Abstract
Bacteriophage Φ6 contains three dsRNA genomic segments L, M, and S. The RNA is located inside a core particle composed of multiple copies of a major structural protein, an RNA-dependent RNA polymerase, a hexameric NTPase, and an auxiliary protein. The virion RNA polymerase in the core particle transcribes segments M and S in vitro. Yet early in infection, the transcription of L is highly active. Late in infection, transcription of L is low, and that of M and S is high. A host protein encoded by yajQ is responsible for the activation of L transcription. Knockout mutants of yajQ do not support the replication of Φ6, although they do support the replication of distantly related members of the Cystoviridae. Φ6 can mutate to independence of YajQ. This requires two mutations in the gene for the RNA-dependent RNA polymerase. YajQ acts indirectly on the polymerase by binding to P1, the major structural protein of the core. Previous studies have shown that the activity of the polymerase in the core is controlled by the conformation of the core particle structure.
Bacteriophage Φ6 is a member of the Cystoviridae, a family of bacteriophages that have genomes of three double-stranded RNA molecules, L, M, and S, packaged inside a polyhedral capsid structure covered by a lipid-containing membrane. Φ6 was the first member of this family to be discovered (1). Cystoviridae infect Gram-negative bacteria, primarily Pseudomonas syringae and its relatives. The phages infect by fusing their membranes with the outer membrane of the host cells and then breaching the cell wall through the action of muramidases and finally having the core of the virion enter the host cell (2). The core contains an RNA-dependent RNA polymerase that synthesizes transcripts that are released from the core to program the synthesis of phage proteins (3). The composition of the transcript population changes during the infection cycle (4). At very early times, there is an almost equal amount of the three transcripts; whereas at later times, transcription is limited to the S and M genomic segments. Isolated cores of the Φ6 virion have very strong transcriptase activity, but it is limited to S and M under normal conditions in magnesium-containing buffers (5). The 5′ 18-base sequences of the three plus strands are identical, with the exception that in S and M, it starts with GG, whereas in L, it starts with GU. This difference is the determinant of the differences in in vitro transcription activity (6). In the presence of manganese ions, the transcription of the L segment is activated (5). The mechanism that results in the temporal control of Φ6 transcription has long been a mystery.
As part of a study of the structure of the Φ6 core, we isolated particles that contained one, two, or three dsRNA genomic segments (7). We found that these particles had proteins P1, P2, P4, and P7 in their procapsid cores; however, the two- and three-segment particles also contained an additional protein (Fig. 1). This protein was isolated, and N-terminal sequencing indicated that it was a host protein encoded by an ortholog of the highly conserved gene yajQ that produces a dispensable protein of unknown function. This article will describe the role of YajQ in the temporal control of Φ6 transcription.
Fig. 1.
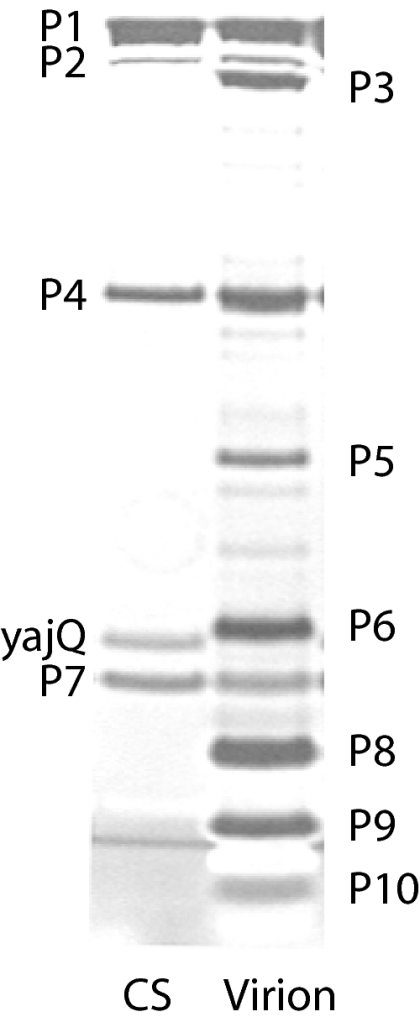
Polyacrylamide gel showing the proteins of the Φ6 virion and the carrier state particle (CS) containing the normal core proteins P1, P2, P4, and P7 along with host protein YajQ (silver stain).
Results and Discussion
Temporal Control of Transcription in Φ6 Infection.
Φ6 and its close relatives have almost identical 5′ sequences of 18 nt in the transcripts of their genomic segments (8). The transcript of segment L in Φ6 begins with GU, whereas those of segments M and S begin with GG (9). The nucleocapsid of Φ6 is composed of a dodecahedral core containing proteins P1, P2, P4, and P7, which is covered by a triangulation number T = 13 shell of protein P8 (10). Protein P2 is an RNA-dependent RNA polymerase that is able to transcribe the three dsRNA genomic segments inside the core. However, transcription in buffers containing magnesium, but not manganese, ions results in the production of only the M and S transcripts (5). This is a consequence of the sequence differences, and if the sequence of L is changed to GG, the transcription of L accompanies that of M and S (6). However, early transcription during Φ6 infection shows considerable quantities of the L transcript, whereas late in infection, the transcript pool is primarily that of M and S (Fig. 2). The nature of the control has been a mystery.
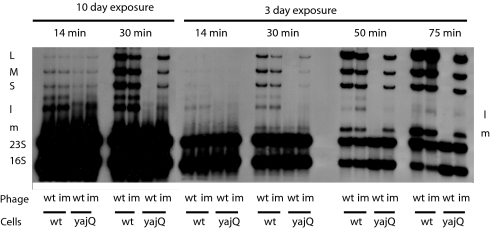
Fig. 2.
Autoradiogram of a polyacrylamide gel displaying H3-labeled RNA of Φ6-infected cells. Phage is wt Φ6 or Φ6 mutant Φ3008 independent (im) of YajQ. Cells are WT P. syringae pv. phaseolicola LM2489 or yajQ knockout strain LM4470 (yajQ). The transcript of S is obscured by 23S RNA. L, M, and S refer to the genomic dsRNAs. Lowercase l and m refer to genomic transcripts.
Involvement of Protein yajQ.
Φ6 can form carrier states in which the virus replicates in host cells without killing and with the expression of reporter genes such as kan for kanamycin resistance (7). It is possible to prepare carrier-state virions that have unique genomic constructions. We prepared such a construction in strain LM4383 wherein the constructed virus genome (10.8 kbp as opposed to 13.4 kbp for wild type) was composed of the normal L segment and a normal S segment that also contained a gene for kanamycin resistance in the 3′ noncoding region. The culture was lysed in a French-press cell, and the lysate was subjected to sucrose-zone and equilibrium-gradient fractionation. Particles were obtained and analyzed for protein and RNA content. It was observed that the core proteins P1, P2, P4, and P7 were accompanied by a previously unobserved protein (Fig. 1). This band was excised from a polyacrylamide gel, and its N-terminal sequence was determined. The sequence was found to be PSFDV. A search in the GenBank database of the proteins of P. syringae pv. phaseolicola NC_005773 produced a match with the N terminus of protein coded by locus PSPPH_4093, which is in the family HMM PF04461. A blast search showed that the protein is highly conserved in bacteria and is similar to protein YajQ of Escherichia coli, a dispensable protein of unknown function but having motifs characteristic of nucleotide or nucleic acid-binding proteins (11, 12). The molecular mass of this protein is 17,836 Da, which is consistent with the migration of the protein in the polyacrylamide gel. We subsequently found that a three-segment carrier-state particle (13.5 kbp) of Φ6 that was deleted for gene 8 also carried the extra protein YajQ. A one-segment carrier-state particle having a smaller genome (7.9 kbp) did not carry this protein.
The yajQ gene of P. syringae pv. phaseolicola was copied and cloned into various plasmids with and without N- and C-terminal His tags. A construction having both N- and C-terminal deletions was cloned into plasmid pET28a (Novagen). This plasmid was used to damage the gene in the host strain to form LM4470. Φ6 was not able to propagate on LM4470. Plaques appeared at a frequency of ≈10−5 compared with those on LM2489, the parent strain. Phages from these plaques were able to plate on LM4470, although the plaque size was somewhat smaller than that found on LM2489 (Fig. 3). The plating of Φ6 on LM4470 was complemented by plasmids coding for YajQ with or without an N-terminal His tag. C-terminal His tags prevented complementation. A representative strain of the YajQ-independent phage is Φ3008. Segment L of Φ3008 was cloned and sequenced, and three changes were identified. They were at nucleotides 1044, 2866, and 5886 of segment L (NC_003715). The first two were in gene 2, K34N and I642V, whereas the third was in gene 1, I632V. The gene 1 mutation was produced in phage propagating on a strain with low levels of YajQ. These mutations were used to reconstruct segment L, and it was found that the mutation in gene 1 alone had a limited effect on plating on LM4470, whereas individually, the others did not. The combination of the 5886 and 2866 mutations resulted in a definite improvement in plating and the combination of all three, Φ3009, closely approximated the behavior of Φ3008. A test of the plating of close relatives of Φ6 showed that those with high sequence identity depended on YajQ for plating, whereas phages Φ8, Φ12, and Φ13 (8) were independent of YajQ.
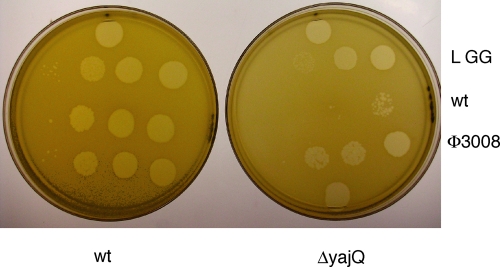
Fig. 3.
Plating of wt Φ6 and yajQ-independent mutant Φ3008 on LM2489 (wt) and LM4470 yajQ knockout strain (ΔyajQ). LGG is Φ6 mutant Φ2057 that has GG as its first two nucleotides on the L transcript. Each row has four samples of 5 μl in a series of hundredfold dilutions, with the lowest titer on the left.
A carrier state can be established in E. coli if Φ6 carries the M segment genes of Φ13, which changes the host range. Such a carrier state was established, but at a very low frequency. Infectious phage Φ3013 was isolated from the carrier-state cells and was found to be independent of YajQ in P. syringae. Mutations were found in gene 2 at nucleotides 1499 and 2750, resulting in K185R and V603A. Apparently, the YajQ of E. coli, although quite similar (46% identity) to that of P. syringae, did not substitute. Empty procapsids can be produced in E. coli, but they do not associate with either YajQ of E. coli or with plasmid-encoded P. syringae YajQ. A plasmid expressing the E. coli yajQ gene did not complement the knockout of yajQ in P. syringae.
Φ6 nucleocapsids are capable of transcribing segments S and M when incubated in magnesium-containing buffers in the presence of ribonucleotide triphosphates. The activity is enhanced by the removal of the protein P8 shell by EGTA (13). We found that nucleocapsids in the presence of N-terminal His tag YajQ had substantial transcription of segment L along with that of S and M (Fig. 4). Nucleocapsids produced from Φ3008 virions showed only a minimal increase in transcription of L in the absence of YajQ. It is also apparent in the early in vivo transcription that the YajQ-independent mutant is not fully independent of YajQ (Fig. 2). It appeared that the role of YajQ might be to promote the transcription of L early in infection.
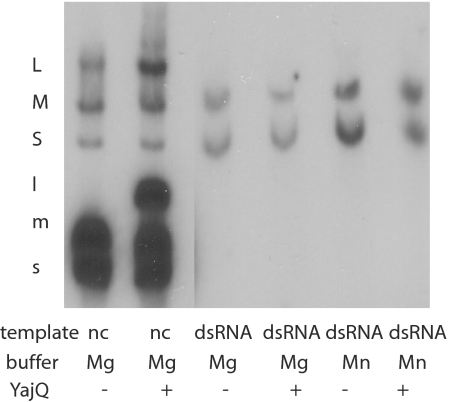
Fig. 4.
A comparison of the effects of YajQ on transcription by nucleocapsids versus transcription of dsRNA by purified Φ6 polymerase protein P2. In vitro transcription with nucleocapsids (nc) in magnesium buffers with and without YajQ. In vitro transcription of equal amounts of dsRNA segments L, M, and S with purified Φ6 RNA polymerase. Samples are untreated or treated with YajQ or manganese ions. Note that Φ6 transcription is semiconservative and results in the labeling of the dsRNA templates.
A test of transcription control in vivo was performed. Infection by wild-type virus was studied in LM2489 (normal cells) and in LM4470 (yajQ knockout). Infection with Φ3008 was also studied. Cells were labeled at 14, 30, 50, and 75 min with tritium-labeled uracil. The results are shown in Fig. 2. It is apparent that early in infection of wild-type cells with wild-type virus, there is appreciable transcription of L and M. In fact, the transcript of L is more abundant than that of M. The transcripts of S cannot be seen because they are masked by the band of 23S RNA. In the yajQ knockout strain, there is production of the M transcript at 14 and 30 min but no evidence of the L transcript. Φ3008 is, however, able to transcribe L, although at a lower rate than in the wild-type strain. At later times, the wild-type virus transcription has shut down completely in the knockout strain, whereas the increase in transcription of M in the wild-type strain can be seen. It seems clear that YajQ is playing a dramatic role in the control of the early transcription of L. It is not clear why the transcription of L diminishes at later times. Perhaps this is due to a change in the relative amount of YajQ versus core particles in the cells or to a difference in the behavior of core particles that enter the cell as compared with those formed during infection. However, Western blot analysis of YajQ content shows that it does not diminish during infection.
If the role of YajQ is to promote the transcription of L, we might predict that a virus with the sequence GG at the 5′ of L might not need YajQ. We have prepared such a phage, Φ2057. It was found that this phage is able to plate on LM4470 with the same titer as on LM2489, although the plaques are smaller (Fig. 3). It might seem advantageous for the phage to have continuous high transcriptase activity integral to the L segment, which codes for the proteins of the viral core particle, but that doesn't seem so. In virtually all temporally regulated viral systems, the abundance of early transcripts is markedly reduced late in infection. Phage Φ8 has identical sequences at the 5′ of each of its three segments, and nucleocapsids transcribe all three at the same rate. However, this strain has a unique method of regulating L transcript late in infection. It actively degrades it through a mechanism that will be described in another report. The Φ6 mutant with GG at the 5′ of segment L produces high levels of L transcript late in infection. This indicates that there is no present mechanism for the specific destruction of the L transcript in Φ6. It appears that some product or condition is responsible for the lack of L transcript late in infection with wild-type virus. The amount of YajQ in the cell does not diminish during infection, as measured by Western blot analysis.
Interaction of yajQ with Core Particles.
P. syringae yajQ with an N-terminal His tag was produced in E. coli. The protein was purified on nickel-containing agarose columns. Nucleocapsids of Φ6 that have had the protein P8 shell removed with EGTA carry out transcription in vitro to produce transcripts of segments S and M in the presence of magnesium ions. If manganese ions are present, transcription of segment L is also seen (5). Addition of YajQ to nucleocapsids incubated in magnesium buffers results in transcription of the L segment (Fig. 4). Purified protein P2, the RNA-dependent polymerase of Φ6 has activity as a transcriptase of dsRNA (14). However, the activity on segment L is minimal in magnesium buffers. The addition of YajQ does not stimulate the transcription of the L segment (Fig. 4). This suggests that the target for YajQ is not directly on the viral polymerase.
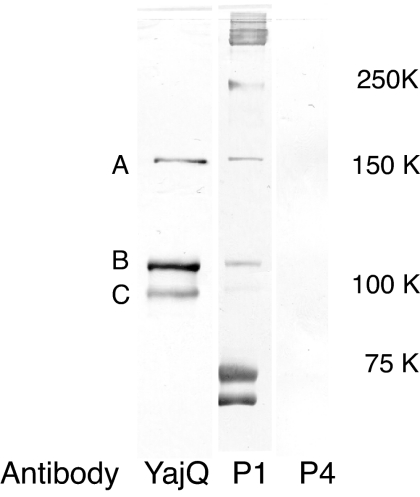
Carrier-state infection was established with virus that contained two genomic segments in cells of P. syringae that were also producing YajQ with N-terminal His tags. The particles were purified on a zonal sucrose gradient and applied to nickel columns. It was found that most of the particles did not adhere to the column, but a fraction did adhere, and this fraction could be eluted with 100 mM imidazole. Most of the YajQ in the particles had been removed by the Ni column. Particles that carried yajQ without His tags did not adhere to the column. It appears that yajQ is bound to filled core particles with moderate affinity in low-salt conditions. In 200 mM NaCl solutions, the protein is removed from the particles. Antibody to YajQ does not bind to cores that have bound YajQ. Cross-linking with formaldehyde of core particles that are carrying N-terminal His-tagged YajQ results in the appearance of cross-links between YajQ and protein P1, the major structural protein of the core. Upon disruption of the cross-linked particles and resolution on Ni columns and polyacrylamide gels, it was found that the major bands associated with YajQ were composed of P1 (Fig. 5).
Fig. 5.
Western blot analysis of proteins eluted from a Ni-NTA-agarose column after disruption of cross-linked carrier-state particles carrying N-terminal His tag YajQ. The first column shows proteins reacting with antibodies to YajQ. The second column shows proteins reacting with antibodies to P1, and the third column shows proteins reacting with antibodies to P4. Molecular mass standards positions are shown to the right, but the major P1 band has a molecular mass of 85 kDa. A, B, and C refer to presumptive cross-linked bands of YajQ and P1.
An unresolved question remains in that we do not yet know how YajQ affects the transcriptional behavior of the virus. We know that the protein binds to the outside of the filled procapsids, but we don't know exactly where it binds or how it influences the behavior of the polymerase. We do know that the Φ6 polymerase inside the core is subject to regulation by the packaging of the genomic segments. Empty procapsids are able to package viral transcripts in the order S, M, L. Only when the packaging is complete does the polymerase begin minus-strand synthesis (15). However, isolated polymerase is able to synthesize minus strands on single plus-strand templates (14). The current idea is that the conformation of the core shell determines the activity of the polymerase. It might be that YajQ binding to the core changes the conformation so as to activate the transcription of L. It seems that YajQ binds only to cores that have fully expanded. A carrier-state particle that has only one segment does not seem to bind YajQ. Empty procapsids do not seem to bind YajQ either. It might be that the binding of YajQ and intermediate shell protein P8 have the same or similar conformational targets in binding to the core particles. The finding that one of the mutations involved in YajQ independence is in gene 1 further supports the idea that YajQ is binding to a domain of protein P1.
Another unanswered question is why transcription of L is low late in infection despite the undiminished presence of YajQ. We consider two major possibilities. The first is that cores derived from mature virions are different in conformation or composition from cores that have not yet acquired the P8 shell and membrane. We have found that cores that have packaged RNA in vitro and have synthesized minus strands are not stimulated by YajQ to transcribe segment L. However, mature virions stripped of membrane and the P8 shell are stimulated by YajQ. It is not yet clear whether the presence or state of P8 is involved, because cores that have been stripped of P8 seem to be more active if P8 remains in solution rather than being eliminated. The second possibility is that there is a change in the chemical environment of the host cell early in infection as opposed to late in infection. We still do not know why P8 is removed and digested from incoming particles (2) but stable on particles formed late in infection. An early study of Φ6 transcription found that chloramphenicol added early in infection allowed transcription of L to continue at late periods when such transcription is usually greatly diminished (4). However, the cause of this effect is not known.
The usage of host proteins in bacteriophage functions is widespread. The functions of GroE in λ-prohead assembly (16) and the incorporation of proteins S1 and EF-Tu in leviviridae polymerase (17) are well known. The cores of the cystoviridae, reoviridae, and totiviridae are unique in their structure and modes of transcription. The manner in which yajQ participates in the temporal control of Φ6 transcription is also unique but might have relevance to transcriptional or replicative control in other viruses that have encapsidated polymerases.
Materials and Methods
Bacterial Strains, Phage and Plasmids.
LM2489 is a rough derivative of P. syringae pv. phaseolicola HB10Y (1) and was used as the primary host for plating Φ6. LM128, a derivative of HB, was also used, and LM2691 is LM128 carrying plasmid pLM1086, which is a derivative of pRK290 (18) and pAR1219 (19) and expresses T7 RNA polymerase in pseudomonads. Plasmid transformation into E. coli used strains JM109 or Stratagene XL1-Blue super competent cells (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)].
Media.
The media used were LC and M8 (20). Ampicillin plates contained 200 μg of ampicillin per milliliter in LC agar. Kanamycin is used at 40 μg/ml.
Reverse Genetics of Φ6.
Mutant forms of the phage were prepared by constructing deletions or modifications in plasmids containing cDNA copies of the genomic segments. These were then electroporated into host cells containing plasmids expressing T7 RNA polymerase (7). In general, thousands of plaques were produced, and samples were purified and their RNA analyzed by gel behavior and by the preparation of DNA copies by RT-PCR, and they were subsequently checked by sequence analysis at the DNA sequencing core facility of University of Medicine and Dentistry of New Jersey.
Labeling of RNA During Phage Infection.
Host cells were diluted from an overnight culture in synthetic M8 medium and grown to 5 × 108 cells per milliliter. One-milliliter aliquots were placed on ice, and phage was added at a multiplicity of 10. The cells were left on ice for 30 min and then transferred to 28°C. Ten microcuries of 3H-5,6-uracil was added at various times. The cultures were spun at 6,000 × g 10 min later and the cells resuspended in 200 μl of lysis buffer [20 mM Tris·HCl (pH 8), 1% SDS, 5 mM EDTA) with 2 M sodium acetate (pH 5.4). The mixture was then frozen at −20°C for 20 min and then spun at 8,000 × g for 10 min at room temperature. The supernatant liquid was extracted twice with phenol-chloroform and precipitated with ethanol. The precipitate was resuspended in 20 μl of DNA buffer and analyzed on 0.8% SeaKem GTG agarose (FMC BioProducts) in TBE with 2 μg/ml ethidium bromide. The bands were visualized with UV light and subjected to autoradiography.
Isolation of Two-Segment Carrier-State Particles.
LM2691 carrying plasmids encoding T7 RNA polymerase was subjected to electroporation with plasmids pLM687 and pLM863 containing cDNA of segment L and S, respectively, to make strain LM4383. The cDNA of segment S contains a gene for kanamycin resistance in the 3′ noncoding region. These plasmids do not replicate in pseudomonads and selection for kanamycin resistance results in the isolation of colonies that are carrier state for Φ6 and contain only two genomic segments per particle. Cultures were grown in LB medium, collected by centrifugation, and lysed in French-press cells. Cell debris was removed by low-speed centrifugation, and the supernates were fractionated on 10–30% sucrose gradients containing 20 mM Tris·HCl (pH8), 150 mM NaCl, and 1 mM MgCl2. The particle-containing band was removed and placed on a 40–60% sucrose gradient overnight, spinning at 75,000 × g. The band was removed and analyzed for RNA and protein content by gel electrophoresis in agarose or polyacrylamide. An unexpected protein band was cut out from the gel and sent to the Protein Core Laboratory at Columbia University for N-terminal sequencing.
Cloning of yajQ.
The yajQ gene of P. syringae was cloned with a hexameric His tag at the N terminus by using olm1106 and olm1116 as primers for PCR. The sequences of the primers are AAACTCGAGTTAGTCGCGGAAGTTGTTGAATTGC and TTTATCTAGAGGAGAACATACATGCACCACCATCATCACCACCCCTCGTTCGACGTAGTG, respectively. The PCR product was inserted into plasmid pET28a (Novagen) XbaI and XhoI sites to form pLM3556. It was also inserted into pLM350 XbaI and HindIII sites to form pLM3555. PLM350 is a shuttle vector that propagates in both E. coli and P. syringae. YajQ was also cloned with primers olm1105 and olm1107. The sequences of these primers are TTTATCTAGAGGAGAACATACATGCCCTCGTTCGACGTAGTGTC and AAACTCGAGTGTCGCGGAAGTTGTTGAATTGC, respectively. The product was inserted into pET28a XbaI and XhoI sites to create a His tag at the C terminus of YajQ. This plasmid, pLM3536, was treated successively with EcoRV and SalI to delete the first 20 aa of YajQ and then with DraIII and BstEII to remove the last 32 aa of YajQ, forming plasmid pLM3559, which was used to prepare a knockout of yajQ in P. syringae.
Knockout of P. syringae yajQ.
P. syringae strain LM2489 was transformed with plasmid pLM3559 and selection performed for kanamycin resistance. The plasmid does not replicate in pseudomonads, and it was assumed that it had recombined with the resident yajQ gene to form LM4470. The strain was then transformed with plasmid pLM3555 that replicates in pseudomonads and expresses YajQ with an N-terminal His tag. Wild-type Φ6 was not able to plate on strain LM4470 that harbored the presumptive yajQ knockout, whereas it was able to plate on strain LM4472 that carried the complementing plasmid. Plaques appeared at a frequency of ≈10−5. These were mutants that are able to plate independently of yajQ. An example is Φ3008.
Preparation of Bacteriophage Φ3009.
Plasmid pLM3569 was constructed by directed mutagenesis of plasmid pLM1413 so as to contain the three mutations found in bacteriophage Φ3008. PLM1413 is a T7 promoter plasmid that contains a cDNA copy of Φ6 segment L with a PstI site at position 6271. Electroporation of pLM1413 along with plasmids containing cDNA copies of segments S and M results in the production of viable wild-type phage. Electroporation of pLM3569 in a similar test results in viable phage, Φ3009, that plates well on the LM4470, the yajQ knockout strain.
In Vitro Transcription with Nucleocapsids or Purified Polymerase.
Nucleocapsids of Φ6 were prepared from purified virions stripped of their lipid-containing membranes by treatment with 2% Triton X-100 (21). Transcription was performed in magnesium buffers with and without added YajQ. Polymerase protein P2 was prepared from BL21DE3 cells carrying plasmid pLM3592, which is a pET28a derivative having a T7 promoter and a C-terminal His tag. The conditions of transcription were those of Makayev and Bamford (14) but with and without manganese ions in the buffer solution. The templates were dsRNA isolated from Φ6 virion with equal amounts of the three genomic segments.
Cross-Linking of YajQ to the Viral Core Structure.
Carrier-state particles were prepared in strain LM4508 that contains the yajQ knockout and the complementing plasmid producing the N-terminal His tag YajQ. The carrier-state particles were formed with plasmids pLM687 and pLM3587. The former codes for the proteins of the core structure, whereas the latter codes for the proteins of segment S, but with a chloramphenicol resistance gene inserted into the EcoRV site in gene 8. The cells were lysed and particles prepared as described above. The purified particles were treated with 0.1% paraformaldehyde for various times. The reaction was terminated by the addition of 50 mM Tris·HCl (pH 8), and the particles were disrupted by 10 cycles of freezing and thawing. The sample was passed through a spin column of Sephadex G-50 to replace Tris with 10 mM sodium phosphate pH 8 and 10 mM imidazole. The material was then applied to Ni-NTA agarose columns, and material that eluted with 250 mM imidazole was concentrated and electrophoresed in 10% polyacrylamide gels, transferred to immobiline sheets, and analyzed as Western blots with antibody to YajQ, P1, and P4. YajQ was seen to be associated primarily with protein P1, the major structural protein of the viral core. Antibody to YajQ was prepared in rabbits by Lampire Biological Laboratories from a purified sample of N-terminal His tag YajQ prepared in E. coli.
Acknowledgments.
This work was supported by National Institutes of Health Grant GM34352.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Vidaver AK, Koski RK, Van Etten JL. Bacteriophage Φ6 : A lipid-containing virus of Pseudomonas phaseolicola. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romantschuk M, Olkkonen VM, Bamford DH. The nucleocapsid of bacteriophage Φ6 penetrates the host cytoplasmic membrane. EMBO J. 1988;7:1821–1829. doi: 10.1002/j.1460-2075.1988.tb03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Etten JL, Vidaver AK, Koski RK, Semancik JS. RNA polymerase activity associated with bacteriophage Φ6. J Virol. 1973;12:464–471. doi: 10.1128/jvi.12.3.464-471.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coplin DL, Van Etten JL, Koski RK, Vidaver AK. Intermediates in the biosynthesis of double-stranded ribonucleic acids of bacteriophage Φ6. Proc Natl Acad Sci USA. 1975;72:849–853. doi: 10.1073/pnas.72.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emori Y, Iba H, Okada Y. Transcriptional regulation of three double-stranded RNA segments of bacteriophage Φ6 in vitro. J Virol. 1983;46:196–203. doi: 10.1128/jvi.46.1.196-203.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frilander M, Poranen M, Bamford DH. The large genome segment of dsRNA bacteriophage Φ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA. 1995;1:510–518. [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Qiao X, Mindich L. Construction of carrier state viruses with partial genomes of the segmented dsRNA bacteriophages. Virology. 2004;319:274–279. doi: 10.1016/j.virol.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Mindich L, et al. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J Bacteriol. 1999;181:4505–4508. doi: 10.1128/jb.181.15.4505-4508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iba H, Watanabe T, Emori Y, Okada Y. Three double-stranded RNA genome segments of bacteriophage Φ6 have homologous terminal sequences. FEBS Lett. 1982;141:111–115. doi: 10.1016/0014-5793(82)80027-6. [DOI] [PubMed] [Google Scholar]
- 10.Huiskonen JT, et al. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure (London) 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Saveanu C, et al. Structural and nucleotide-binding properties of YajQ and YnaF, two Escherichia coli proteins of unknown function. Protein Sci. 2002;11:2551–2560. doi: 10.1110/ps.0217502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teplyakov A, et al. Crystal structure of the YajQ protein from Haemophilus influenzae reveals a tandem of RNP-like domains. J Struct Funct Genomics. 2003;4:1–9. doi: 10.1023/a:1024620416876. [DOI] [PubMed] [Google Scholar]
- 13.Olkkonen VM, Ojala P, Bamford DH. Generation of infectious nucleocapsids by in vitro assembly of the shell protein onto the polymerase complex of the dsRNA bacteriophage Φ6. J Mol Biol. 1991;218:569–581. doi: 10.1016/0022-2836(91)90702-8. [DOI] [PubMed] [Google Scholar]
- 14.Makeyev EV, Bamford DH. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 2000;19:6275–6284. doi: 10.1093/emboj/19.22.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frilander M, Gottlieb P, Strassman J, Bamford DH, Mindich L. Dependence of minus strand synthesis upon complete genomic packaging in the dsRNA bacteriophage Φ6. J Virol. 1992;66:5013–5017. doi: 10.1128/jvi.66.8.5013-5017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopoulos CP, Hohn B. Identification of a host protein necessary for bacteriophage morphogenesis (the groE gene product) Proc Natl Acad Sci USA. 1978;75:131–135. doi: 10.1073/pnas.75.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown S, Blumenthal T. Reconstitution of Qβ RNA replicase from a covalently bonded elongation factor Tu-Ts complex. Proc Natl Acad Sci USA. 1976;73:1131–1135. doi: 10.1073/pnas.73.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system for Gram-negative bacteria; construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinclair JF, Cohen J, Mindich L. The isolation of suppressible nonsense mutants of bacteriophage Φ6. Virology. 1976;75:198–208. [PubMed] [Google Scholar]
- 21.Van Etten JL, Lane L, Gonzalez C, Partridge J, Vidaver A. Comparative properties of bacteriophage Φ6 and Φ6 nucleocapsid. J Virol. 1976;18:652–658. doi: 10.1128/jvi.18.2.652-658.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]