Abstract
The Aire transcription factor plays an important role in immunological self-tolerance by mediating the ectopic expression of peripheral self-antigens by thymic medullary epithelial cells (MECs), and the deletion of thymocytes that recognize them. In Aire-deficient humans or mice, central tolerance is incomplete and multiorgan autoimmune disease results. We examined the variability of Aire's effects on ectopic transcription among individual mice of three different inbred strains. Aire's function was, overall, quite similar in the three backgrounds, although generally stronger in C57BL/6 than in BALB/c or NOD mice, and a minority of Aire-regulated genes did show clear differences. Gene expression profiling of wild-type MECs from single mice, or from the two thymic lobes of the same mouse, revealed significantly greater variability in Aire-controlled ectopic gene expression than in Aire-independent transcripts. This “noisy” ectopic expression did not result from parental or early developmental imprinting, but from programming occurring after the formation of the thymic anlage, resulting from epigenetic effects or from the stochastic nature of Aire activity. Together, genetic and nongenetic variability in ectopic expression of peripheral antigens in the thymus make for differences in the portion of self determinants presented for tolerance induction. This variable self may be beneficial in preventing uniform holes in the T-cell repertoire in individuals of a species, but at the cost of variable susceptibility to autoimmunity.
Keywords: gene expression, immunological tolerance, microarray, thymus, autoimmune regulator
Imposition of self-tolerance on the T-cell repertoire entails the exposure of differentiating thymocytes to a large variety of self-antigens. For self-antigens characterized by expression that is normally restricted to one or a few organs (peripheral tissue antigens [PTA]), exposure takes place through their expression by epithelial cells of the thymus. A segment of this ectopic gene expression is under the control of the Aire transcriptional regulator, a segment of great import because the absence of Aire leads to multiorgan autoimmunity in both knockout (KO) mice and human patients. Aire is expressed primarily by thymic medullary epithelial cells (MECs) (1–4). The connection between Aire and PTA expression was formally demonstrated by analysis of the global gene-expression profiles of MECs isolated from Aire-KO mice. Relative to those of wild-type (WT) controls, Aire-deficient MECs displayed absent or reduced expression of mRNAs encoding such PTAs as insulin, salivary gland proteins, and caseins (4, 5).
Aire-KO mice exhibit an autoimmune disease similar to human autoimmune-polyendocrinopathy-candidiasis-ectodermal-dystrophy (APECED), characterized by lymphocytic infiltrates of, and autoantibodies against, numerous peripheral tissues (4, 6). APECED disease manifestations vary widely from patient to patient, with even siblings exhibiting disparate disease profiles (7). In mice, the situation seems similar, in that the original KO strain also displayed heterogeneity of disease severity and target tissues (4, 6). Part of this variability may be due to environmental influences, although a recent study uncovered no evidence of a significant role for innate immune system stimulation (8). Another part of the variability is undoubtedly of genetic origin. For instance, Aire-deficient mice on the NOD genetic background developed an extreme exocrine pancreatitis that was not found on the C57BL/6 (B6) or BALB/c backgrounds. NOD and BALB/c Aire-KOs also developed severe autoimmune gastritis, which was observed only rarely on the B6 background. The modifier genes controlling these disease phenotypes were traced to several genomic regions, different for each target organ, including both MHC and non-MHC elements (9). In human patients, there is also evidence that APECED disease manifestations are linked to HLA genotypes (10). Some of the disease heterogeneity is also likely to be of epigenetic or stochastic origin. For example, in mice, there is substantial individual variation even among animals of the same inbred strain housed together (9).
There are many possible ways in which genetics may influence the disease caused by Aire deficiency. One interesting possibility is that PTA expression by MECs is somehow influenced by the genetic background. Experiments in several murine models have demonstrated that genetically regulated thymic expression levels of antigens such as insulin, myelin P0 and interphotoreceptor retinoid-binding protein correlate with tolerance to each particular antigen (11–15). Similarly, in humans, polymorphisms in the promoter region of the insulin gene that correlate with susceptibility to type 1 diabetes also correlate with levels of insulin gene transcripts in the thymus (16, 17).
Taubert et al. recently reported that expression levels of Aire and PTAs in MECs varied widely from person to person. For insulin and several other PTAs, mRNA levels correlated closely with those of Aire itself (18). Given the outbred nature of the human population, it is impossible to know whether these variations reflect genetic, epigenetic, or environmental differences between individuals.
Here, we tackled this question in mice, by examining Aire's impact on ectopic gene expression in MECs of individual mice from several inbred strains, allowing us to parse out the fraction of variation due to genetic effects from that due to interindividual variability.
Results
Gene-Expression Profiling.
We chose to profile MEC gene expression in three strains wherein the phenotypic effects of the Aire-KO were quite different. B6 can be considered a reference strain, in which the autoimmune manifestations provoked by the Aire KO mutation were quite mild, dominated by retinal degeneration (9, 15). In contrast, the disease induced by Aire deficiency on the NOD/Lt background was severe, even lethal, by 10–12 weeks of age, and resulted in much higher titers of autoantibodies as well as destruction of the exocrine pancreas and lung (9, 19). Aire KO-BALB/c mice were again different, exhibiting a florid stomach disease (9, 19). All mice examined were males of 3–4 weeks of age, before the onset of any extensive pathology. MECs were sorted from thymi of individual mice (≈20,000 MECs per mouse; 3 to 4 independent KO/wild-type littermate pairs for each strain); and RNA was prepared, amplified, labeled and hybridized to Affymetrix Mouse Genome 430 2.0 microarrays, representing ≈30,000 genes. Data (NCBI GEO accession no. GSE8564.) were processed and normalized in the GenePattern suite using the RMA algorithm (20, 21). For interstrain comparisons, mean expression values were calculated by averaging the replicates within each strain.
Similar Overall Patterns of Aire-Controlled Gene Expression in MECs of Different Mouse Strains.
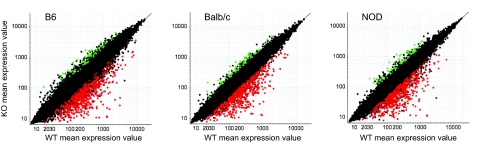
Overall, the effects of Aire on gene expression in MECs of the various strains were quite similar (Fig. 1). Comparing WT and KO expression values in MECs of each strain showed many genes to exhibit higher expression in WT MECs, (i.e., were Aire activated), whereas fewer had elevated levels of expression in Aire-deficient MECs (Aire repressed). The probes highlighted in Fig. 1 are those commonly regulated by Aire, with a WT/KO fold-change (FC) ratio greater than 2 (red) or less than 0.5 (green) in all three strains, demonstrating substantial overlap among them. A “consensus signature” of changes shared between all three strains accounted for 96.8%, 96.7%, and 99.4% of alterations on the B6, NOD and BALB/c backgrounds, respectively (for an arbitrary FC of >3 in the primary strain, and of >1.5 in the other two strains). Direct comparison of Aire's effects in the three strains by plotting the WT/KO FCs against each other supported this notion (Fig. 2A). The overall diagonal pattern of the probes in each strain-to-strain comparison confirmed that most of the same genes were regulated by Aire in all three strains.
Fig. 1.
Similarities of Aire effects on gene expression in MECs from mice with different genetic backgrounds. Gene expression profiles were determined for MECs of mice from three inbred lines. The scatter plots display gene expression values (averaged from three replicates per strain) for all probes on the M430 array, comparing expression in KOs and WT littermate controls. Red and green dots indicate probes with a WT/KO FC value >2, or <0.5, in all 3 strains.
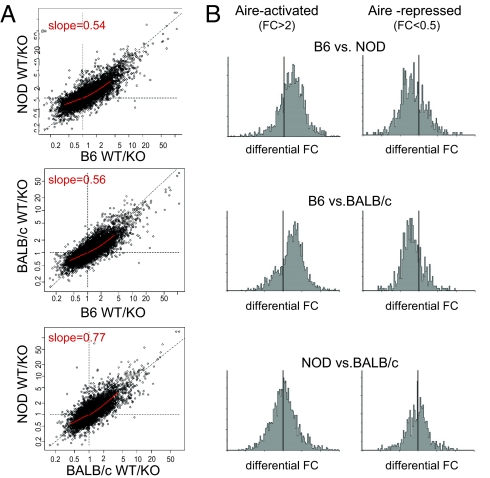
Fig. 2.
Fine differences in Aire's regulation of gene expression in different genetic backgrounds. (A) Comparisons of WT/KO ratios in MECs from the three strains. The red line on each plot shows the results of a locally smoothed regression (lowess). The average slope derived from the lowess is shown on each graph. (B) Histograms of the difference between FC values determined for Aire-induced or Aire-repressed genes, comparing strains pairwise. These differences were calculated for each gene by subtracting the log2(FC) values.
Subtle Differences in Aire's Regulation of MEC Gene Expression in the Three Strains.
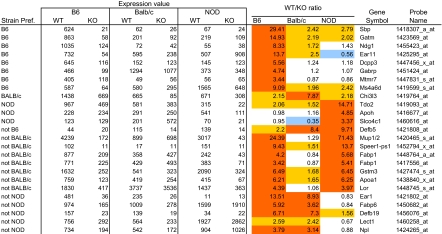
Closer examination of Figs. 1 and 2 revealed that even though the bulk of gene regulation by Aire was the same on the B6, BALB/c, and NOD backgrounds, there were also rare but clear differences. First, there was a minority of genes whose regulation by Aire was specific to, or specifically missing in, one strain. These genes fall along the FC = 1 horizontal or vertical dotted lines of Fig. 2A, and the most specific examples are listed in Table 1; a more comprehensive list of strain-specific probes can be found in supporting information (SI) Table S1. Genes with this characteristic were quite rare overall: for instance, only 53 probes were uniquely activated by Aire in B6 but not BALB/c or NOD mice (using FC thresholds of 2 and 1.25). We queried genomic and gene ontology databases to see whether these strain-specific genes might be connected to the strain-specific autoimmune manifestations of Aire deficiency (9). However, we failed to uncover any clear patterns: they were distributed over all of the chromosomes, were expressed peripherally in a broad spectrum of tissues, and were a mix of intracellular, secreted and membrane proteins. In particular, no gene specific to the exocrine pancreas was found in the NOD-only list; nor was any stomach-specific gene found in the BALB/c-only list.
Table 1.
Genes regulated by Aire uniquely in one strain
Second, for the majority of genes affected in all strains, Aire's impact was not equivalent in the different strains. This feature was evidenced by diversion from the x = y diagonal of the cloud of genes in the comparisons in Fig. 2A, with a greater degree of Aire-related changes in B6 than in the two other strains. Fitting by local regression indicated an average slope of 0.54 and 0.56 in the NOD/B6 and BALB/c/B6 plots, respectively. This trend was confirmed by calculating, for all genes affected by Aire, the ratio of FCs (calculated for simplicity by subtracting the log2-transform of the FCs). The histograms of Fig. 2B indicate that this feature was indeed a general trend, globally affecting Aire-responsive genes: the bulk of Aire-activated genes were more strongly up-regulated in B6 than in NOD or BALB/c mice, and repressed genes were also more strongly down-regulated in B6 than in the other strains (mean negative value for the difference in FCs in the latter case). In general, and as might be expected, the stronger effect of Aire in B6 MECs was reflected in higher expression values for Aire-activated genes in B6 than in NOD or BALB/c MECs. Of the 1815 genes induced more than twofold by Aire in at least one strain, 1206 were expressed at a higher level in B6 than in NOD MECs, and 1055 were expressed at a higher level in B6 than in BALB/c MECs. This difference was no longer apparent when the KOs were compared (data not shown), substantiating the conclusion that Aire had strong activating influences on the B6 background.
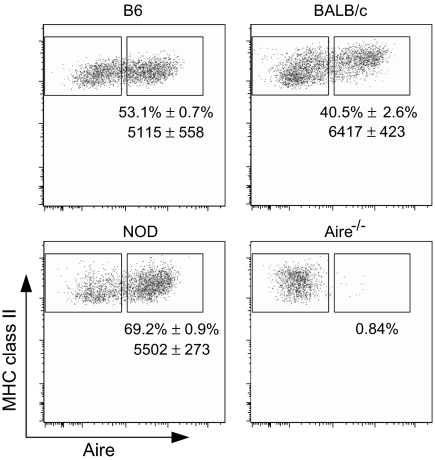
The overall differences in the strength of Aire's effect in the various strains could be explained quite simply by different levels of Aire expression, or by different proportions of Aire-expressing cells among the sorted MHC class IIhi MECs. To determine whether the strength of Aire regulation correlated with Aire protein levels, we performed flow cytometric analysis after intracellular staining of MECs from the three different strains (Fig. 3). In MECs gated as CD45−Ly51intG8.8+MHCIIhi, only a proportion of MHC class IIhi MECs expressed Aire, as expected (22). Interestingly, and perhaps counterintuitively given the more severe phenotype of the KO, the proportion of Aire+ MHCIIhi MECs was higher in NOD mice than in the other two strains. The proportion in B6 animals fell between that of NOD and BALB/c mice. The intensity of Aire expression was similar in the MHC class IIhi MECs off all three strains.
Fig. 3.
Aire protein levels do not vary among strains. Flow cytometric analysis of Aire expression in MECs of different genetic backgrounds (Aire vs MHC-IIhi profiles of gated CD45−G8.8+Ly51intMHC-IIhi MECs; representative of three independent mice for each inbred strain). Numbers are the mean percentage of Aire+ cells among class IIhi MECs, and the mean anti-Aire fluorescence intensity, ± SD, of Aire staining.
Interindividual Variation in Aire-Regulated Genes.
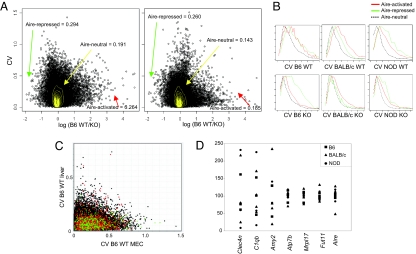
In addition to examining differences in Aire's function on different genetic backgrounds, another goal of these experiments was to determine the extent of interindividual variability in Aire-regulated PTA transcription in the thymus. This question relates to the work of Taubert et al., who demonstrated a high degree of variability between human individuals for the expression of Aire and several of the ectopic transcripts it controls (18). In that study, like any such analysis in the human system, it was not clear how much of the variability could be attributed to genetic differences between individuals, or to other factors—environmental, stochastic or epigenetic. Here, the genetic background of the littermates examined was homogeneous, and environmental influences were not expected to vary for mice housed under controlled conditions in the same SPF facility. Thus, we were in a good position to analyze stochastic or epigenetic variability. To that end, the coefficient of variation (CV) between replicates for WT and KO mice of each strain was calculated for each gene. As illustrated in Fig. 4A Left), Aire-activated genes with a high WT/KO FC showed, as a group, a higher CV than the bulk of Aire-neutral genes represented on the array. This observation was confirmed, for all three strains, in the upper three histograms of Fig. 4B, which compare the distribution of CVs for Aire-affected genes to those of a randomly-selected set of Aire-neutral genes with a matched distribution of expression values.
Fig. 4.
Expression of Aire-regulated genes varies among individuals. (A) CV vs. FC plots of all genes on the M430 array, from the expression values in the three B6 WT (Left) or B6 KO (Right) individual mice. Also indicated on each plot are the mean CVs of Aire-activated transcripts (WT/KO FC >2), Aire-repressed (WT/KO FC <0.5), or Aire-neutral (WT/KO FC between 0.77 and 1.3 and chosen to have the same distribution of expression values as the combined Aire-activated and Aire-repressed lists). Analysis of the BALB/c and NOD strains yielded similar results (Table 2). (B) Histograms of CVs for Aire-activated (red), Aire-repressed (green), and Aire-neutral (black) probes in WT (Top) and KO (Lower) MECs. (C) Scatter plot comparing CVs in M430 datasets from liver or MECs (B6 strain). Red and green dots are probes with a WT/KO FC value >2, or <0.5, in all three strains. (D) Expression values for several Aire-activated and Aire-neutral genes. Each point represents the expression level in an individual mouse, with shapes indicating the different strains. The Aire-activated genes shown are representative of those genes having a WT/KO FC >2 in MECs, and an interindividual CV of at least 0.9. Aire-neutral genes had a WT/KO FC between 0.77 and 1.3 and an interindividual CV at the median, 0.16.
We then asked whether the increased variability in Aire-induced genes was an inherent property of those particular genes or, rather, was caused by Aire itself. Comparing the CVs in MECs from Aire-WT and -KO thymi (Fig. 4A, compiled in Table 2) showed a clear drop in the distribution of CVs for Aire-activated genes in KO thymi, although not quite to the same level as the Aire-neutral gene set. Thus, the residual expression of these genes was less variable than when Aire was present, i.e., Aire was directly contributing to the variability.
Table 2.
Aire's effect on interindividual variability in gene expression
| Type of mouse | Aire-neutral |
Aire-activated |
Aire-repressed |
||
|---|---|---|---|---|---|
| Mean CV | Mean CV | -log10 (p-) | Mean CV | -log10 (p-) | |
| B6 WT | 0.191 | 0.264 | 42.8 | 0.294 | 27.1 |
| B6 KO | 0.143 | 0.185 | 18.4 | 0.260 | 48.6 |
| NOD WT | 0.132 | 0.264 | 94.4 | 0.291 | 57.7 |
| NOD KO | 0.118 | 0.202 | 52.1 | 0.200 | 36.8 |
| BALB/c WT | 0.172 | 0.307 | 133.0 | 0.215 | 16.5 |
| BALB/c KO | 0.146 | 0.213 | 76.8 | 0.199 | 21.4 |
| liver WT | 0.069 | 0.076 | 0.077 | ||
If so, one might predict that the variability in Aire-activated genes would be a specific feature of MECs, and would not be found in peripheral tissues where these genes are normally expressed. Thus we analyzed the variability of the same set of genes in another tissue, the liver, by downloading from NCBI GEO repository the microarray datasets reported by Ackert-Bicknell et al. (23), generated from liver samples of three individual B6 mice (accession #GSE5959). In the liver, there was no difference in mean CV between Aire-activated genes (0.076) and Aire-neutral genes (0.069; Fig. 4 C and Table 1).
Do the higher mean CV observed for Aire-activated genes translate into meaningful interindividual differences in expression values of individual genes? Indeed, many Aire-activated genes with high CVs exhibited broad distributions of expression value, with ranges as large as 10- to 20-fold, as illustrated for a few such genes in Fig. 4D. This wide range of expression was found in mice of all three strains. Thus, the particular variability in Aire-regulated transcripts in MECs leads to quantitative variations that are likely to have a direct impact on tolerance induction.
Origin of the Interindividual Variability in Aire-Regulated Genes.
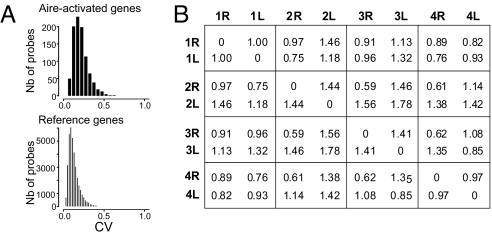
Several hypotheses could account for the variability of ectopic expression for Aire-regulated genes and for the stochastic patterns observed in individual mice: 1) transmissible epigenetic programming, imprinted at the parental level; 2) imprinting at an early stage of embryonic development (e.g., variable patterns of DNA remethylation in the first few days of development); 3) epigenetic patterns set during the differentiation of the thymic epithelial cells, around embryonic day11, after lateralization of the body plan; 4) a continuous reshuffling of ectopic expression patterns, each epithelial precursor or clone thereof adopting a defined pattern. The thymus of an adult mouse is composed of two lobes, whose stroma develops independently from endodermal tissue of pharyngeal pouches, around embryonic day 11 (24). The hypotheses outlined above lead to testable predictions concerning ectopic expression in the two thymic lobes of a mouse: according to #1 and 2, both lobes should have related profiles, but for #3 and 4 both lobes should behave independently, and be no more similar that lobes from different mice. We thus generated a new set of expression profiles, where MECs were purified independently from the right and left lobes of four wild-type B6 mice. Eight microarray profiles were generated as above. A higher degree of variability was observed in Aire-regulated genes, relative to Aire-neutral genes, across the datasets as a whole, confirming the previous observations (CV = 0.184 vs. 0.103 for 783 Aire-induced genes at a FC >2.5; Fig. 5A). We then used an aggregate measure of Euclidian distance, encompassing 783 Aire-induced genes, to compare each lobe to all others (Fig. 5B). Quite clearly, the 2 lobes from the same mouse were no closer to each other than to lobes of other mice; this conclusion also applied when when calculating the distance based on Aire-induced genes that had the highest intrinsic CV (154 genes, CV >0.3; data not shown). Principal component analysis also failed to uncover components that would specifically distinguish the two lobes from the same mouse. Thus, the data are not consistent with hypotheses 1 or 2; rather, the pattern of ectopic gene expression is set independently in each thymic lobe.
Fig. 5.
Patterns of Aire-induced expression vary independently in each thymic lobe. MECs were purified from left and right thymic lobes of four wild-type B6 mice, and each (A) Histogram of genewise CVs across all datasets, for Aire-induced genes (FC >2.5) or all other genes on the array. (B) Relative Euclidian distances were calculated between each individual lobe, integrating expression values for all Aire-induced genes.
Discussion
These data indicate that the transcriptional programs regulated by Aire in MECs of a given individual are conditioned by genetic factors and by non-genetic “noise”: genetic polymorphism affects the levels and fine specificity of ectopic transcripts, and interindividual variability in these ectopic transcripts further shuffles the outcome.
These results are of interest in light of the work of Taubert et al., who described extensive variability in Aire and ectopic transcription in MECs from different humans (18). Our data would suggest that both genetic and nongenetic elements are involved. We did not reproduce, in the mouse, the very wide range of transcript abundance for Aire (up to 50-fold), the variations we observed being much more modest. The range of variability may be different between species. Also, the interpretation of the human data may be complicated by variation in the proportion of Aire+ cells in the samples of human thymus.
In this setting, it was of interest to estimate the relative influence of these factors, using the CV as a metric for variation. Aire-regulated genes had a mean CV of 0.40 when all interindividual and genetic variation was encompassed; interindividual variability alone gave a CV of 0.25 (average of the interindividual CVs within each strain); the genetic component, estimated by calculating a CV between the expression means for the three strains, scored 0.33. For a random sample of Aire-neutral genes with a comparable spread of expression values, the three CVs were 0.18, 0.14 and 0.12, respectively. Taking into account an estimated experimental CV of ≈0.13, we estimated that the variance in Aire-regulated transcription between any two Mus musculus individuals was approximately two-thirds genetic and one-third nongenetic in origin.
Although the genes regulated by Aire were mostly similar in different strains, Aire-activated genes were expressed at a higher level in B6 MECs than in MECs of BALB/c and NOD animals, which pointed to an impact of the genetic background in modulating Aire's function as a transcriptional inducer. There are several ways through which this control may occur. We have already ruled out one possibility, that Aire protein levels are intrinsically higher in B6 MECs than in the other strains. Secondly, examination of the Perlegen sequence database turned up no differences in the Aire coding regions between the three strains. It is possible that the levels or functional activities of one or more of Aire's partner factors are higher or stronger in B6 MECs, or that proteins that inhibit Aire's function are less effective on the B6 background. Transcription may also differ at a more basic level, with widespread alterations in the accessibility to chromatin remodeling in different inbred strains.
We did not find, within the rare instances of strain-specific (or strain-preferential) Aire control, explanations for the particular tropisms of autoimmunity in the different KOs (9, 25). For example, there was no evidence for effects of the Aire KO particularly on genes active in the exocrine pancreas for NOD mice, nor on genes active in the stomach for BALB/c animals. It seems likely, then, that tissue tropism stems from the combination of MHC restriction and tissue-specific factors. One might speculate, however, that the more robust effect of Aire on transcription profiles observed in B6 thymi might contribute to the general autoimmune-resistant nature of this strain.
In addition to the strain-to-strain differences in Aire's function, we found marked variability in the expression of Aire-activated genes from one individual to another within the same strain, and between the two thymic lobes of the same mouse. This interindividual variability significantly distinguished Aire-activated genes from other Aire-neutral genes in the genome. This augmentation was specific to MECs, as it was not observed in the liver for the same set of genes. Neither did we observe it with a different transcription factor also involved in controlling autoimmunity, Foxp3. The results of the dual lobe profiling show that the variability is not set by parental or early embryonic imprinting, but is set independently in each thymic lobe. This variability may reflect stochastic mechanisms involved in ectopic transcription in MECs. We and the Kyewski group have recently observed that Aire-controlled expression of ectopically expressed genes in a single MEC is inherently probabilistic, any given cell transcribing a subset of PTA genes, following gene-specific probabilities (26 and accompanying paper). One would have assumed that the integration of these probabilities over the 15,000 Aire+ MECs of a thymic lobe would smooth out stochastic effects, resulting in uniform patterns of ectopic expression. This proved not be the case, and the stochastic aspect of ectopic expression at the single cell level somehow extends to the single organ level. This lack of smoothing and persistent stochasticism in a large cell pool suggests that the MECs present in a thymic lobe at a given time must descend from a small number of precursors, in which the accessibility to Aire of a given locus is determined in a stable and inheritable manner, before MEC clonal expansion and Aire expression. Such a view is consistent with current notions of population dynamics of thymic epithelial cells (8, 27, 28) where Aire+ MECs appear derived from a limited number of precursors, after a phase of cell division and expansion.
Noise in gene expression is an unavoidable consequence of the size and complexity of regulatory pathways and of the low number of molecules involved (29, 30). The immune system may be exploiting this noise to increase individuality in the tolerance to self within a species. Since tolerance to self-antigens can also lead to “holes” in the repertoire of responses to nonself, there would be a distinct advantage, at the population level, in some diversity of the molecular drive to self-tolerance. Such variability would ensure a greater diversity of potential responses within the species. In this respect, interindividual variability in ectopic PTA expression may function as does the wide diversity of MHC alleles, helping to ensure that at least some of the individuals will be apt to survive novel pathogenic challenges. Of course, varying susceptibility to autoimmune deviation is the price to pay for this diversity in the education to self, and the phenomenon may contribute to the proportion of nongenetic determinism of autoimmune diseases. Susceptibility to autoimmune diseases has a strong genetic component, but one whose influence is modulated by nongenetic factors (illustrated by the frequent discordance of diabetes or multiple sclerosis in monozygotic twins). The present results suggest that epigenetic modulation of the degree and specificity of self-tolerance may account, at least in part, for this individual re-pegging of disease risk.
Materials and Methods
Mice.
Aire-deficient mice (4) used in this study were backcrossed to the B6, NOD and BALB/c strains for at least nine generations before intercrossing. For each knockout line, heterozygotes were bred to each other, and resulting KO and WT littermate control animals were used, unless otherwise noted. All experiments were approved by the HMS IACUC (protocol 2954).
Thymus Digestion and MEC Sorting.
Thymic epithelial cells for microarray analysis were prepared according to (4). Individual thymi from young adult (3–4.5- week-old) animals were digested, separated by density-gradient centrifugation, and the thymic epithelial cell-enriched fraction was harvested, stained and sorted as CD45−G8.8+Ly51intI-Abhi. RNA was prepared from sorted cells using TRIzol.
RNA Amplification and Microarray Hybridization.
Total MEC RNA was amplified using a T7 polymerase-based method. First- and second-strand cDNA synthesis, in vitro transcription of complementary RNA, and a second round of first- and second-strand cDNA synthesis were performed using the MessageAmp aRNA Kit (Ambion, Austin, TX). The resulting cDNA was used in a second round of in vitro transcription, performed using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, NY), incorporating biotinylated ribonucleotides into the aRNA. The labeled aRNA was purified, fragmented (94° for 35 min), and hybridized to Affymetrix M430v2 chips.
Microarray Data Analysis.
The microarray data are available at GEO under accession no. GSE8563. The raw probe-level chip data (.CEL files) were normalized by the robust multiarray average (RMA) algorithm (21) using the ExpressionFileCreator module of the GenePattern 2.0 software package (20), and analyzed using the MultiPlot module in GenePattern. Random datasets were generated from a Gaussian distribution extrapolated from log-transformed values for each gene. This distribution was centered on the genewise mean across all samples, with a standard deviation that was the mean of the per-class standard deviations for that gene. The false discovery rate was extrapolated from the frequency at which a given FC value is equal or higher in the random data, for FC increments of 0.1. Genewise P values were calculated using Welch's modified t test. Calculations of genewise CVs on filtered gene sets were performed in S+ (Insightful).
Supplementary Material
Acknowledgments.
We thank Vincent Auyeung for the splenic Treg microarray data and Dr. Hamish Scott for the anti-Aire antibody. This work was supported by a National Institutes of Health Grant RO1 DK60027, Young Chair funds (to D.M. and C.B.), Joslin Diabetes Center's NIDDK-funded DERC core facilities, and National Institutes of Health Training Grant T32 DK07260 (to E.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808070105/DCSupplemental.
References
- 1.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 2.Heino M, et al. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem Biophys Res Commun. 1999;257:821–825. doi: 10.1006/bbrc.1999.0308. [DOI] [PubMed] [Google Scholar]
- 3.Heino M, et al. RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse. Eur J Immunol. 2000;30:1884–1893. doi: 10.1002/1521-4141(200007)30:7<1884::AID-IMMU1884>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 5.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsey C, et al. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- 7.Ishii T, et al. Novel mutations of the autoimmune regulator gene in two siblings with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Clin Endocrinol Metab. 2000;85:2922–2926. doi: 10.1210/jcem.85.8.6726. [DOI] [PubMed] [Google Scholar]
- 8.Gray DHD, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med. 2007 doi: 10.1084/jem.20070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, et al. Modifier loci condition autoimmunity provoked by Aire deficiency. J Exp Med. 2005;202:805–815. doi: 10.1084/jem.20050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halonen M, et al. AIRE mutations and human leukocyte antigen genotypes as determinants of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy phenotype. J Clin Endocrinol Metab. 2002;87:2568–2574. doi: 10.1210/jcem.87.6.8564. [DOI] [PubMed] [Google Scholar]
- 11.Chentoufi AA, Polychronakos C. Insulin expression levels in the thymus modulate insulin-specific autoreactive T-cell tolerance: The mechanism by which the IDDM2 locus may predispose to diabetes. Diabetes. 2002;51:1383–1390. doi: 10.2337/diabetes.51.5.1383. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto K, Miyake S, Schachner M, Yamamura T. Heterozygous null mutation of myelin P0 protein enhances susceptibility to autoimmune neuritis targeting P0 peptide. Eur J Immunol. 2003;33:656–665. doi: 10.1002/eji.200323677. [DOI] [PubMed] [Google Scholar]
- 13.Thebault-Baumont K, et al. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. J Clin Invest. 2003;111:851–857. doi: 10.1172/JCI16584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liston A, et al. Gene dosage-limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J Exp Med. 2004;200:1015–1026. doi: 10.1084/jem.20040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devoss J, et al. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J Exp Med. 2006;203:2727–2735. doi: 10.1084/jem.20061864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vafiadis P, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet. 1997;15:289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 17.Pugliese A, et al. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 18.Taubert R, Schwendemann J, Kyewski B. Highly variable expression of tissue-restricted self-antigens in human thymus: Implications for self-tolerance and autoimmunity. Eur J Immunol. 2007;37:838–848. doi: 10.1002/eji.200636962. [DOI] [PubMed] [Google Scholar]
- 19.Gavanescu I, et al. Loss of Aire-dependent thymic expression of a peripheral tissue antigen renders it a target of autoimmunity. Proc Natl Acad Sci USA. 2007;104:4583–4587. doi: 10.1073/pnas.0700259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Gray DH, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med. 2007 doi: 10.1084/jem.20070795. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SH, et al. Selection and expansion of CD8alpha/alpha(1) T cell receptor alpha/beta(1) intestinal intraepithelial lymphocytes in the absence of both classical major histocompatibility complex class I and nonclassical CD1 molecules. J Exp Med. 1999;190:885–890. doi: 10.1084/jem.190.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollander G, et al. Cellular and molecular events during early thymus development. Immunol Rev. 2006;209:28–46. doi: 10.1111/j.0105-2896.2006.00357.x. [DOI] [PubMed] [Google Scholar]
- 25.Niki S, et al. Alteration of intra-pancreatic target-organ specificity by abrogation of Aire in NOD mice. J Clin Invest. 2006;116:1292–1301. doi: 10.1172/JCI26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derbinski J, et al. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci USA. 2008;105:657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodewald HR, et al. Thymus medulla consisting of epithelial islets each derived from a single progenitor. Nature. 2001;414:763–768. doi: 10.1038/414763a. [DOI] [PubMed] [Google Scholar]
- 28.Bleul CC, et al. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature. 2006;441:992–996. doi: 10.1038/nature04850. [DOI] [PubMed] [Google Scholar]
- 29.Spudich JL, Koshland DE., Jr Non-genetic individuality: Chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 30.Kaufmann BB, van Oudenaarden A. Stochastic gene expression: From single molecules to the proteome. Curr Opin Genet Dev. 2007;17:107–112. doi: 10.1016/j.gde.2007.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.