Abstract
Bacterial populations are subject to complex processes of diversification that involve mutation and horizontal DNA transfer mediated by transformation, transduction, or conjugation. Tracing the evolutionary events leading to genetic changes allows us to infer the history of a microbe. Here, we combine experimental and in silico approaches to explore the forces that drive the genome dynamics of Streptococcus agalactiae, the leading cause of neonatal infections. We demonstrate that large DNA segments of up to 334 kb of the chromosome of S. agalactiae can be transferred through conjugation from multiple initiation sites. Consistently, a genome-wide map analysis of nucleotide polymorphisms among eight human isolates demonstrated that each chromosome is a mosaic of large chromosomal fragments from different ancestors suggesting that large DNA exchanges have contributed to the genome dynamics in the natural population. The analysis of the resulting genetic flux led us to propose a model for the evolutionary history of this species in which clonal complexes of clinical importance derived from a single clone that evolved by exchanging large chromosomal regions with more distantly related strains. The emergence of this clone could be linked to selective sweeps associated with the reduction of genetic diversity in three regions within a large panel of human isolates. Up to now sex in bacteria has been assumed to involve mainly small regions; our results define S. agalactiae as an alternative paradigm in the study of bacterial evolution.
Keywords: conjugation, evolution, homologous recombination, population genomics, horizontal gene transfer
The development of population genetics for bacterial species has revealed that although bacteria reproduce clonally, their genomes evolve not only by point mutations but also by DNA exchange through homologous recombination. It is assumed that bacterial recombinational exchange involves the replacement of a small region of the genome of a recipient cell by the corresponding region from another isolate of the same species (1). Homologous recombination follows unidirectional horizontal DNA transfer mediated by transformation, transduction, or conjugation (2, 3). These mechanisms are regarded properly as parasexual, because none involves wholesale exchanges of chromosomal genetic material as occurs in sexual reproduction of eukaryotic organisms. In natural populations, transformation and phage-mediated transduction probably are the most relevant mechanisms (4). Because recombination is not linked to bacterial reproduction, its rate varies greatly among bacterial species. Differences in the ratio of genetic changes caused by recombination relative to de novo mutation have led to the identification of a spectrum of bacterial population structures from clonal to panmictic (5).
Each type of evolutionary event, whether it be mutation, recombination, or selection, leaves a characteristic pattern that can be revealed by molecular population genetics. The analysis of polymorphisms among bacterial populations thus has allowed reconstruction of the evolutionary history of different human pathogens such as Helicobacter pylori (6), Mycobacterium leprae (7), and Salmonella typhimurium (8). In the last decade, the development of the multilocus sequence typing (MLST) of housekeeping genes has allowed us to account for recombination as well as point mutation in genealogical inference. However, because MLST studies focus mainly on only a few genetic markers, they describe the diversity of structure without identifying genome-level changes that drive differentiation and lineage divergence. The ever-increasing number of complete genome sequences for single bacterial species now makes it possible to search for recombinations across entire genomes (9, 10).
The population structure of the opportunistic pathogen Streptococcus agalactiae has been studied by various methods including MLST. This bacterial pathogen was described initially in 1887 as an animal pathogen (11). Human infections caused by this bacterium were reported in the 1930s (12), and since the 1970s it has been recognized as the leading neonatal pathogen in the developed world (13). Analyses of MLST data from different geographic regions showed that isolates infecting and colonizing neonates cluster mainly in five clonal complexes (CCs), of which CC17 is a major hyperinvasive neonatal clone (14). A marked diversity observed within CCs suggested the importance of recombinational exchanges (15–17). Accordingly, it was proposed that the horizontal transfer of genes involved in the synthesis of the capsular polysaccharide, a major virulence factor, occurs under natural conditions (18). These putative recombinational exchanges make the relationships between CCs difficult to discern. The recent analysis of eight genome sequences has shown that strain-classification methods such as MLST do not reflect the real genetic diversity described by whole-genome analysis, probably because they do not take into account the variable genome of the species (16, 19). Consistently, the high variability of the dispensable genome led to the concept of an open pan-genome for the species S. agalactiae (16). In this study, we investigated whether recombinational exchanges could be observed under laboratory conditions. Strikingly, we demonstrated that S. agalactiae strains exchange large fractions of their chromosome, probably through cis and trans mobilization by conjugative elements. To infer the contribution of such large recombinational exchanges to the dynamics of the S. agalactiae chromosome, we analyzed the distribution of nucleotide polymorphisms among the eight sequenced genomes. This analysis provided evidence that large chromosomal exchanges are major contributors to the genome dynamics of S. agalactiae and allowed us to propose an evolutionary scenario for the species.
Results and Discussion
Demonstration of Large Chromosomal Exchanges Under Laboratory Conditions.
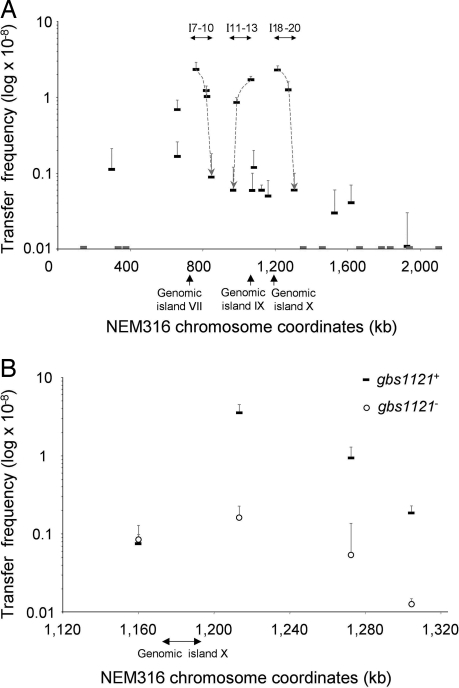
Several studies (15–17) have suggested a role of recombinational replacements in the genome evolution of S. agalactiae, although such exchanges never have been reported under laboratory conditions. To test whether chromosomal fragments could be transferred and exchanged between different S. agalactiae isolates, we assessed the transfer of an erythromycin marker (ErmR) inserted at 30 different locations (I-1 to I-30) in the chromosome of strain NEM316 belonging to CC23. These insertions were distributed over the chromosome, outside putative integrated mobile genetic elements. Mating experiments were carried out using each of the NEM316 derivatives as the donor and the distantly related CC17 BM110 strain as recipient. Marker transfers were obtained at frequencies ranging from 1.1 × 10−10 to 2.4 × 10−8 for 20 donors, whereas no transfer (frequency < 10−11) was detected with the 10 remaining donors [Fig. 1A and supporting information (SI) Table S1]. For each successful transfer, the location of the ErmR marker was identical in the donor and the selected transconjugants, indicating that the DNA region carrying the ErmR marker was transferred. We accurately mapped the transferred regions by taking advantage of the polymorphism between the donor and the recipient strains. SNP mapping in 10 transconjugants obtained with different donors confirmed that the transferred DNA corresponds to a single segment surrounding the ErmR marker (Table 1). The length of exchanged regions ranged from 21 kb to more than 334 kb.
Fig. 1.
Transfer frequency of the ErmR marker. (A) Transfer frequency of the 30 insertions (I-1 to I-30) distributed throughout the chromosome independently of any predicted mobile genetic element. Strain BM110 was used as recipient. All mating assays were performed twice, and standard deviations are indicated. Dashes on the x axis indicate insertions for which no transconjugant was detected. Dashed lines and arrows indicate putative gradients of transfer frequencies observed for three regions. The three arrows under the x-axis indicate putative mobile genetic elements predicted in the genome of strain NEM316 (20). (B) Transfer frequency of four insertions (I-17 to I-20) in the vicinity of genomic island X. Strain A909RF was used as recipient. All mating assays were performed three times, and standard deviations are indicated. Thick dashes indicate transfer frequency for gbs1121+ donors. Circles indicate transfer frequency for gbs1121− donors.
Table 1.
Estimation of the size of the transferred DNA segments in transconjugants
| Transconjugant no.* | ErmR insertion site† | Limits of the transferred segment |
Minimal size of the transferred segment, kb | |
|---|---|---|---|---|
| First detected SNP identical to donor† | Last detected SNP identical to donor† | |||
| TC2 | 314 | 308 | 341 | 33 |
| TC6–1 | 658 | 466 | 810 | 334 |
| TC6–2 | 658 | 565 | 810 | 252 |
| TC10 | 847 | 830 | 858 | 28 |
| TC12 | 986 | 972 | 993 | 21 |
| TC16 | 1,124 | 1,012 | 1,117 | 105 |
| TC17 | 1,160 | 1,127 | 1,225 | 99 |
| TC18 | 1,213 | 1,211 | 1,225 | 14 |
| TC19 | 1,272 | 1,211 | 1,497 | 286 |
| TC24 | 1,619 | 1,570 | 1,619 | 49 |
*Number indicates the NEM316 derivative used as donor strain.
†Coordinates on the NEM316 chromosome in kb.
Because homologous DNA adjacent to the marker was exchanged in the transconjugants, we investigated the involvement of homologous recombination by comparing the transfer efficiencies of the ErmR marker from the I-8 derivative (Fig. 1A) to the recA+ strain A909RF or its recA− derivative. Transconjugants were obtained at a frequency of 2.1 × 10−8 with the recA+ recipient, but none were recovered with the recA− strain (frequency < 10−11). This result demonstrated that homologous recombination is required for the incorporation of exogenous DNA by the recipients.
The Chromosome Is Mobilized from Multiple Sites by Conjugation.
S. agalactiae is not thought to be naturally transformable, but we cannot rule out the possibility that it may be competent in certain growth conditions (20). To exclude this possibility, we showed that the presence of DNase I in the mating medium did not affect the transfer frequencies (data not shown). Transduction also is unlikely because the donor strain is devoid of prophages. Conjugation therefore is the most likely mechanism to account for the observed transfers of chromosomal DNA.
Conjugative transfer of chromosomal DNA generally is attributed to integrative conjugative elements carrying an origin of transfer (oriT). Chromosomal transfer is initiated from the oriT by a relaxase through the introduction of a specific nick. Because transfer is unidirectional, it leads to a gradient of transfer in which the genes closest to the oriT are transferred at higher frequencies than those that are more distally located. This process was described first in Escherichia coli strains with a high frequency of recombination (Hfr) carrying an integrated F plasmid (21). In our experiments, transfer frequencies were dependent on the location of the marker, suggesting that transfer could be initiated from multiple sites (Fig. 1A). Three regions corresponding to insertions I-7 to I-10, I-11 to I-13, and I-18 to I-20 showed putative gradients of transfer frequencies for the ErmR marker. These regions are located near previously described genomic islands (20) including genomic island X, an integrative conjugative element carrying an oriT and encoding the corresponding relaxase, Gbs1121 (unpublished data, M.B. and P.G.). We further investigated the involvement of this relaxase by comparing the transfer frequencies of I-17 to I-20 insertions using NEM316 gbs1121+ or gbs1121− derivatives as donors and strain A909RF as recipient (Fig. 1B). When using gbs1121+ donors, a unidirectional gradient of transfer frequency was observed from I-18 to I-20 (P < 0.05). When matings were performed with gbs1121− donors, the transfer frequency of I-18 to I-20 was significantly reduced (P < 0.05), but the transfer frequency of I-17 was not affected. Furthermore, the genomic island X was not detected among 40 transconjugants selected for the transfer of I-18. These data suggest that an Hfr-type chromosomal mobilization was initiated from the oriT of genomic island X, emphasizing the role of mobile genetic elements in chromosomal mobilization. Conversely, transfers of I-17 probably are not initiated within genomic island X, because this island was detected in 9 of the 20 I-17 transconjugants tested. Moreover, several insertions that transferred at low frequencies were not located in the vicinity of putative mobile genetic elements. These transfers may result from the recognition of ectopic oriTs by mobilization proteins encoded by conjugative elements present in NEM316, as had been shown previously in an unrelated conjugative system (22). Other mechanisms different from classic Hfr chromosomal DNA transfer also may be involved in these large chromosomal replacements, as has been demonstrated in Mycobacterium smegmatis (23).
A Genome-Wide Map of Diversity Brings Evidence of Extensive Chromosomal Exchanges Among Natural Isolates.
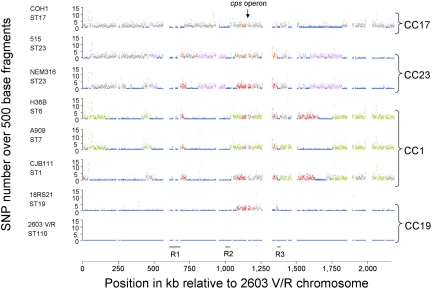
The Hfr-mediated conjugation events that promote exchange of E. coli chromosomal genes in the laboratory are considered to be of little consequence within natural populations and are not thought to lead to a random association of the alleles at different neutral loci (24, 25). To test whether the large chromosomal replacements observed under laboratory conditions contribute to the genome dynamics of S. agalactiae, we looked for such replacements among the eight available genome sequences. A direct way of identifying historical recombination events is to study SNPs in the core genome of related isolates (10). To identify patterns of recombination, nucleotide substitutions between strains were counted in 500-bp windows across the previously defined core chromosome (16) representing 28 possible pairwise comparisons (Fig. 2). Strikingly, each pairwise comparison revealed an alternate pattern of highly conserved regions (<0.05% polymorphism on average) and less-conserved regions (>0.7% polymorphism), suggesting the occurrence of recombinational exchanges. The abrupt transition between conserved and divergent blocks allowed us to predict putative recombination points with DNA exchanges involving regions of up to hundreds of kilobases. In addition to these large exchanges, a limited number of putative replacements involving regions of less than 5 kb were observed also but are not considered here. The identification of such large exchanges within the species makes it possible to trace each chromosomal segment back to its origin.
Fig. 2.
Distribution of SNPs along the genome sequences of eight S. agalactiae strains. The number of SNPs (y-axis) is plotted according to the position of the corresponding 500-bp fragment on the strain 2603 V/R chromosome (x-axis). For each coordinate, identical colors indicate regions highly conserved between the corresponding strains (<0.05% polymorphism); gray denotes a high percentage of polymorphism with the seven other chromosomes (>0.7%). Empty sites represent previously described genomic islands larger than 5 kb (16); the black lines R1, R2, and R3 indicate the three regions displaying a low level of polymorphism in the eight genomes. Right brackets indicate clonal complexes; black arrow indicates the locus encoding the capsular polysaccharide. ST, sequence type.
Large Chromosomal Replacements Have Driven the Emergence of Major Clinical Clonal Complexes.
The eight genome sequences published belong to four major clinical CCs (16, 20, 26). This diversity allows the analysis of multiple genome sequences separated by different genetic distances and provides a means of inferring the population structure at the genome level. We distinguished four major groups according to the number and the lengths of regions that are highly conserved among strains (Fig. 2). Our genome classification fits with the clustering defined by MLST (15) because genomes that belong to the same CCs show a large extent of highly conserved regions (Table 2 and Table S2). However, a comparison of strains from the same CC revealed that they also are distinguished by long regions with significant levels of polymorphism. For example, strains 515 and NEM316 showed significant divergence in 12 regions spanning 662 kb, even though they both belong to the sequence type, ST23. Similarly, the closely related strains 2603 V/R and 18RS21 diverge significantly along one 148-kb region that includes the cps genes involved in the synthesis of the capsular polysaccharide, explaining why they express different capsular types. Interestingly, the region surrounding the cps locus showed the highest density of recombination points among the eight genomes (Fig. 2). Thus it is probable that the exchange of this locus encoding an immunodominant component is selected by the host immune response.
Table 2.
The number and size of highly conserved regions among eight S. agalactiae strains
| Strain | COH1 | 515 | NEM316 | H36B | A909 | CJB111 | 18RS21 | 2603 V/R | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC19 | 2603 V/R | 536* | 8† | 558 | 11 | 753 | 14 | 789 | 7 | 900 | 6 | 854 | 8 | 1,603‡ | 2 | — | |
| 18RS21 | 572 | 11 | 591 | 14 | 815 | 14 | 824 | 9 | 926 | 8 | 890 | 10 | — | — | |||
| CC1 | CJB111 | 245 | 7 | 216 | 10 | 483 | 9 | 1,079‡ | 11‡ | 1,048‡ | 9 | — | — | — | |||
| A909 | 357 | 6 | 251 | 10 | 445 | 12 | 1,471 | 5‡ | — | — | — | — | |||||
| H36B | 248 | 7 | 224 | 10 | 426 | 11 | — | — | — | — | — | ||||||
| CC23 | NEM316 | 259 | 11 | 1,089‡ | 13 | — | — | — | — | — | — | ||||||
| 515 | 254 | 9 | — | — | — | — | — | — | — | ||||||||
| CC17 | COH1 | — | — | — | — | — | — | — | — | ||||||||
*Total length in kilobases of the highly conserved regions.
†Number of highly conserved regions.
‡There is a high extent of conserved regions within these clonal complexes.
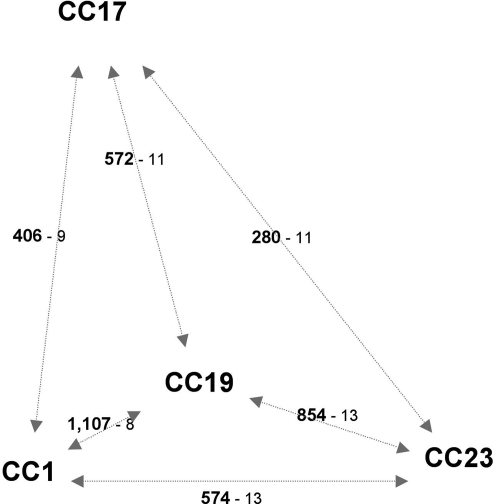
Strains belonging to distant CCs also share highly conserved regions, and we determined that all CC pairs share at least 280 kb of DNA that are highly conserved and scattered over 8 to 14 regions (Fig. 3). This distribution of many highly conserved regions among distant CCs can be explained by the emergence of a clone that evolved either by importing large chromosomal DNA fragments from a pool of unrelated strains or by exporting large DNA fragments to unrelated strains. Given the extent of recombinational exchanges, the relationships between CCs are difficult to define, because neither trees nor networks provide appropriate models. However, strains from CC19 share the highest number of highly conserved regions with all other CCs (Fig. 3). Consistently, except two regions, any chromosomal piece from strains belonging to this CC is highly conserved in at least one strain of the other CCs, but the reverse is not the case (Fig. 2). This asymmetrical distribution of highly conserved regions suggests that the clone that has spread throughout the population is closely related to CC19. The high conservation level of regions that are likely to be derived from this emerging clone (i.e., <0.05% polymorphism) indicates a probably recent dissemination within the population. This global genetic flux analysis demonstrates that in S. agalactiae CCs associated with human carriage and neonatal infections are not unrelated and probably are derived from an emerging clone that has exchanged large chromosomal segments with unrelated isolates.
Fig. 3.
Central position of CC19 according to the extent of highly conserved regions between the different CCs. The numbers in bold type indicate the total length in kilobases of the highly conserved regions between two CCs. The numbers in lightface type indicate the number of these highly conserved regions.
A similar pattern of SNP distribution was observed when comparing S. enterica Typhi and Paratyphi A (9). For three-quarters of their genomes they are distantly related, but the remaining quarter is much more conserved with an average nucleotide divergence of 0.18% instead of 1.2%. A convergent model was proposed involving 124 genetic importations of fragments with an average size of 6.4 kb. However, these genetic exchanges probably occurred within a limited time span and involved only these two human-restricted serovars. Among the eight S. agalactiae strains, we observed progressive levels of divergence suggesting that exchanges do not correspond to a single rapid burst of recombination between a pair of lineages.
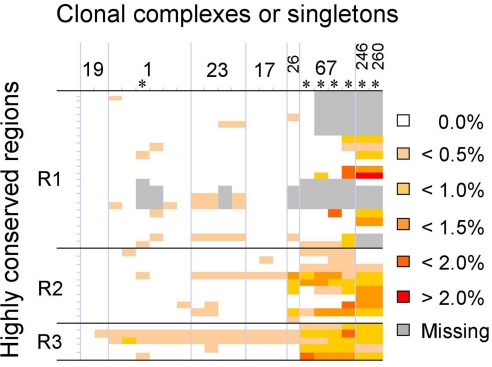
Regional Variations in Nucleotide Diversity Identify Putative Selective Sweeps.
Our evolutionary model raises the question of what genetic basis might have led to the emergence of this dominant clone. The genome-wide map of nucleotide diversity revealed that, in addition to the seven sets of ribosomal RNA operons, three regions are highly conserved in the eight strains (Fig. 2). These regions were termed “R1,” “R2,” and “R3” and encompass gbs0587-gbs0655, gbs1019-gbs1041, and gbs1416-gbs1425, respectively. In organisms in which recombination occurs readily, the natural selection at loci encoding selective advantages produces selective sweeps associated with a reduction in genetic diversity around these loci. Therefore we further characterized the nucleotide diversity of these three regions by sequencing 37 loci in an additional set of 14 strains from various origins representing 15,119 kb. Sequence analysis showed that these three regions are highly conserved in all studied human isolates (Fig. 4). Interestingly, these regions were more diverse in CC67 and singletons ST246 and ST260 that are specific to animal strains (15, 17). Because these regions are highly conserved only in human isolates, they may be linked to selective advantages in this host that led to the emergence of the dominant clone.
Fig. 4.
Polymorphism levels of regions R1 to R3 in 22 strains from various origins. The sequence of 37 loci (representing 15,119 kb) was compared using strain 2603 V/R as a reference. Colors indicate the level of nucleotide divergence. Gray boxes indicate absence of the locus. The marks on the left vertical line indicate the limits of the 37 loci. Asterisks indicate strains isolated from animals. Strain designations (left to right): 2603 V/R, 18RS21, CJB111, Mad74, 553–08, H36B, A909, CCH268, NEM316, CCH350, 515, CCH263, COH1, NEM1575, Ban29, Dak30, 411–07, 304–36, 043–14, 501–18, 2–22, SS1218. SNPs are detailed in Tables S5 and S6.
These three highly conserved regions encompass 127 kb, and therefore it is difficult to predict the functional basis of selection for these chromosomal segments. Some hypotheses may be proposed, however. Region R1 is of particular interest, because it encodes three loci involved in the synthesis of virulence factors or putative colonization factors including the β-hemolysin, a pilus-like structure, and an ABC exporter coupled to a two-component regulatory system. The latter is located in a remnant mobile element inserted at a tRNAArg locus (17) and is highly similar to the VncRS locus from Streptococcus pneumoniae (27, 28). The whole mobile element is absent only in CCs specific to animals (17). Interestingly, VncRS-related systems, exhibiting 92% and 93% identity, were identified in a conjugative transposon related to Tn916 in Enterococcus faecalis (29) and in CTn5 in Clostridium difficile (30), respectively. The CTn5 transposon recently was shown to belong to a conjugative transposon family amplified in human gut microbiomes (31). Because, like S. agalactiae, these two bacterial species are inhabitants of the human intestine, it is possible that the horizontal acquisition of this VncRS-related system contributes to the adaptation to this niche.
Conclusions
Recombinational exchanges in bacterial populations were thought to involve the transfer of small pieces of genomes, primarily through natural transformation or phage-mediated transduction (5). Conjugative transfer and the replacement of hundreds of kilobases of bacterial chromosomes already have been described under laboratory conditions, but their impact on natural populations remained largely unknown (32). This work demonstrates that large conjugal exchanges have contributed significantly to the genome dynamics of S. agalactiae and strengthen the role of integrative conjugative elements in the dynamics of bacterial chromosomes (33, 34). The outcome of such extensive exchanges probably depends on complex selection processes allowing transconjugants to outcompete the parental strains. Consistently, the analysis of resulting gene flux points to particular regions under different selective pressures that probably are involved in the adaptation to the host. In the near future, the nascent era of population genomics should provide further clues about the genetic mechanisms and the evolutionary forces that shape bacterial genomes.
Methods
Bacterial Strains and Growth Conditions.
The S. agalactiae strains used in this study are listed in Table S3 and were cultured in Todd-Hewitt broth or agar (Difco) at 37°C. Antibiotics were used at the following concentrations: erythromycin, 10 μg/ml; rifampicin, 100 μg/ml; fusidic acid, 10 μg/ml; tetracycline, 10 μg/ml. When specified, 10 units of DNase I were added during mating assays.
General DNA Techniques.
The oligonucleotide primers were designed with Primer3 software and are listed in Table S4. Sequencing reactions were performed on PCR products using the ABI BigDye Terminator cycle-sequencing kit and the ABI 3700 capillary DNA sequencer (Applied Biosystems). The insertion sites of the ErmR markers were determined by direct sequencing with chromosomal DNA template using forward and reverse himar oligonucleotides (Table S4). Sequence data were analyzed using phredPhrap, BLAST (36), and MULTALIGN programs.
Construction of Bacterial Strains.
NEM316 derivatives were generated by transposon mutagenesis, using the temperature-sensitive plasmid pCAM45 as previously described (35). The plasmid-borne Himar1-derived minitransposon contains an ErmR marker and an R6Kγ origin of replication flanked by the transposon inverted repeats. In almost all cases, pCAM45 mutagenesis leads to the unique insertion of the 1.4-kb minitransposon into the chromosome. The mutants generated are stable because the inserted fragment is segregated from the transposition functions carried by the vector.
Insertional inactivations of recA and of the relaxase encoded by genomic island X were carried out by single cross-over using the forward and reverse “recA” and “gbs1121” oligonucleotides (Table S4). The resulting EcoRI-BamHI amplicons were cloned into the thermosensitive shuttle plasmid pG+host8 (37). Electroporation was performed, and transformants and mutants were selected as previously described (38).
Bacterial Conjugations.
Donors and recipients were spread onto a hydrophobic edge membrane filter (Millipore). Donor-to-recipient ratios were calculated to obtain a ratio of approximately 1:1 after 24-h mating assays. Transconjugants were selected with appropriate antibiotics. When specified, 10 units of desoxyrobonuclease I (Roche) were added during filter mating. The frequency of transfer was determined by dividing the number of transconjugants by the number of donor cells. To map the transferred regions, transconjugants were characterized by sequencing 48 loci scattered along the chromosome (Table S4).
Identification of Large Regions Conserved Between Strains and Clonal Complexes.
To identify patterns of recombination, the genome sequence of each strain was fragmented in contiguous and nonoverlapping 500-bp segments, and each fragment then was compared with the seven other genomes by using the Blastn algorithm (36), representing 28 possible pairwise comparisons. For each best-score, the number of SNPs was counted when query and match lengths were identical. Sequences present in multiple copies such as insertion sequences were not considered, nor were the 69 genomic islands that are absent in at least one of the genomes (16). Boundaries between large regions (>5 kb) that are either highly similar or highly divergent between the two interrogated strains were identified by visual inspection. The extent of highly conserved regions between CCs deduced from MLST was based on regions highly conserved between at least two strains belonging to different CCs.
Supplementary Material
Acknowledgments.
We thank Carmen Buchrieser, Didier Mazel, and Michel-Yves Mistou for many stimulating discussions. We thank Ciarán Condon, Gunnar Lindahl and Isabelle Rosinski-Chupin for critical reading of the manuscript. Plasmid pGhost8+ and strain A909 were provided by Emmanuelle Maguin and by the Collection de l'Institut Pasteur (CIP), respectively. Financial support was received from the Institut Pasteur, the Centre National de la Recherche Scientifique, the French-Israeli Network in Bioinformatics, and the European contract LSHB-CT-2005–512061. M.B. received support from the Caisse Nationale du Régime Social des Indépendants.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803654105/DCSupplemental.
References
- 1.Smith JM, Dowson CG, Spratt BG. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 2.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 4.Spratt BG. Exploring the concept of clonality in bacteria. Methods Mol Biol. 2004;266:323–352. doi: 10.1385/1-59259-763-7:323. [DOI] [PubMed] [Google Scholar]
- 5.Spratt BG, Maiden MC. Bacterial population genetics, evolution and epidemiology. Philos Trans R Soc Lond B Biol Sci. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 7.Monot M, et al. On the origin of leprosy. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 8.Roumagnac P, et al. Evolutionary history of Salmonella Typhi. Science. 2006;314:1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Didelot X, Achtman M, Parkhill J, Thomson NR, Falush D. A bimodal pattern of relatedness between the Salmonella Paratyphi A and Typhi genomes: Convergence or divergence by homologous recombination? Genome Res. 2007;17:61–68. doi: 10.1101/gr.5512906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nocard M. Sur une mammite contagieuse des vaches laitières [On a bovine mastitis] Annales de l'Institut Pasteur. 1887;1:109–127. [Google Scholar]
- 12.Fry R. Fatal infections by hemolytic Streptococcus group B. Lancet. 1938;1:199–201. [Google Scholar]
- 13.Schuchat A. Epidemiology of group B streptococcal disease in the United States: Shifting paradigms. Clin Microbiol Rev. 1998;11:497–513. doi: 10.1128/cmr.11.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones N, et al. Multilocus sequence typing system for group B Streptococcus. J Clin Microbiol. 2003;41:2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisharat N, et al. Hyperinvasive neonatal group B Streptococcus has arisen from a bovine ancestor. J Clin Microbiol. 2004;42:2161–2167. doi: 10.1128/JCM.42.5.2161-2167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.”. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brochet M, et al. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 2006;8:1227–1243. doi: 10.1016/j.micinf.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Luan SL, et al. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J Clin Microbiol. 2005;43:3727–3733. doi: 10.1128/JCM.43.8.3727-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brochet M, Couvé E, Glaser P, Guedon G, Payot S. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J Bacteriol. 2008 doi: 10.1128/JB.00824–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaser P, et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 21.Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 22.Becker EC, Meyer RJ. Relaxed specificity of the R1162 nickase: A model for evolution of a system for conjugative mobilization of plasmids. J Bacteriol. 2003;185:3538–3546. doi: 10.1128/JB.185.12.3538-3546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Karnati PK, Takacs CM, Kowalski JC, Derbyshire KM. Chromosomal DNA transfer in Mycobacterium smegmatis is mechanistically different from classical Hfr chromosomal DNA transfer. Mol Microbiol. 2005;58:280–288. doi: 10.1111/j.1365-2958.2005.04824.x. [DOI] [PubMed] [Google Scholar]
- 24.Whittam TS, Ochman H, Selander RK. Geographic components of linkage disequilibrium in natural populations of Escherichia coli. Mol Biol Evol. 1983;1:67–83. doi: 10.1093/oxfordjournals.molbev.a040302. [DOI] [PubMed] [Google Scholar]
- 25.Levin BR. Periodic selection, infectious gene exchange and the genetic structure of Escherichia coli populations. Genetics. 1981;99:1–23. doi: 10.1093/genetics/99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tettelin H, et al. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci USA. 2002;99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novak R, Charpentier E, Braun JS, Tuomanen E. Signal transduction by a death signal peptide: Uncovering the mechanism of bacterial killing by penicillin. Mol Cell. 2000;5:49–57. doi: 10.1016/s1097-2765(00)80402-5. [DOI] [PubMed] [Google Scholar]
- 28.Haas W, Sublett J, Kaushal D, Tuomanen EI. Revising the role of the pneumococcal vex-vncRS locus in vancomycin tolerance. J Bacteriol. 2004;186:8463–8471. doi: 10.1128/JB.186.24.8463-8471.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsen IT, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 30.Sebaihia M, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 31.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson DA, Enright MC. Evolution of Staphylococcus aureus by large chromosomal replacements. J Bacteriol. 2004;186:1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochhut B, et al. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol Microbiol. 2006;61:584–595. doi: 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- 34.Rice LB, Carias LL, Hutton-Thomas R, Rudin S. Interaction of related Tn916-like transposons: Analysis of excision events promoted by Tn916 and Tn5386 integrases. J Bacteriol. 2007;189:3909–3917. doi: 10.1128/JB.00859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.May JP, Walker CA, Maskell DJ, Slater JD. Development of an in vivo Himar1 transposon mutagenesis system for use in Streptococcus equi subsp equi. FEMS Microbiol Lett. 2004;238:401–409. doi: 10.1016/j.femsle.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswas I, Gruss A, Ehrlich SD, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dramsi S, et al. Assembly and role of pili in group B Streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.