Abstract
Thymic medullary epithelial cells (MECs) express a broad repertoire of peripheral-tissue antigens (PTAs), many of which depend on the transcriptional regulatory factor Aire. Although Aire is known to be critically important for shaping a self-tolerant T cell repertoire, its role in MEC maturation and function remains poorly understood. Using a highly sensitive and reproducible single-cell PCR assay, we demonstrate that individual Aire-expressing MECs transcribe a subset of PTA genes in a probabilistic fashion, with no signs of preferential coexpression of genes characteristic of particular extrathymic epithelial cell lineages. In addition, Aire-dependent PTA genes in MECs are transcribed monoallelically or biallelically in a stochastic pattern, in contrast to the usually biallelic transcription of these same genes in the relevant peripheral cells or of Aire-independent genes in MECs. Expression of PTA genes in MECs depends on transcriptional regulators and uses transcriptional start sites different from those used in peripheral cells. These findings support the “terminal differentiation” model of Aire function: as MECs mature, they transcribe more and more PTA genes, culminating in a cell population that is both capable of presenting antigens (MHCIIhi, CD80hi) and can draw on a large repertoire of antigens to present.
Keywords: AIRE, autoimmunity, gene expression, immunological tolerance, T cell tolerance
The thymus is responsible for producing a repertoire of α:β T cells that is both operational and innocuous. It is divided into distinct regions, each consisting of an array of cell types that interact with differentiating thymocytes, promoting either their continued maturation or their death. The thymic medulla is composed largely of dendritic cells (DCs), macrophages, and medullary epithelial cells (MECs), which together provide a unique environment for nurturing the final stages of T cell maturation and for eliminating certain self-reactive specificities from the repertoire. Initially, as a surprise, MECs were found to express transcripts encoding a large and diverse repertoire of antigens usually associated with particular peripheral tissues [peripheral-tissue antigens (PTAs)] (1–3), raising the question of precisely how MECs and their PTAs promote tolerance induction.
Important clues came from studies on the human multiorgan autoimmune disorder, autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) (4, 5). The gene underlying this disease was named AutoImmune REgulator (AIRE) (6, 7). Mice with a null-mutation in the murine equivalent, Aire, also exhibited multiorgan autoimmunity (8, 9). Comparison of gene-expression profiles of MECs from Aire+/+ and Aire−/− animals revealed that Aire controls the ectopic expression of a large fraction of the PTAs in thymic MECs (8). Precisely how it accomplishes this task is not yet known, although its structure and activities argue for a role in transcriptional regulation at some level (10, 11). Bioinformatic analyses demonstrated that Aire-regulated genes are clustered (12, 13); intriguingly, a given cluster might contain genes up-regulated, down-regulated, or unaffected by Aire, suggesting that it might operate by opening large, contiguous regions to the influence of other positive and negative regulators.
The precise role of PTAs in the maturation and function of MECs remains controversial. Currently, two models vie for acceptance. The “terminal differentiation” model proposes a hierarchy of PTA transcript expression based on the state of MEC differentiation: as these cells mature from an Aire−CD80loMHC-IIlo (MEClo) stage to the end-stage Aire+CD80hiMHC-IIhi (MEChi), they would transcribe more and more PTA genes, each MEChi expressing a large and diverse subset of the full repertoire, in a more or less random fashion (3). Arguing for this scenario was gene-expression profiling on purified MEC subsets, which showed an increase in PTA diversity according to what was presumed to be the sequence of differentiation (13). The “developmental model,” in contrast, contends that immature MECs transcribe the greatest number and diversity of PTA genes; i.e., can be thought of as “multipotential.” As a consequence of Aire expression, the cells would be provoked to differentiate according to standard epithelial cell programs, each MEC following one particular pathway, and PTA expression would be progressively confined to adhere to that program (14). The original version of this model was formulated on the basis of microscopic observations of discreet epithelial (in particular, thyroid) structures in the thymic medulla (15). More recent support came from the demonstration that MEChi have a higher mitotic index than MEClo (16), which was taken as evidence that the former, which express Aire and a rich repertoire of PTAs, are the less mature subset. However, as discussed below, more recent data have challenged this interpretation (17–19).
The “terminal differentiation” and “developmental” models are clearly distinguished by their predictions concerning the repertoire of PTAs expressed by the most mature MECs. The former predicts that single terminal-stage MECs would express a broader set of PTAs, not reflecting any particular epithelial cell type. In contrast, the latter argues that individual end-stage MECs should express a set of PTAs reflective of a diversity of extrathymic cell types, with transcriptional initiation sites and reliance on specific transcription factors that mimic those seen in the corresponding peripheral cell. We have used diverse approaches to address this issue. The results paint a portrait of ectopic gene expression that is very different from that of the same genes being addressed in their “home” tissues.
Results
Single-Cell RT-PCR Assay for PTA Transcripts in MECs.
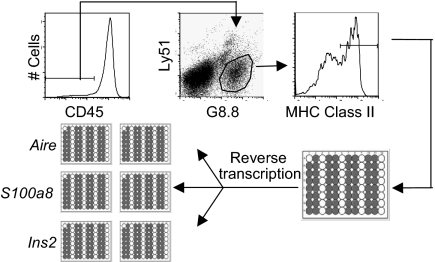
In an initial set of experiments, we sought to define how the expression of PTA transcripts is distributed in individual Aire+ and Aire− MEChi cells. CD45−G8.8+Ly51intMHC-IIhi (hereafter, “MEChi”) from wild-type C57BL/6 (B6) mice were sorted individually into 96-well plates, lysed in situ, and their RNA reverse-transcribed. Each single-cell cDNA was then split into several wells and used as template for parallel, seminested PCR assays of Aire and PTA transcripts (Fig. 1). For optimal efficacy, the single-cell PCRs were run in parallel wells and were not multiplexed. The single-cell RT-PCR assay routinely detected the presence of Aire transcripts in 40–50% of single MEChi cells [supporting information (SI) Fig. S1], a fraction consistent with the proportion of Aire+ cells among the MEChi population according to flow cytometric analysis (17). In comparison, β-actin transcripts were detected in 80–90% of sorted MEChi cells (data not shown). The reproducibility in detecting transcripts in these single-cell RT-PCRs was determined by conducting two independent PCRs for the same gene in parallel, after splitting the cDNA from single MECs. Concordance of detection of Aire transcripts was high, ranging from 70% to 90% among the different experiments (Fig. S1). We also tested MEClo (CD45−G8.8+Ly51+MHC-IIlo), but very few of these expressed either Aire or PTA transcripts (data not shown), and so they were not analyzed further.
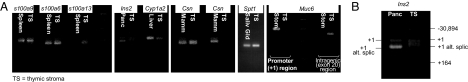
Fig. 1.
RT-PCR assay. CD45−G8.8+Ly51intClass IIhi MECs (MEChi) were sorted from 3- to 4-week-old wild-type B6 mice into 96-well plates containing gene-specific primers. Transcripts provided template for several independent, seminested PCR assays of the individual PTA genes and Aire. Single-cell PCRs were conducted in duplicate in parallel.
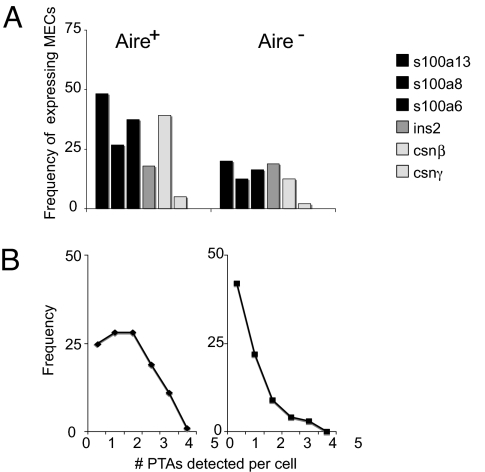
To assess the frequency and distribution of Aire and PTA gene expression in MEChi, we quantified transcripts from Aire, genes within the S100 cluster, the Csnβ-γ diad, and Ins2, by the single-cell RT-PCR assay, in several hundred individual MEChi from three to five wild-type B6 mice. Of the S100 cluster, we focused on S100a6, -8, and -13, chosen because they represent a group of PTAs mapping closely in the genome (within 200 kb), and because they exhibit different responsiveness to Aire in microarray analyses: expression of S100a8 was Aire-activated, S100a6 was Aire-repressed, and S100a13 was largely unaffected by Aire. The genes encoding insulin and casein were selected as highly specific loci transcribed in single epithelial cell types. As illustrated in Fig. 2A, each PTA transcript was expressed in only a fraction of MEChi, ranging from 48% for S100a13 to 5% for Csnγ. As expected, the fraction of transcripts was lower in Aire-negative MEChi. We also performed a limited single-cell analysis of MEChi from Aire-deficient mice: fewer MECs expressing Aire-activated PTA transcripts were detected (5% for S100a8 and none for Ins2), as expected from our earlier microarray data (8), and confirming the specificity of the assay (data not shown).
Fig. 2.
Aire-positive MEChi express a higher frequency of PTAs. (A) Single-cell RT-PCR conducted on single MEChi were analyzed, and the numbers of Aire-positive (Left) and Aire-negative (Right) MEChi expressing S100a13, S100a6, S100a8, Ins2, Csnβ, and Csnγ were determined. Numbers represent percentage of cells positive for a given PTA among Aire-positive or Aire-negative MEChi (also see Table 1). (B) The frequency of cells expressing 0–5 of the PTA genes assessed was counted among Aire-positive (Left) and Aire-negative (Right) MEChi.
Table 1.
Concordance of single-cell RT-PCR assays and percentage of PTA expression in Aire-positive and Aire-negative MEChi
| MEChi | Gene | No. cells tested | Positive |
Concordance |
Concordance, % | Percent positive (corrected) | |||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | 2+ | 1+ | 2− | |||||
| Aire-positive | S100a13 | 150 | 87 | 58 | 52 | 35 | 63 | 60 | 66 |
| S100a8 | 150 | 44 | 29 | 31 | 13 | 106 | 70 | 40 | |
| S100a6 | 150 | 75 | 50 | 44 | 31 | 75 | 59 | 60 | |
| Ins2 | 117 | 20 | 17 | 6 | 14 | 97 | 30 | 46 | |
| Csnβ | 62 | 38 | 61 | 26 | 12 | 24 | 68 | 67 | |
| Aire-negative | S100a13 | 170 | 30 | 18 | 8 | 22 | 140 | 27 | 48 |
| S100a8 | 170 | 16 | 9 | 3 | 13 | 154 | 19 | 46 | |
| S100a6 | 170 | 50 | 29 | 26 | 24 | 120 | 52 | 46 | |
| Ins2 | 139 | 29 | 21 | 6 | 23 | 110 | 21 | 52 | |
| Csnβ | 66 | 11 | 17 | 8 | 3 | 55 | 73 | 28 | |
When portrayed as the number of transcripts detected in each Aire+ or Aire− MEChi (Fig. 2B), the data indicated that most Aire-positive MEChi expressed only one to three of the five PTA transcripts tested, and that only a very minor proportion expressed all five transcripts. Aire-negative MEChi expressed fewer PTA transcripts per cell on average, as expected. Clearly, then, there is a marked heterogeneity in ectopic expression of PTA transcripts in individual MECs.
Aire-Expressing MEChi Transcribe Diverse Combinations of PTAs.
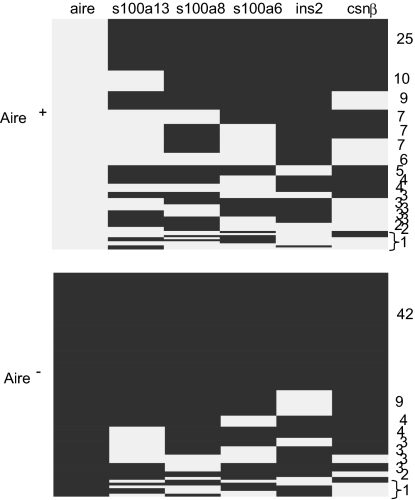
Because expression of PTAs in MECs was variable and argued for a heterogeneous population of cells, we asked whether there were any dominant patterns of PTA transcript coexpression in single cells. The answer would be a direct test of the predictions of the two current models of Aire participation in MEC function. The plot of Fig. 3 orders the different patterns of PTA transcript coexpression observed for 192 individual Aire+ MEChi. A large number of combinations were observed, whose frequency varied mainly in proportion to the frequency of the individual transcripts. There were no obligate associations between loci: expression of S100a13 did not automatically entail expression of S100a8 or S100a6, nor did it rule out expression of Csnβ or Ins2; and many cells expressed various assortments of the S100 genes, Csnβ and Ins2. Because these particular transcripts cover a diverse range of tissue and cell types, the combinations documented are not consistent with the predictions made by the developmental model. Rather, whether a single cell expressed a given PTA transcript appeared to follow a probabilistic determinism.
Fig. 3.
Aire-positive MEChi express a diverse combination of PTA transcripts. Each row represents the combination of genes within the S100 cluster, Ins2, and Csnβ expressed among Aire-positive (Upper) and Aire-negative (Lower) MEChi. Expression of a gene is represented as gray squares, and no expression is represented as black squares. Numbers to the right of the graph represent the frequency of cells with a particular combination of PTA gene expression.
Bioinformatic analyses of gene-expression profiling showed that the Aire-dependent PTAs expressed in MECs tend to emanate from clusters of genes mapping relatively close to each other in the genome (12, 13). Although diverse sets of PTA transcripts were found in individual MECs, we asked whether there might still be some pattern of preferential coexpression of clustered PTA genes. A “relative risk” (RR) was calculated, akin to the metric commonly applied in genetics; i.e., whether a cell expressing transcript A has an increased probability of expressing transcript B. An estimate of the significance of the results was obtained by randomly permuting (10,000 times) the table of results, and asking how many times the observed associations were observed by chance. Relative risks for all gene combinations are tabulated in Fig. S2, combining the results of three independent experiments. Some significant departures from the random (RR = 1) were found among Aire+ MEChi cells, such as the preferential coexpression of S100 family members or, conversely, the disjunction between S100a13 and Ins2. These results support the conclusion that in Aire-expressing MEChi, genes within an Aire-regulated cluster tend to be coexpressed more frequently than genes outside of the cluster and suggest that Aire may be responsible for imposing this coexpression pattern, even though coexpression is not the rule and still follows a stochastic pattern.
Stochastic Activity of Aire-Regulated Genes Reflects Independent Allelic Expression.
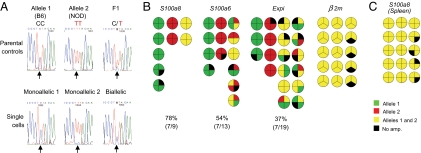
The patterns of ectopic PTA transcript expression prompted us to ask whether the probabilistic element operates at the level of the whole cell or on individual chromosomes. In the first scenario, each cell would express a set of transcription factors that specifies its distinct transcriptional profile, and copies of the genes on both chromosomes would be active. In the second scenario, each locus would be active or inactive in a probabilistic manner, independently of the other copy of the same gene, resulting in a degree of monoallelic transcription. For example, stochastic gene expression of this nature appears to be the mechanism underlying the monoallelic gene transcription pattern observed for Il4 and Il13 in differentiated T-helper type-2 cells (20, 21), which can be attributed largely to the degree of accessibility of each allele. We examined the allelic expression patterns of S100a8, S100a6, and Expi [a gene normally expressed in the mammary gland, whose ectopic expression in the thymus is enhanced by Aire (8, 13)]; β2m was chosen as a control to portray constitutive gene expression. We searched the Perlegen database (Perlegen Sciences, http://mouse.perlegen.com/mouse) for single-nucleotide polymorphisms (SNPs) present in the coding regions of several PTA transcripts in inbred mice, and bred the appropriate F1 animals to enable identification of the chromosome origin of the transcripts. Single MECs were isolated from these F1 mice, cDNA was generated and split into four parallel wells for PCR, and the products were sequenced. Sequencing four times from the same cell greatly diminishes potential artifacts due to limiting amounts of mRNA (22), and only those cells that yielded amplicons in at least three of the four wells were sequenced (Fig. 4A).
Fig. 4.
Single MECs express PTAs monoallelically or biallelically in a probabilistic fashion. (A Upper) Representative sequences of S100a8 from B6, NOD, and F1 parental controls. (A Lower) Representative S100a8 sequences from single MECs sorted from F1 (B6×NOD) thymi. (B) Allelic patterns of S100a8, S100a6, Expi, and β2m in single MEChi. Allelic patterns from single cells were assessed after conducting four independent PCRs from cDNA split from single cells. Only cells giving at least three out of four PCR products were considered informative. Each quadrant represents the allelic-expression pattern observed in each PCR plate. (C) Patterns obtained in Gr1+CD11b+ spleen cells.
The results are summarized in Fig. 4B, where each cell is represented by a small pie chart wherein each quadrant represents the sequence of one of the four PCR products. Red and green denote sequences purely of one or the other of the parental alleles, and yellow signifies mixed sequences. Many of the cells used only one of the chromosomal loci, particularly in the case of the S100 genes, for which only a minority of cells expressed both alleles (78% and 53% monoallelic expression for S100a8 and S100a6, respectively). Alleles of either type were represented in these monoallelic cells, indicating that the loci or the two chromosomes could be transcribed with the same general efficacy. There was also no evidence for parental imprinting, because alleles inherited from either parent were represented equally. Cells expressing a single allele of Expi were also detected, although there was a slightly higher proportion of biallelic transcription in this instance (37% monoallelic). In contrast, both alleles of β2m were expressed in every cell tested.
To determine whether monoallelic expression was a unique feature of ectopic transcription in MECs or rather was an intrinsic characteristic of the S100 genes, we assayed isolated splenic Gr1+CD11b+ neutrophils, the main cell type that expresses S100a8 outside of the thymus (23). Single-cell amplification and sequencing showed S100a8 to be biallelically expressed in these cells (Fig. 4C). Thus, monoallelic expression of S100a8 appears to be a peculiarity of ectopic transcription in thymic MECs, not a property of S100a8 itself.
These data argue that MEC expression of PTA transcripts happens probabilistically for any ectopically expressed locus on any chromosome. This finding is clearly incompatible with the developmental model and seems to refine the terminal differentiation model, one version of which implies that Aire would have a deterministic impact on PTA gene transcription.
Ectopic Transcription of PTA Genes in MECs Utilizes Distinct but Overlapping Transcription Start Sites.
We then used another approach to compare expression of PTAs in the thymus to that of the peripheral tissues. According to the developmental model, one would expect the fine specificity of transcriptional control to be similar in the two cases and, therefore, the initiation of transcription to reflect similar initiation complexes forming at the promoter regions and thus the transcriptional start sites (TSSs) to be conserved. Therefore, we mapped the 5′ ends of a selected set of transcripts in total RNA isolated from sorted MECs and from peripheral tissues using 5′RLM-RACE (24). The transcript patterns were compared by gel electrophoresis, and the cap sites identified by DNA sequencing. Several different behaviors were observed (Fig. 5A). For some genes, the initiation sites were indistinguishable in the two tissues, as for s100a9 or Spt1, the TSSs were identical, down to the base pair, in thymic MECs vs. spleen or salivary gland. For other loci, the same region of initiation was used, but there were minor shifts in the relative preference for multiple TSSs over a short range (e.g., s100a13 and Csnβ). For still others, such as Muc6 and Mup4, the patterns were completely different in MECs and the “home” tissues: as concerns the former, the major TSS was not used; instead transcription was initiated in the region of an internal TSS located in exon 20, where the TSS pattern was again different. The case of Ins2 was also of some interest, given its potential to control immunological tolerance in the context of diabetes (25). As highlighted in Fig. 5B and Fig. S3, the major β cell TSS at position +1 was used in MEC RNA (including a transcript with an alternative splice of intron 1), but it was accompanied by equivalent use of alternative TSSs at position +164 (in exon 2, just upstream of the translation initiation codon) and in the far upstream region at −30894 (a short exon splicing directly onto the normal exon 2). Thus, ectopic gene expression in thymic MECs is characterized by a substantially different distribution of TSSs, suggesting that the transcription factors that participate in the initiation complex may be different.
Fig. 5.
PTA genes in MECs vs. peripheral tissues use distinct but overlapping transcription start sites. (A) 5′RLM-RACE of RNA from purified MECs and peripheral tissues. (B) The major TSS for Ins2 and two alternative TSSs (at position +164 and −30894) are used in MECs.
Ectopic Transcription of Ins2 in MECs Is Independent of Transcription Factors That Control Its Usual Expression in the Pancreas.
Should ectopic PTA expression reflect cell differentiation akin to that of extrathymic tissues, as posited by the developmental model, one would expect that these differentiating cells would emulate the transcriptional hierarchies found in peripheral epithelial lineages and thereby require that MECs would express and use the same transcription factors needed for the development of peripheral tissues and/or expression of tissue-specific genes.
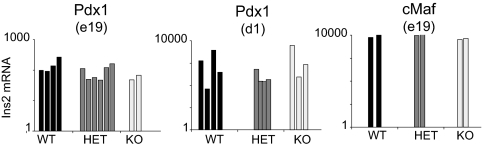
To test this prediction, we determined whether the expression of Ins2 in MECs is dependent on the expression of transcription factors known to play a role in development of the pancreas, differentiation of β cells, and transcription of Ins2. Several transcription factors are necessary for pancreas development and Ins2 gene transcription, including Pdx1 and the Maf family of transcriptional regulators (Fig. S4) (26, 27). The role of Pdx1 in pancreas organogenesis and in the differentiation of β cells is well described (28); its absence in mice prevents formation of the pancreas and results in perinatal diabetes. The cMaf transcription factor, a member of the β cell-specific large-Maf family of basic leucine-zipper (bZIP) transcription factors, is known to bind the RIPE3b/C1-A2 element in the Ins2 enhancer (29, 30), and cMaf-deficient β cells express reduced levels of Ins2 transcripts. Should differentiating MECs follow the same transcriptional hierarchies driving the development of the pancreas and the transcription of Ins2, the absence of Pdx1 or cMaf would be expected to result in a concomitant decrease in ectopic Ins2 transcripts in MECs. Therefore, we quantitated the expression of Ins2 in whole thymi from mice lacking Pdx1 or cMaf. As illustrated in Fig. 6, Ins2 expression was readily detectable, with no significant difference in mRNA levels between the genotypes. Hence, ectopic expression of Ins2 in MECs does not require transcription factors essential for this gene's expression in pancreatic islet β cells.
Fig. 6.
Expression of Ins2 does not depend on the expression of Pdx1 or c-Maf. Quantitative RT-PCR of Ins2 expression from whole thymi derived from wild-type, heterozygous, and knockout mice deficient for Pdx1 at embryonic day 19 and postnatal day 1, and wild-type, heterozygous, and cMaf-deficient mice at embryonic day 19.
Discussion
Our recently published and present data make a strong case that PTA expression in thymic MECs is best portrayed by the “terminal differentiation” model of Aire function; i.e., that more and more Aire-dependent PTAs are expressed as MECs differentiate, culminating in a fully mature population that both is equipped to present antigens (MHC-IIhiCD80hi) and has a broad repertoire of antigens to offer. We have reported that Aire-negative MEClo ultimately give rise to Aire-positive MEChi (17), consistent with and extending recent findings from two other groups (18, 19), and that the latter cells represent a postmitotic end-stage population about to die (17). Here, we demonstrated that Aire-positive MEChi are the class of MECs that transcribe the greatest number and diversity of PTA genes, consistent with earlier speculation and data from the Kyewski group (3, 13). Individual MECs expressed only a subset of PTAs, but there was no indication that the expression profiles of single MECs mimicked transcriptional programs characteristic of extrathymic epithelial cell types. Rather, PTAs appeared to be expressed in a probabilistic manner in individual cells, in no discernable pattern. These data are in overall agreement with the recent report of Derbinski et al. (31). Our study went on to demonstrate that the transcription of PTA genes in MECs, but not in the relevant peripheral tissues, can be monoallelic or biallelic, in a probabilistic manner, again more in tune with the terminal differentiation model. We also showed that the transcription factors required for PTA expression in the thymus differed from those needed for the same transcript in the relevant peripheral tissue. The latter conclusion differs from that of Gillard et al. (32), who found transcription factors involved in extrathymic epithelial lineages to be reduced in Aire−/− MECs but did not test their functional relevance, leaving open the possibility that they are simply components of the Aire-dependant PTA repertoire. Perhaps not surprisingly, then, different transcriptional start sites were used in the thymus and peripherally. The behavior of the ins2 gene, normally expressed in pancreatic β cells, serves nicely to summarize these points: it was expressed in 10–20% of Aire+ MECs, in no particular association with either S100 or csn genes (Fig. 2); its transcription in MECs did not require Pdx1 or cMaf, transcription factors regulating its expression in pancreatic β cells (Fig. 6); and perhaps not surprisingly, then, different start sites were used in the initiation of transcription (Fig. 5).
Stochastic gene expression in prokaryotic or eukaryotic cells has been reported widely in the literature. In model eukaryotic systems such as Saccharomyces cerevisiae, the variability in expression at the single-cell level highly depends on the rate of transcription, which can potentially be explained by changes in upstream events such as slow transitions between promoter states (33). For example, using strains carrying mutations in the TATA box of the PHO5 promoter or in its upstream activating sequence (UAS), Raser and O'Shea (34) found that the highest source of variability stemmed from mutations in the latter, which is thought to influence the rate of promoter activation through recruitment of chromatin-remodeling factors. Slow transitions between active and inactive promoter states in eukaryotic systems have been correlated to the relatively large amount of energy required to generate open and accessible regions of the genome (35). Thus, it is logical that these slow transition states could be an important source of stochastic gene expression in eukaryotes (33). The probabilistic gene expression seen in single MEChi might be due to variations in the accessibility of PTAs and differences in the local concentration of chromatin remodeling factors or of other transcriptional regulators, or of Aire itself.
Also emerging from our results was the fact that Aire-regulated genes often showed monoallelic expression in individual MECs, in contrast to the biallelic pattern of expression found both for the same Aire-dependent genes in peripheral tissues and for the tested Aire-independent genes in MECs. Monoallelic expression of genes can offer the cell a discriminatory advantage, such as in allelic exclusion of T cell receptors, NK cell receptors, or odorant receptors (36–38). Most recently, a series of autosomal genes has been reported to be monoallelically expressed in a random manner; e.g., cytokine expression in T cells and Pax5 (39, 40). Although the exact mechanism by which random monoallelic expression of autosomal genes occurs is unknown, this phenomenon has been associated with asynchronously replicating genes (41) and differences in chromatin modifications (42). The stochastic usage of Aire-dependent PTA gene alleles may reflect a limiting factor necessary for their expression in MECs: either Aire itself or some other transcriptional regulator, or the accessibility of a particular gene or genomic region, could be too low to support efficient transcription on both alleles. Therefore, the probability of expressing a particular PTA from a given allele likely depends on the accessibility of the gene in the single MEC and/or the local concentration of transcription or chromatin remodeling factors.
The biological advantage of partial and probabilistic expression of PTAs by MECs almost certainly lies in ensuring a thymic environment that efficiently supports negative selection of self-reactive thymocytes. Were each MEC to express all PTAs, it is unlikely that many of them would attain a high enough representation within the limited number of MHC molecules displayed at the surface at any one time to promote effective clonal deletion of the relevant TCR specificities. This situation is greatly ameliorated by having the individual MECs each display a random subset of PTAs. An additional factor to consider is the time element: it may be that the PTA repertoire of an individual MEC changes over time, perhaps even oscillating between different expression states. However, the fact that MECs have such a short lifetime, dying within days after Aire expression (17), suggests that the time element may not really be so important.
Methods
For additional details related to these methods, see SI Methods and Tables S1–S5.
Mice.
For single-cell RT-PCR experiments, 3- to 5-week-old B6 mice (Charles River Laboratories) were used. Aire-deficient mice were derived and genotyped as described in ref. 8. Mice used for SNP analysis and sequencing were F1(B6×NOD) derived in our laboratory and F1(B6xDBA) (The Jackson Laboratory). PdxtTA mice were kindly provided by Doug Melton (Harvard University), and were derived and genotyped as described in ref. 43. cMaf-deficient mice were kindly provided by Arun Sharma (Joslin Diabetes Center), and were derived and genotyped as described in ref. 44.
Thymic Epithelial Cells.
Thymic epithelial cells were prepared as described in ref. 8.
Single-Cell Sorting, Reverse Transcription, and PCR.
Thymic MECs were isolated as described in ref. 8. Single-cell sorting and RT-PCR were conducted as described in ref. 45.
Single-Cell Allelic Discrimination and Sequencing.
Single-cell RT-PCR was conducted as described above, and cDNA was split into four independent, parallel single-cell PCRs. Five microliters of the second-round PCR products was visualized on a 2% agarose gel. Those cells containing positive PCR products in three out of four PCRs were purified by using the QIAquick PCR purification kit (Qiagen).
Real-Time PCR.
Total RNA was prepared by using TRIzol (Invitrogen) from fetal whole thymi from Pdx1-tTA or cMaf-deficient mice according to the manufacturer's protocol. cDNA was made from total RNA primed with random hexamers, and real-time PCR using TaqMan was performed as described in ref. 8. Primers and probes are listed in Table S4.
5′-RLM-RACE.
Total cytosolic mouse RNA was prepared by using TRIzol (Invitrogen) from isolated MECs, spleen, salivary glands, pancreas, liver, mammary glands, and stomach according to the manufacturer's protocol. Additional details appear in SI Methods.
Supplementary Material
Acknowledgments.
We thank Jasmine Perez for excellent technical support. This work was supported by National Institutes of Health Grant R01 DK60027 and Young Chair funds (to D.M. and C.B.), and by Joslin's National Institutes of Diabetes and Digestive and Kidney Diseases-funded Diabetes and Endocrinology Research Center core facilities. J.V. and W.B. received support from National Institutes of Health Training Grant T32 DK07260.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808069105/DCSupplemental.
References
- 1.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D. Peripheral-antigen-expressing cells in thymic medulla: Factors in self-tolerance and autoimmunity. Curr Opin Immunol. 1998;10:656–662. doi: 10.1016/s0952-7915(98)80085-x. [DOI] [PubMed] [Google Scholar]
- 3.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 4.Leonard M. Chronic idiopathic hypoparathyroidism with superimposed Addison's disease in a child. J Clin Endocrinol Metab. 1946;6:493–495. doi: 10.1210/jcem-6-7-493. [DOI] [PubMed] [Google Scholar]
- 5.Thorpe E, Handley H. Chronic tetany and chronic mucelial stomatitis in a child aged four and one half years. Am J Dis Child. 1929;28:328–338. [Google Scholar]
- 6.Aaltonen J, et al. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 7.Nagamine K, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 8.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey C, et al. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- 10.Pitkanen J, et al. Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem Biophys Res Commun. 2005;333:944–953. doi: 10.1016/j.bbrc.2005.05.187. [DOI] [PubMed] [Google Scholar]
- 11.Giraud M, et al. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448:934–937. doi: 10.1038/nature06066. [DOI] [PubMed] [Google Scholar]
- 12.Johnnidis JB, et al. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci USA. 2005;102:7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillard GO, Farr AG. Contrasting models of promiscuous gene expression by thymic epithelium. J Exp Med. 2005;202:15–19. doi: 10.1084/jem.20050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farr AG, Dooley JL, Erickson M. Organization of thymic medullary epithelial heterogeneity: Implications for mechanisms of epithelial differentiation. Immunol Rev. 2002;189:20–27. doi: 10.1034/j.1600-065x.2002.18903.x. [DOI] [PubMed] [Google Scholar]
- 16.Gray DH, et al. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood. 2006;108:3777–3785. doi: 10.1182/blood-2006-02-004531. [DOI] [PubMed] [Google Scholar]
- 17.Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med. 2007;204:2521–2528. doi: 10.1084/jem.20070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamazaki Y, et al. Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol. 2007;8:304–311. doi: 10.1038/ni1438. [DOI] [PubMed] [Google Scholar]
- 19.Rossi SW, et al. RANK signals from CD4+3− inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L, Hu-Li J, Paul WE. Probabilistic regulation of IL-4 production in Th2 cells: Accessibility at the Il4 locus. Immunity. 2004;20:193–203. doi: 10.1016/s1074-7613(04)00025-1. [DOI] [PubMed] [Google Scholar]
- 21.Guo L, Hu-Li J, Paul WE. Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity. 2005;23:89–99. doi: 10.1016/j.immuni.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Rhoades KL, et al. Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr Biol. 2000;10:789–792. doi: 10.1016/s0960-9822(00)00565-0. [DOI] [PubMed] [Google Scholar]
- 23.Nacken W, Lekstrom-Himes JA, Sorg C, Manitz MP. Molecular analysis of the mouse S100A9 gene and evidence that the myeloid specific transcription factor C/EBPepsilon is not required for the regulation of the S100A9/A8 gene expression in neutrophils. J Cell Biochem. 2001;80:606–616. [PubMed] [Google Scholar]
- 24.Dike S, et al. The mouse genome: Experimental examination of gene predictions and transcriptional start sites. Genome Res. 2004;14:2424–2429. doi: 10.1101/gr.3158304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois-Lafforgue D, et al. Proinsulin 2 knockout NOD mice: A model for genetic variation of insulin gene expression in type 1 diabetes. Diabetes. 2002;51(Suppl 3):S489–S493. doi: 10.2337/diabetes.51.2007.s489. [DOI] [PubMed] [Google Scholar]
- 26.Kataoka K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem (Tokyo) 2007;141:775–781. doi: 10.1093/jb/mvm105. [DOI] [PubMed] [Google Scholar]
- 27.Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- 28.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura W, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic β-cells. Dev Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olbrot M, Rud J, Moss LG, Sharma A. Identification of β-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci USA. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derbinski J, et al. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci USA. 2008;105:657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillard GO, et al. Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J Immunol. 2007;178:3007–3015. doi: 10.4049/jimmunol.178.5.3007. [DOI] [PubMed] [Google Scholar]
- 33.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: From theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 34.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paldi A. Stochastic gene expression during cell differentiation: Order from disorder? Cell Mol Life Sci. 2003;60:1775–1778. doi: 10.1007/s00018-003-23147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergman Y. Allelic exclusion in B and T lymphopoiesis. Semin Immunol. 1999;11:319–328. doi: 10.1006/smim.1999.0188. [DOI] [PubMed] [Google Scholar]
- 37.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 38.Held W, Roland J, Raulet DH. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- 39.Calado DP, Paixao T, Holmberg D, Haury M. Stochastic monoallelic expression of IL-10 in T cells. J Immunol. 2006;177:5358–5364. doi: 10.4049/jimmunol.177.8.5358. [DOI] [PubMed] [Google Scholar]
- 40.Nutt SL, et al. Independent regulation of the two Pax5 alleles during B-cell development. Nat Genet. 1999;21:390–395. doi: 10.1038/7720. [DOI] [PubMed] [Google Scholar]
- 41.Ensminger AW, Chess A. Coordinated replication timing of monoallelically expressed genes along human autosomes. Hum Mol Genet. 2004;13:651–658. doi: 10.1093/hmg/ddh062. [DOI] [PubMed] [Google Scholar]
- 42.van Rietschoten JG, et al. Differentially methylated alleles in a distinct region of the human interleukin-1α promoter are associated with allele-specific expression of IL-1α in CD4+ T cells. Blood. 2006;108:2143–2149. doi: 10.1182/blood-2006-01-021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holland AM, et al. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci USA. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JI, et al. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong J, et al. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.