcription activation domain. When a RNA-interacting protein expressed from the cDNA library binds to the bait RNA and brings together a transcription activation complex, the reporter genes will be activated, allowing the identification of positive cDNA clones by the expression of the reporter genes.
RBM5 Promotes Casp-2 Exon 9 Exclusion.
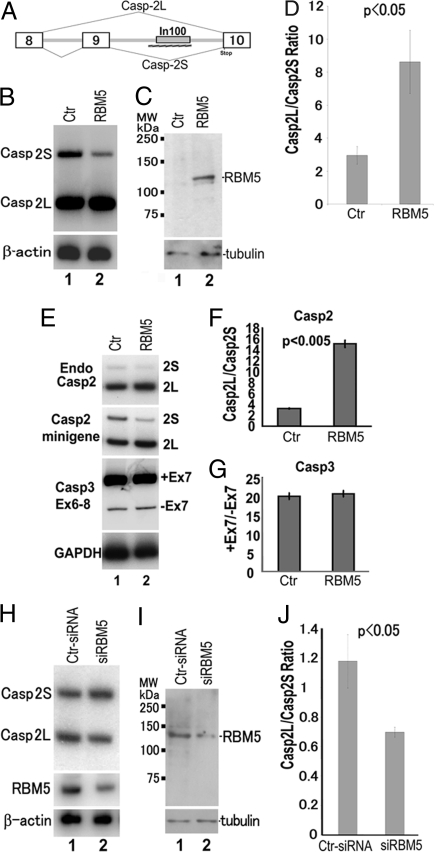
RBM5 was originally identified as a candidate tumor suppressor gene in the genomic region deleted in human lung cancer (23–25). RBM5 encodes a RNA recognition motif (RRM)-containing protein that plays a role in both cell cycle regulation and apoptosis (26–29). We examined effects of RBM5 on Casp-2 splicing by using the casp-2 minigene system that we established (20, 21, 30). C2 casp-2 minigene was transfected together with the vector control or a plasmid expressing Myc-tagged RBM5 into HEK293 cells. Casp-2 alternative splicing was examined by RT-PCR as described (20, 21) (Fig. 2). The expression of Myc-tagged RBM5 was detected by Western blotting using both anti-Myc (data not shown) and a specific anti-RBM5 antibody (Fig. 2C). RBM5 overexpression significantly increased Casp-2L and decreased Casp-2S, increasing the Casp-2L/Casp-2S ratio (Fig. 2 B–D). As shown in Fig. 2 E and F, RBM5 overexpression also altered the Casp-2L/Casp-2S ratio of the endogenous Casp-2 gene. However, RBM5 did not affect alternative splicing of several other apoptosis genes examined, including Bcl-x (data not shown) and caspase 3 (exon 6–8, Fig. 2 E and G). This demonstrates that RBM5 does not nonspecifically affect alternative splicing of all genes involved in cell death.
Fig. 2.
RBM5 overexpression increases whereas RBM5 knockdown decreases the Casp-2L/Casp-2S ratio. (A) Diagram of the casp-2 minigene illustrating the formation of Casp-2L and Casp-2S splicing isoforms. The intronic splicing element In100 is depicted by the gray box, and the hatched bar underneath indicates the RNA region used as bait in the yeast RNA–protein interaction screening (see Fig. 1). (B–J) RBM5 overexpression increases the Casp-2L/Casp-2S ratio without affecting Casp-3 exon 6–8 alternative splicing. Casp-2 minigene C2 was transfected into HEK293 cells together with the vector control plasmid or the plasmid expressing myc-tagged RBM5. (B) Casp-2 splicing isoforms and expression of β-actin (as a RNA loading control) were detected by RT-PCR with the Casp-2 minigene-specific primers or β-actin primers. (C) Expression of RBM5-myc or tubulin (as a loading control) proteins was detected by Western blotting with the respective antibodies. (E–G) RBM5 overexpression not only affects casp-2 minigene alternative splicing, but also increases the Casp-2L/Casp-2S ratio produced from the endogenous casp-2 gene (E and F) without affecting caspase 3 exon 7 alternative splicing (E and G). (H–J). RBM5 knockdown decreases the Casp-2L/Casp-2S ratio. Casp-2 minigene C2 was transfected into HEK293 cells together with the vector control siRNA or RBM5-specific siRNA. (H) Expression of Casp-2 splicing isoforms, RBM5, and β-actin was detected by RT-PCR using corresponding primers. (I) Expression of endogenous RBM5 or tubulin proteins was detected by Western blotting with respective antibodies. (J) Quantification of casp-2 isoform ratio in the siRNA experiment. (D, F, G, J) Quantification represents data from three independent experiments.
To investigate the role of the endogenous RBM5 gene in regulating Casp-2 alternative splicing, we used specific siRNA to down-regulate RBM5 expression. After testing different conditions for siRNA transfection in HEK293 cells, we were able to reduce RBM5 expression by ≈60–70%, as detected by RT-PCR with RBM5-specific primers (Fig. 2H Middle) and by Western blotting using specific anti-RBM5 antibody (Fig. 2I). Compared with the control-siRNA, RBM5-siRNA significantly reduced the Casp-2L/Casp-2S ratio when cotransfected with the Casp-2 minigene (Fig. 2J), demonstrating that the endogenous RBM5 protein is involved in regulating Casp-2 alternative splicing. Both RBM5 overexpression and knockdown experiments support that RBM5 regulates Casp-2 alternative splicing and promotes the formation of proapoptotic Casp-2L isoform.
RBM5 Overexpression Induces Apoptosis.
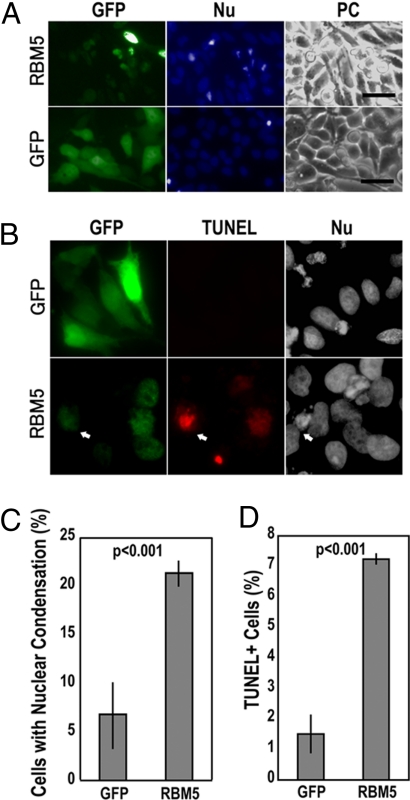
To test the role of RBM5 in cancer cell death, we examined effects of RBM5 overexpression in HeLa cells. Similar to our data (30), ≈5% of HeLa cells showed apoptotic cell death under our culture conditions, and the vector control plasmid transfection did not affect cell death (Fig. 3). However, RBM5 expression significantly increased cell death as assayed by both nuclear morphological changes and TUNEL (Fig. 3). Approximately 20% of RBM5-GFP-expressing cells showed condensed or fragmented nuclei compared with ≈5% in the GFP control group (Fig. 3 A and C). Consistently, TUNEL staining showed significantly increased TUNEL-positive cells compared with the control group (Fig. 3 B and D). These data indicate that RBM5 overexpression is sufficient to induce HeLa cancer cell death.
Fig. 3.
Induction of cell death by RBM5 overexpression. (A and B) Micrographs of GFP, nuclear morphology (Nu), or phase-contrast image (PC), or TUNEL staining of HeLa cells transfected with GFP-tagged RBM5 or GFP-vector control. Cell death was examined by nuclear staining with Hoechst dye (A) and TUNEL staining (B). (Scale bar: 50 μm.) (C and D) Quantification of cell death in transfected cells as determined by the percentages of cells showing condensed or fragmented nuclei (C) or cells showing positive TUNEL staining (D; see Materials and Methods). The arrow in B marks a typical GFP-RBM5-expressing cell that shows positive TUNEL staining. We noticed that many GFP-RBM5-positive cells undergoing apoptosis became detached during washing procedures in the TUNEL staining, providing an explanation of the overall low level of TUNEL-positive cells photographed and scored. Approximately 1,000 cells were examined in each group, and the data represent three independent experiments.
RBM5 Interacts with Casp-2 Pre-mRNA in a Sequence Upstream of In100 Element.
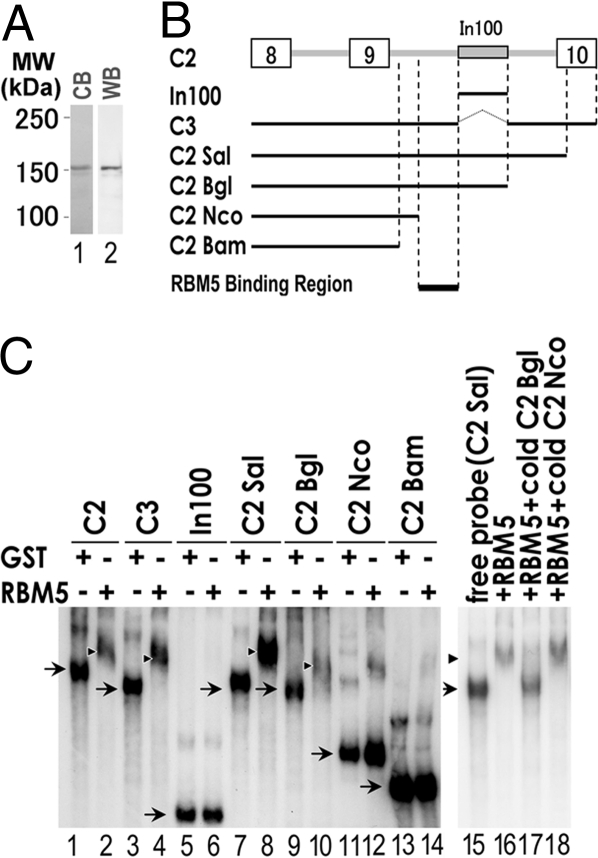
To investigate mechanisms underlying RBM5 activity in regulating Casp-2 alternative splicing, we examined the interaction of RBM5 with Casp-2 pre-mRNA by using purified recombinant RBM5 protein (Fig. 4A). We set up a gel-mobility shift assay by using the wild-type Casp-2 pre-mRNA transcript prepared from C2 construct or different mutant Casp-2 RNA transcripts containing deletions in different regions (Fig. 4B). Interestingly, both the RNA transcript containing In100 (C2) and the RNA lacking In100 element (C3) interacted with RBM5, resulting in specific gel-shift bands (Fig. 4C, lanes 1–4). Specific gel-shift signals were detected with C3, C2Sal, and C2Bgl RNAs, but not with C2Nco or C2Bam RNA transcripts (Fig. 4C, lanes 1–4 and lanes 7–14). In addition, In100 RNA failed to bind to RBM5 (Fig. 4C, lanes 5 and 6). These results indicate that the RBM5-binding site is inside intron 9 but outside of the In100 element. Similar gel-shift experiments were performed in the presence of competing nonradiolabeled RNA transcripts. C2Bgl but not C2Nco RNA was able to compete with the C2Sal RNA in RBM5 binding (Fig. 4C, lanes 15–18), demonstrating that the RNA region between In100 and NcoI inside intron 9 contained the specific RBM5-binding site.
Fig. 4.
RBM5 interacts with Casp-2 pre-mRNA. (A) Purified recombinant RBM5 protein as detected by Coomassie blue (CB) or Western blotting (WB) using specific anti-RBM5 antibody. (B) Series of Casp-2 pre-mRNA transcripts used for testing RBM5 binding. (C) Gel-shift assay to map RBM5-binding region in Casp-2 pre-mRNA. The radiolabeled RNA transcripts were incubated with either GST control protein or purified GST-RBM5 protein, and the RNA protein complexes were separated by native gel electrophoresis followed by autoradiography (lanes 1–14). The arrowheads and arrows indicate RNA–RBM5 complexes and free probes, respectively. RBM5 interacts with C3, C2Sal, and C2Bgl RNAs, but not In100, C2Nco, or C2Bam RNAs. Lanes 15–18 contain the gel-shift reactions in the presence of excess amounts of corresponding RNA transcript not radiolabeled as a competitor. C2Bgl but not C2Nco was capable of competing with the C2Sal RNA in forming RBM5 complex. These results indicate the RBM5-binding site is located between the NcoI and BglII sites but outside of In100 region, namely, the region immediately upstream of In100 and downstream of the NcoI site.
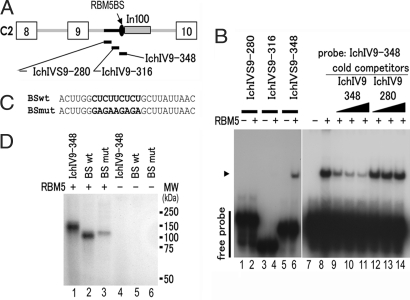
To define the RBM5-binding site further, we used RNA oligomers IchIVS9–280, IchIV9–316, and IchIV9–348, covering the entire sequence between the NcoI site and In100 in intron 9 (Fig. 5A). These RNA oligomers were radiolabeled and incubated with the purified GST-RBM5 protein in the gel-mobility shift assay. Only IchIV9-348 RNA, but not other RNA oligomers, showed the RBM5-binding signal (Fig. 5B, lane 6). Using unlabeled RNA oligomers, we demonstrated that IchIV9-348, but not IchIV9-280, competed with RBM5 binding (Fig. 5B, lanes 8–12), indicating that the IchIV9-348 fragment contained the RBM5-binding site.
Fig. 5.
RBM5 binds to the U/C-rich intronic sequence immediately upstream of In100. (A) RNA oligomers corresponding to the RBM5-binding region in intron 9 of Casp-2. (B) The corresponding RNA oligomers were end-labeled with [γ-32]ATP and used in the gel-mobility shift assay as in shown Fig. 4. Only IchVS9-348 RNA forms a complex with the purified RBM5 protein. Cold IchIV9-348 RNA, but not IchIV9-280 RNA, competed for binding to RBM5. The arrowhead indicates the RNA–RBM5 complex. (C) RNA oligomers containing either the wild-type sequence (BSwt) or mutated RBM5-binding site (BSmut). (D) UV-cross-linking assay was performed by using end-labeled RNA oligomers in the presence (lanes 1–3) or absence (lanes 4–6) of the purified RBM5 protein. Compared with BSwt, BSmut showed significantly reduced interaction with RBM5.
The interaction between RBM5 and Casp-2 pre-mRNA was further characterized by using UV-cross-linking assay with IchIV9-348 or shorter RNA oligonucleotides containing either the wild-type RBM5-binding sequence (BSwt) or mutant sequence in which U/C-rich sequence was mutated to G/A sequence (BSmut), as illustrated in Fig. 5C. Both IchIV9-348 and BSwt RNAs showed comparable interaction with purified RBM5 protein (Fig. 5D, lanes 1 and 2). However, changing the U/C sequence to G/A significantly reduced, although not completely eliminated, RBM5 binding (Fig. 5D, compare lane 3 with lane 2). This demonstrates that the U/C-rich element upstream of In100 interacts with RBM5.
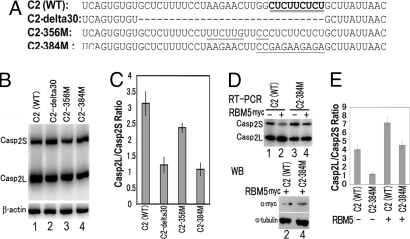
Mutating the RBM5-Binding Site Alters Casp-2L/Casp-2S Ratio.
To investigate the functional significance of RBM5 binding in Casp-2 alternative splicing, we constructed a series of minigenes containing different mutations in the IchIV9-348 region (Fig. 6A). Compared with the wild-type C2 minigene, C2-356M only showed a slightly reduced Casp-2L/Casp-2S ratio. However, either deleting 30-nucleotide (C2-delta30) or changing the U/C-rich sequence to G/A (C2-384M) significantly reduced Casp-2L/Casp-2S ratio (Fig. 6 B and C). When the RBM5 plasmid was cotransfected with the wild-type C2 minigene, the formation of Casp-2S was reduced with a concurrent increase in the Casp-2L level. This RBM5-induced activity was significantly decreased when the C2-384M minigene was used, although the RBM5 expression levels were similar (Fig. 6 D and E). The RBM5 response was not completely eliminated, consistent with the residual RBM5 binding to C2-384M RNA (Fig. 5D). These results demonstrate that the U/C-rich RBM5-binding site plays a role in regulating the ratio of Casp-2 alternative splicing.
Fig. 6.
The U/C-rich RBM6-binding sequence regulates Casp-2 alternative splicing. (A) Sequences of Casp-2 minigenes used in transfection and RT-PCR assay, with C2-delta30, 356M, and 384M containing different mutations in the RBM5-binding region. (B and C) Splicing of different Casp-2 mutants after transfection into HEK293 cells as detected by RT-PCR. (C) Quantification of the Casp-2S/Casp-2L ratio shown in B. Either deleting 30 nucleotides (C2-delta30) or mutating the U/C-rich element significantly decreased the Casp-2L/Casp-2S ratio by ≈3-fold. (D and E) C2-384 mutant shows compromised response to RBM5 activity in suppressing Casp-2S and enhancing Casp-2L production, compared with C2 (WT). (Lower) Equivalent level of RBM5-myc overexpression by Western blotting. (C and E) Quantification of data representing three independent experiments.
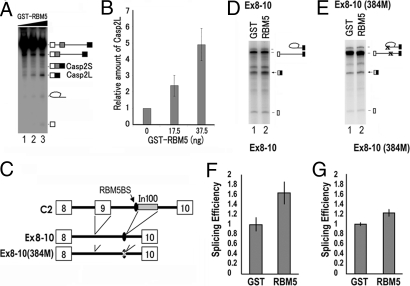
RBM5 Stimulates Casp-2 Exon 8 to Exon 10 Splicing in Vitro.
To examine further the role of RBM5 in regulating Casp-2 splicing regulation, we used an in vitro splicing assay with HeLa cell nuclear extract (NE) and the radiolabeled Casp-2 pre-mRNA. After incubation with HeLaNE, the splicing products, Casp-2S and Casp-2L, were detected by gel electrophoresis followed by autoradiography as described in ref. 21. The Casp-2L level was increased by the addition of the purified RBM5 protein in a dose-dependent manner (Fig. 7 A and B). To test whether the RBM5 activity depended on the presence of exon 9, we prepared an exon 8–10 minigene construct (Ex8–10) in which exon 9 and In100 sequences were deleted, but the RBM5-binding site was retained, as illustrated in Fig. 7C. When the Ex8–10 pre-mRNA was used in the in vitro splicing assay in the absence and presence of exogenous RBM5 protein, it was clear that RBM5 enhanced the efficiency of splicing between exon 8 and exon 10 (Fig. 7 D and F). When the RBM5-binding site was mutated, the resulting Ex8–10(384M) splicing substrate showed significantly reduced response to RBM5 (compare Fig. 7 E and G with D and F). RBM5 did not affect beta-globin pre-mRNA splicing [supporting information (SI) Fig. S1]. Together, these results demonstrate that RBM5 acts to stimulate the splicing between exon 8 and exon 10 independently of exon 9 and In100 sequences and that the U/C-rich RBM5-binding site upstream of In100 is important for RBM5 splicing stimulation.
Fig. 7.
RBM5 increases Casp-2L production in vitro by stimulating exon 8–10 splicing, and mutation of RBM5-binding sites significantly reduced the RBM5 activity. (A) Radiolabeled C2 pre-mRNA transcript was incubated with HeLa nuclear extract in the presence and absence of recombinant RBM5 protein under the splicing conditions. The in vitro splicing products were detected by autoradiography after gel electrophoresis. (B) Quantification demonstrating a dose-dependent stimulation of Casp-2L formation by RBM5. (C–G) In vitro splicing assay carried out using the Casp-2 exon 8–10 mutant minigenes illustrated in C. (D and E) Autoradiographs of splicing products of reactions carried out in the presence of control GST or recombinant RBM5 protein. (F and G) Quantification of splicing efficiency as determined by using a PhosphorImager. The splicing efficiency is measured as the ratio of ligated exons to the pre-mRNA; and the efficiency of splicing reaction containing GST control protein was set as 1. (B, F, G) Quantification of data representing three independent experiments.
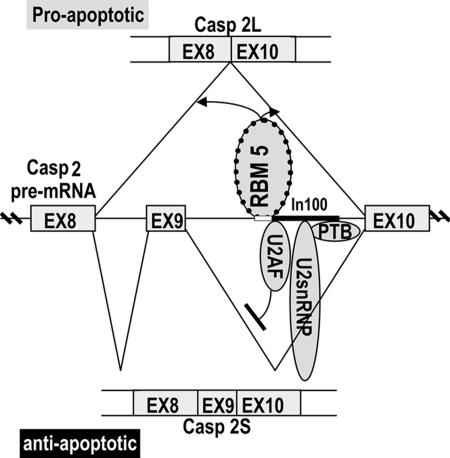
Model for RBM5 Function in Splicing Regulation.
Our overexpression and knockdown experiments indicate that RBM5 promotes the formation of proapoptotic Casp-2L isoform (Fig. 2). The in vitro splicing assay shows that activation of Casp-2L splicing does not depend on exon 9 or In100 sequences (Fig. 7). Deleting or mutating the RBM5-binding site affects the activity of RBM5 in regulating casp-2 splicing. These results led us to propose a model for RBM5 function in regulating Casp-2 alternative splicing as depicted in Fig. 8: RBM5 binds to a U/C-rich sequence immediately upstream of In100 and promotes the formation of proapoptotic Casp-2L by activating the splicing between the 5′ splice site of intron 8 and the 3′ splice site of intron 9. Consistent with this model, RBM 5 overexpression induced apoptosis in HeLa cells (Fig. 3). Together with previous studies, our results suggest that one mechanism for RBM5 tumor suppressor activity is to facilitate the production of proapoptotic isoforms of gene products, including CASP-2L.
Fig. 8.
A model for RBM5 function in Casp-2 alternative splicing regulation. RBM5 binds to a U/C-rich element (shown by the white box) immediately upstream of In100 (as demarcated by the black box inside intron 9) and activates splicing between exon 8 and 10 of Casp-2 pre-mRNA. RBM5 acts by promoting splice site pairing/association between exon 8 and exon 10. In100 recruits U2AF and U2 snRNP and forms pseudospliceosome complex with U1snRNP on 5′ splicing site of intron 9 to block splicing between exon 9 and 10 (20, 21). Both the RBM5-binding sites and the In100 element contribute to caspase-2 alternative splicing regulation.
Discussion
To investigate the role of alternative splicing in regulating cell death genes, we have examined both cis-elements and transacting splicing regulators involved in regulating Casp-2 splicing (20–22, 30). One cis-regulatory sequence identified from our previous study is In100, the intronic element containing a decoy 3′ splice site and polypyrimidine tract-binding (PTB) protein-binding sites (20, 21). Recent experiments suggest that in addition to U2snRNP, U2 auxiliary factor and PTB, other splicing regulators may interact with this region (K.F. and J.Y.W., unpublished observation). We used an intronic fragment containing In100 and flanking sequences as bait to screen for proteins interacting with this intronic regulatory region (Fig. 1). One factor identified by this strategy is the candidate tumor suppressor gene, RBM5/LUCA-15/H37. The gel-mobility shift and UV-cross-linking assays show that purified RBM5 protein interacts directly with Casp-2 pre-mRNA (Figs. 4 and 5). Systematic deletion and mutagenesis experiments indicate that RBM5 does not interact with In100 per se, but binds to the U/C-rich sequence immediately upstream of In100 (Figs. 5 and 6). RBM5 stimulates splicing between exon 8 and exon 10 in an In100-independent manner (Fig. 7).
Several lines of evidence support that RBM5 has tumor suppressor activities. RBM5 was originally identified as one of genes deleted in human lung cancer in the 3p21.3 chromosomal region (23) together with two single-nucleotide polymorphism changes (24). RBM5 expression is repressed in 70–80% of lung cancers (25). RBM5 overexpression causes cell cycle arrest, apoptosis, and inhibition of tumor growth (25–27, 29), sensitizing cells to apoptosis induced by death receptor ligands, FAS, TNF-α, and TRAIL (31, 32). However, the mechanism for this sensitization was not known. An antisense transcript, Je2, negatively regulates RBM5 expression (28, 33).
RBM5 is an RNA-binding protein containing two RNA recognition motifs (26, 34). However, the cellular targets for RBM5 had not been reported. Our results indicate that at least one target gene that RBM5 regulates at the posttranscriptional level is casp-2, an initiator caspase gene critical for cell death regulation. Our experiments have revealed that RBM5 binds to a U/C-rich sequence in intron 9 of casp-2 and that this RBM5-binding sequence is functionally important for RBM5 activity in regulating casp-2 alternative splicing (Figs. 5 and 6). Our observation that RBM5 overexpression in HeLa cells causes cell death (Fig. 3), together with the previous report that RBM5 stable transformants of lung cancer cell lines undergo apoptosis (29), supports the role of RBM5 as an apoptosis regulator in cancer cells. RBM5 is localized in nuclei and associated with spliceosomal complexes (26, 35, 36). Our work here indicates that RBM5 binds to casp-2 pre-mRNA and regulates the balance of casp-2L versus casp-2S splicing isoforms, uncovering a previously unknown activity of RBM5 as a splicing regulator. It is conceivable that other RBM5-like proteins also interact with the casp-2 RBM5-binding site and influence casp-2 alternative splicing. Two RBM5 homologs have been identified: RBM6, located adjacent to RBM5 on chromosome 3 at 3p21.3, and RBM10, on X chromosome at Xp11.23. RBM6 and RBM10 have 30% and 50% sequence homology to RBM5, respectively (37). It is possible that RBM5, RBM6, and RBM10 have overlapping function, although the biochemical functions of RBM6 and RBM10 are still not clear (37). In addition, several splicing isoforms of RBM5 have been identified, and the functional differences among different RBM5 splicing variants remain unclear (27, 28, 33, 38). Interestingly, it was recently found that alternative splicing of two other apoptosis genes, FAS and c-FLIP, was altered only when RBM5, 6, and 10 were all depleted and that RBM5 promoted sequence-specific pairing of the distal splice sites in FAS gene (54). Together with previous studies showing that RBM5 regulates multiple apoptosis pathways (28), it is clear that the role of RBM5 in regulating apoptosis may involve multiple genes.
More than 70% of human genes undergo alternative splicing to generate splicing isoforms. Many genes show distinct splicing patterns in tumor and normal tissues, including genes crucial for tumorigenesis such as oncogenes and tumor suppressor genes (6, 39–42). Furthermore, many genes important for cell death, including casp-2, are regulated by alternative splicing (for example, 43–45). These studies reveal the high level of complexity of alternative splicing regulation of genes involved in tumorigenesis and suggest that alternative splicing is an extremely powerful mechanism for the genetic/proteomic diversity associated with cancer.
Several splicing regulators have been implicated in tumor development including TLS-ERG (translocation liposarcoma gene fused to the ETS-related gene), SRp20, SRm160, SF2/ASF, and hnRNP A1 (46–51). Our previous work has reported that hnRNP A1 promotes the formation of antiapoptotic casp-2S isoform (30), consistent with its oncogenic activity (51). Our experiments reported here reveal a previously unknown link between the candidate tumor suppressor gene and alternative splicing regulation. Alternative splicing patterns of many genes change in tumorigenesis, suggesting splicing regulation as an important aspect in cancer development (6, 39–42). It is conceivable that multiple mechanisms can affect alternative splicing in cancer cells, such as changes in the expression of spliceosomal components or splicing regulators, and mutations and chromosomal aberrations affecting transcriptional and posttranscriptional regulation of splicing factors. Although many tumor suppressor genes have been cloned, there were no reported cases of tumor suppressor(s) that act by regulating alternative splicing of cell death genes. Our study thus has provided insight for understanding tumor suppressor gene function. Stimulating the formation of proapoptotic splicing isoforms might provide therapeutic benefits in cancer treatment.
Materials and Methods
Yeast RNA–Protein Interaction Expression Cloning Screening.
The RNA-protein Hybrid Hunter system (Invitrogen) was used according to the manufacturer's instructions to screen for proteins interacting with the Casp-2 intron 9 splicing regulatory sequence. The bait plasmid was constructed by fusing In100 element with flanking sequence to the MS2-binding site in the pRH5′ plasmid (Invitrogen). The detailed screening information is provided in SI Materials and Methods.
Cell Culture, Transfection, and Splicing Assays.
HEK293 and HeLa cells were cultured and transfected by using a modified calcium phosphate method as described (30, 52). In the siRNA experiments, 150 pmol of either control siRNA or RBM5-specific siRNA (Ambion) and 1 μg of Casp-2 minigene were cotransfected by Lipofectamine 2000 (Invitrogen) as described in the manual. After transfection, splicing products were detected by using RT-PCR as described (15, 30), with details provided in SI Materials and Methods.
Recombinant Protein Purification.
GST-control or GST-RBM5 protein was purified from Escherichia coli by using an AKTA PURIFIER system (GE Healthcare) with the detailed protocol described in SI Materials and Methods.
Plasmids and Antibodies.
Casp-2 minigene plasmids C2, C3, Ex8–10, and In100 were described (20, 21). C2 mutant minigenes were constructed using PCR (see SI Materials and Methods). Monoclonal antibodies anti-myc and anti-tubulin were from purchased Covance. The polyclonal Ab anti-RBM5 was described in ref. 28.
Cell Death Assays.
GFP-tagged RBM5 or the control vector was transfected into HeLa cells. Twenty-four hours after transfection, cells were stained with 5 μM membrane-permeable Hoescht dye and examined under a fluorescent microscope (Axiovert S100; Zeiss), as described (30, 53). Cells with condensed or fragmented nuclei were scored as apoptotic cells, and mitotic cells were excluded from scoring. TUNEL assay was performed by using an in situ cell death kit (Roche) following the manufacturer's instructions. More than 1000 cells were scored in the GFP vector control or GFP-RBM5 groups. Three independent experiments were carried out for quantification.
Gel-Mobility Shift and in Vitro Splicing Assays.
The wild-type Casp-2 minigene constructs were described before (21). α-32P-labeled RNA prepared from C2, C3, Ex8–10, and In100 constructs were used for the gel-shift assay as described in ref. 21 with details provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We are grateful to Dr. J. Valcarcel (Institució Catalana de Recerca i Estudis Avançats and Centre de Regulació Genòmica, Spain) for sharing reagents and communicating unpublished results. We thank the members of Wu laboratory for helpful discussion and critical reading of the manuscript. This work was supported by National Institutes of Health Grants GM070967 and EY014576 (to J.Y.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805569105/DCSupplemental.
References
- 1.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 2.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen GS, Abrams JM. Caspase activation: Stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- 4.Bonzon C, Bouchier-Hayes L, Pagliari LJ, Green DR, Newmeyer DD. Caspase-2-induced apoptosis requires bid cleavage: A physiological role for bid in heat shock-induced death. Mol Biol Cell. 2006;17:2150–2157. doi: 10.1091/mbc.E05-12-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavrik IN, Golks A, Baumann S, Krammer PH. Caspase-2 is activated at the CD95 death-inducing signaling complex in the course of CD95-induced apoptosis. Blood. 2006;108:559–565. doi: 10.1182/blood-2005-07-007096. [DOI] [PubMed] [Google Scholar]
- 6.Lin CF, et al. Sequential caspase-2 and caspase-8 activation upstream of mitochondria during ceramide- and etoposide-induced apoptosis. J Biol Chem. 2004;279:40755–40761. doi: 10.1074/jbc.M404726200. [DOI] [PubMed] [Google Scholar]
- 7.Paroni G, Henderson C, Schneider C, Brancolini C. Caspase-2-induced apoptosis depends on caspase-9, but its processing during UV- or tumor necrosis factor-dependent cell death requires caspase-3. J Biol Chem. 2001;276:21907–21915. doi: 10.1074/jbc.M011565200. [DOI] [PubMed] [Google Scholar]
- 8.Poh TW, Pervaiz S. LY294002 and LY303511 sensitize tumor cells to drug-induced apoptosis via intracellular hydrogen peroxide production independent of the phosphoinositide 3-kinase-Akt pathway. Cancer Res. 2005;65:6264–6274. doi: 10.1158/0008-5472.CAN-05-0152. [DOI] [PubMed] [Google Scholar]
- 9.Shin S, et al. Caspase-2 primes cancer cells for TRAIL-mediated apoptosis by processing procaspase-8. EMBO J. 2005;24:3532–3542. doi: 10.1038/sj.emboj.7600827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamm C, Zhivotovsky B, Ceccatelli S. Caspase-2 activation in neural stem cells undergoing oxidative stress-induced apoptosis. Apoptosis. 2008;13:354–363. doi: 10.1007/s10495-007-0172-7. [DOI] [PubMed] [Google Scholar]
- 11.Tu S, et al. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat Cell Biol. 2006;8:72–77. doi: 10.1038/ncb1340. [DOI] [PubMed] [Google Scholar]
- 12.Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene. 2006;25:5683–5692. doi: 10.1038/sj.onc.1209569. [DOI] [PubMed] [Google Scholar]
- 13.Wagner KW, Engels IH, Deveraux QL. Caspase-2 can function upstream of bid cleavage in the TRAIL apoptosis pathway. J Biol Chem. 2004;279:35047–35052. doi: 10.1074/jbc.M400708200. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, Kinoshita M, Noda M. Characterization of a mammalian cell death gene Nedd2. Leukemia. 1997;11:385–386. [PubMed] [Google Scholar]
- 15.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 16.Droin N, Beauchemin M, Solary E, Bertrand R. Identification of a caspase-2 isoform that behaves as an endogenous inhibitor of the caspase cascade. Cancer Res. 2000;60:7039–7047. [PubMed] [Google Scholar]
- 17.Martinet W, Knaapen MW, De Meyer GR, Herman AG, Kockx MM. Overexpression of the antiapoptotic caspase-2 short isoform in macrophage-derived foam cells of human atherosclerotic plaques. Am J Pathol. 2003;162:731–736. doi: 10.1016/S0002-9440(10)63869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parent N, Sane AT, Droin N, Bertrand R. Procaspase-2S inhibits procaspase-3 processing and activation, preventing ROCK-1-mediated apoptotic blebbing and body formation in human B lymphoma Namalwa cells. Apoptosis. 2005;10:313–322. doi: 10.1007/s10495-005-0805-7. [DOI] [PubMed] [Google Scholar]
- 19.Bergeron L, et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cote J, Dupuis S, Jiang Z, Wu JY. Caspase-2 pre-mRNA alternative splicing: Identification of an intronic element containing a decoy 3′ acceptor site. Proc Natl Acad Sci USA. 2001;98:938–943. doi: 10.1073/pnas.031564098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cote J, Dupuis S, Wu JY. Polypyrimidine track-binding protein binding downstream of caspase-2 alternative exon 9 represses its inclusion. J Biol Chem. 2001;276:8535–8543. doi: 10.1074/jbc.M008924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havlioglu N, et al. An intronic signal for alternative splicing in the human genome. PLoS ONE. 2007;2:e1246. doi: 10.1371/journal.pone.0001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: Identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p213 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 24.Oh JJ, Koegel AK, Phan DT, Razfar A, Slamon DJ. The two single-nucleotide polymorphisms in the H37/RBM5 tumour suppressor gene at 3p21.3 correlated with different subtypes of non-small cell lung cancers. Lung Cancer. 2007;58:7–14. doi: 10.1016/j.lungcan.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh JJ, West AR, Fishbein MC, Slamon DJ. The two single-nucleotide polymorphisms in the H37/RBM5 tumour suppressor gene at 3p21.3 correlated with different subtypes of non-small cell lung cancers. Cancer Res. 2002;62:3207–3213. [Google Scholar]
- 26.Edamatsu H, Kaziro Y, Itoh H. LUCA15, a putative tumour suppressor gene encoding an RNA-binding nuclear protein, is down-regulated in ras-transformed Rat-1 cells. Genes Cells. 2000;5:849–858. doi: 10.1046/j.1365-2443.2000.00370.x. [DOI] [PubMed] [Google Scholar]
- 27.Mourtada-Maarabouni M, Sutherland LC, Meredith JM, Williams GT. Simultaneous acceleration of the cell cycle and suppression of apoptosis by splice variant delta-6 of the candidate tumour suppressor LUCA-15/RBM5. Genes Cells. 2003;8:109–119. doi: 10.1046/j.1365-2443.2003.00619.x. [DOI] [PubMed] [Google Scholar]
- 28.Mourtada-Maarabouni M, Sutherland LC, Williams GT. Candidate tumour suppressor LUCA-15 can regulate multiple apoptotic pathways. Apoptosis. 2002;7:421–432. doi: 10.1023/a:1020083008017. [DOI] [PubMed] [Google Scholar]
- 29.Oh JJ, et al. 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res. 2006;66:3419–3427. doi: 10.1158/0008-5472.CAN-05-1667. [DOI] [PubMed] [Google Scholar]
- 30.Jiang ZH, Zhang WJ, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci USA. 1998;95:9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rintala-Maki ND, Abrasonis V, Burd M, Sutherland LC. Genetic instability of RBM5/LUCA-15/H37 in MCF-7 breast carcinoma sublines may affect susceptibility to apoptosis. Cell Biochem Funct. 2004;22:307–313. doi: 10.1002/cbf.1106. [DOI] [PubMed] [Google Scholar]
- 32.Rintala-Maki ND, Sutherland LC. LUCA-15/RBM5, a putative tumour suppressor, enhances multiple receptor-initiated death signals. Apoptosis. 2004;9:475–484. doi: 10.1023/B:APPT.0000031455.79352.57. [DOI] [PubMed] [Google Scholar]
- 33.Sutherland LC, et al. LUCA-15-encoded sequence variants regulate CD95-mediated apoptosis. Oncogene. 2000;19:3774–3781. doi: 10.1038/sj.onc.1203720. [DOI] [PubMed] [Google Scholar]
- 34.Drabkin HA, et al. DEF-3(g16/NY-LU-12), an RNA binding protein from the 3p21.3 homozygous deletion region in SCLC. Oncogene. 1999;18:2589–2597. doi: 10.1038/sj.onc.1202601. [DOI] [PubMed] [Google Scholar]
- 35.Behzadnia N, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YI, et al. Proteomic analysis of in vivo assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? J Cell Biochem. 2005;94:5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- 38.Rintala-Maki ND, et al. Expression of RBM5-related factors in primary breast tissue. J Cell Biochem. 2007;100:1440–1458. doi: 10.1002/jcb.21134. [DOI] [PubMed] [Google Scholar]
- 39.Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Gardina PJ, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole-genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C, et al. Cell type and culture condition-dependent alternative splicing in human breast cancer cells revealed by splicing-sensitive microarrays. Cancer Res. 2006;66:1990–1999. doi: 10.1158/0008-5472.CAN-05-2593. [DOI] [PubMed] [Google Scholar]
- 42.Zhang C, et al. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics. 2006;7:202. doi: 10.1186/1471-2105-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu JY, Tang H, Havlioglu N. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog Mol Subcell Biol. 2003;31:153–185. doi: 10.1007/978-3-662-09728-1_6. [DOI] [PubMed] [Google Scholar]
- 44.Izquierdo JM, et al. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Izquierdo JM, Valcarcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J Biol Chem. 2007;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- 46.Fushimi K, Osumi N, Tsukahara T. NSSRs/TASRs/SRp38s function as splicing modulators via binding to pre-mRNAs. Genes Cells. 2005;10:531–541. doi: 10.1111/j.1365-2443.2005.00855.x. [DOI] [PubMed] [Google Scholar]
- 47.Komatsu M, Kominami E, Arahata K, Tsukahara T. Cloning and characterization of two neural-salient serine/arginine-rich (NSSR) proteins involved in the regulation of alternative splicing in neurones. Genes Cells. 1999;4:593–606. doi: 10.1046/j.1365-2443.1999.00286.x. [DOI] [PubMed] [Google Scholar]
- 48.Yang L, Embree LJ, Hickstein DD. TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins. Mol Cell Biol. 2000;20:3345–3354. doi: 10.1128/mcb.20.10.3345-3354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W, Itoyama T, Chaganti RS. Splicing factor SRP20 is a novel partner of BCL6 in a t(3;6)(q27;p21) translocation in transformed follicular lymphoma. Genes Chromosomes Cancer. 2001;32:281–284. doi: 10.1002/gcc.1191. [DOI] [PubMed] [Google Scholar]
- 50.McGarvey T, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon–product complexes. J Cell Biol. 2000;150:309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miura M, Zhu H, Rotello R, Hartwieg EA, Yuan J. Induction of apoptosis in fibroblasts by IL-1β-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 54.Bonnal S, et al. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell. 2008 doi: 10.1016/j.molcel.2008.08.008. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.