Abstract
Interferon regulatory factor (IRF) 4 is a member of the IRF family of transcription factors and plays critical roles in the development of CD4+ T cells into Th2 and Th17 cells. Using the infection model of Nippostrongyrus brasiliensis, we have confirmed the critical roles of IRF-4 in Th2 development in vivo by using IRF-4−/− BALB/c mice. However, naïve IRF-4−/−CD4+ T cells produced Th2 cytokines, including IL-4, IL-5, and IL-10, but not IL-2 or IFN-γ, at levels higher than wild-type BALB/c CD4+ T cells in response to T cell receptor stimulation. In contrast, effector/memory IRF-4−/−CD4+ T cells did not exhibit increased production of Th2 cytokines. Knockdown of IRF-4 expression by using small interfering RNA promoted IL-4 production in naïve CD4+ T cells but inhibited it in effector/memory CD4+ T cells. These results indicate that IRF-4 plays differential roles in the regulation of Th2 cytokine production in naïve CD4+ T cells and effector/memory CD4+ T cells. IRF-4 inhibits Th2 cytokine production in naïve CD4+ T cells, whereas it promotes Th2 cytokine production in effector/memory CD4+ T cells.
Keywords: siRNA, IL-4, Nippostrongyrus brasiliensis
CD4+ T cells play critical roles in the generation of protective immunity against a variety of pathogens by dictating the type of immune response that is effective against each pathogen encountered. The three types of effector CD4+ T cells, Th1, Th2, and Th17, are characterized by their ability to produce signature cytokines IFN-γ, IL-4, and IL-17, respectively (1, 2). Th2 cells produce IL-4, IL-5, and IL-13 and are responsible for humoral immunity and host immune responses against extracellular parasites. Differentiation of helper T cells is determined after encounter of naïve CD4+ T cells with antigen (3, 4). Initiation of Th2 differentiation is potentiated by IL-4 during encounter of naïve CD4+ T cells with antigen, but the early source of IL-4 that is important for Th2 differentiation under physiological conditions is unclear (5). Although innate immune cells such as basophils and natural killer (NK) T cells can produce IL-4, it has been shown that Th2 cells can develop from naïve CD4+ T cells independently of IL-4 produced by non-T cells (6). Naïve CD4+ T cells themselves can produce small amounts of IL-4 after antigen stimulation, which is sufficient for Th2 differentiation under certain conditions (7). Therefore, IL-4 production by naïve CD4+ T cells must be tightly regulated to coordinate differentiation of effector helper CD4+ T cells. Differentiation of helper CD4+ T cells to Th1 or Th2 is genetically controlled, and the BALB/c strain possesses a genetic predisposition toward the development of Th2 cells (8). These strain differences appear to be controlled at several different levels, and the underlying mechanisms are not clearly understood (9–11).
IFN regulatory factors (IRFs) are a family transcription factors that bind to a specific DNA motif known as the IFN-stimulated response element (ISRE) and play critical roles in a variety of immune processes (12). One of the members, IRF-4, is expressed specifically in lymphocytes and macrophage/dendritic cells (13–17). In contrast to other IRF family members, the expression of IRF-4 in lymphocytes is induced by stimulation of the antigen receptor and plays critical roles for the differentiation of naïve lymphocytes to effectors (13, 18). In T cells, IRF-4 plays a critical role in the differentiation of CD4+ T cells to Th2 and Th17 effectors (19–22). However, IRF-4 is dispensable for Th1 development of CD4+ T cells, and IRF-4−/−CD4+ T cells can develop protective immunity during the early phase of Leishmania major infection. In these studies, IRF-4−/− mice of C57BL/6 (B6) genetic background, a Th1-biased strain, were used, and it was not clear whether mice with a Th2-biased genetic background also show defects in Th2 development in the absence of the IRF-4 gene.
To examine the role of IRF-4 in effector T cell development under a Th2-biased genetic background, we have backcrossed IRF-4−/− mice to the BALB/c strain. These mice did not develop Th2 immune responses even under strong Th2-biased conditions. Surprisingly, however, naïve IRF-4−/−CD4+ T cells produced Th2 cytokines at levels much higher than BALB/c wild-type T cells, suggesting that IRF-4 negatively regulates production of IL-4 in naïve CD4+ T cells. Further study showed that IRF-4 plays differential roles in the regulation of Th2 cytokine production by naïve vs. effector/memory CD4+ T cells. IRF4 inhibits IL-4 production in naïve CD4+ T cell, whereas it promotes IL-4 production in effector/memory CD4+ T cells.
Results
IRF-4−/− Mice Are Sensitive to Nippostrongyrus brasiliensis Infection.
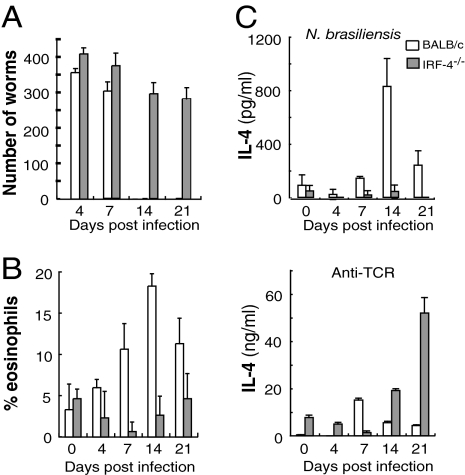
IRF-4 knockout (KO) mice were backcrossed to BALB/c mice (IRF-4−/− mice) to examine the role of IRF-4 in T cell function under a Th2-biased genetic background. We investigated the response of BALB/c and IRF-4−/− mice to infection with N. brasiliensis, which normally induces strong Th2-biased immune responses. In BALB/c mice, expulsion of the adult worms occurred within 2 weeks after infection. In contrast, IRF-4−/− mice maintained similar numbers of intestinal worms for >3 weeks (Fig. 1A). In addition, IRF-4−/− mice did not show any signs of eosinophilia, a hallmark of the Th2 response, during the course of N. brasiliensis infection, whereas BALB/c mice exhibited eosinophilia with a peak at 2 weeks after infection (Fig. 1B). Expulsion of intestinal adult worms is critically dependent on IL-4 and IL-13 produced by T cells (23, 24). CD4+ T cells were prepared from the draining lymph nodes of the infected mice, and their ability to produce IL-4 was determined by ELISA. CD4+ T cells of BALB/c mice produced IL-4 in response to N. brasiliensis antigen, whereas those from IRF-4−/− mice did not (Fig. 1C). The lack of IL-4 production was not caused by a defect in antigen presentation by IRF-4−/− antigen-presenting cells (APCs) during the culture because IRF-4−/−CD4+ T cells did not produce IL-4 when cultured with wild-type BALB/c APCs pulsed with N. brasiliensis antigen (data not shown). In addition, IRF-4−/− mice showed Th1-based protective immune responses against infection with L. major [supporting information (SI) Fig. S1]. Collectively, these studies established that the Th2 response in vivo is critically dependent on IRF-4, even in mice genetically biased to Th2.
Fig. 1.
IRF-4−/− mice are susceptible to N. brasiliensis infection. (A) BALB/c (open bars) and IRF-4−/− mice (filled bars) were infected with N. brasiliensis (500 organisms) s.c. at the base of the tail. Worm burden was analyzed after longitudinal dissection of the small intestine. The data represent the mean ± SD with three mice per group. (B) Eosinophils in the peripheral blood were counted under a microscope. (C) Mesenteric lymph node cells (1 × 105) were collected on the indicated days after infection and were cultured in the presence of N. brasiliensis antigen (Upper) or on plates coated with anti-TCR mAb (Lower) for 48 h. The IL-4 levels in the supernatant were determined by ELISA. Representative results of three independent experiments are shown.
IRF-4−/−CD4+ T Cells Produce High Levels of Th2 Cytokines.
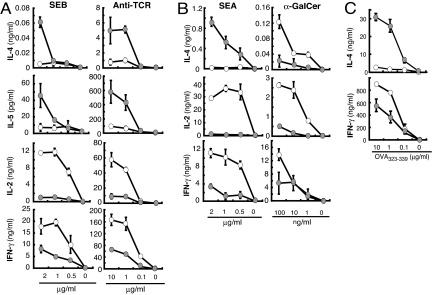
During the course of the study, we evaluated the ability of CD4+ T cells to produce IL-4 in response to anti-T cell receptor (TCR) signals. Unexpectedly, CD4+ T cells from IRF-4−/− mice produced IL-4 at levels higher than that produced by CD4+ T cells from BALB/c mice when stimulated with anti-TCR mAb despite the lack of their antigen-specific IL-4 production (Fig. 1C). Therefore, we examined whether conventional CD4+ T cells expressing α/β TCR that recognize MHC/peptide antigens mediate the high IL-4 responses. CD4+ T cells from BALB/c and IRF-4−/− mice were stimulated with APC pulsed with staphylococcal enterotoxin B (SEB) or with plate-coated anti-TCR mAb, and their ability to produce cytokines was determined (Fig. 2A). IRF-4−/−CD4+ T cells produced IL-2 and IFN-γ at levels lower than BALB/c CD4+ T cells, consistent with previous studies (18, 20, 21). These T cells, however, produced IL-4 and IL-5 at levels much higher than BALB/c CD4+ T cells when stimulated with SEB-pulsed APC or anti-TCR mAb (Fig. 2A). To determine whether conventional CD4+ T cells or NK T cells produce high levels of IL-4 and IL-5, we stimulated these cells with staphylococcal enterotoxin A (SEA), which stimulates T cells expressing Vβ1, 3 10, 11, 12, and 17; or with α-galactocylceramide (α-GalCer), which stimulates Vα14+ NK T cells to determine whether conventional CD4+ T cells or NK T cells produce high levels of IL-4 and IL-5 (Fig. 2B) (25). IRF-4−/−CD4+ T cells produced higher levels of IL-4 and lower levels IL-2 and IFN-γ in response to SEA compared with BALB/c CD4+ T cells. To confirm that IRF-4−/− naïve conventional CD4+ T cells produce higher levels of IL-4, we have backcrossed DO11.10 TCR transgenic mice with IRF-4−/−BALB/c mice and examined the cytokine production of naïve DO11.10 IRF-4−/−CD4+ T cells in response to APC pulsed with OVA323–339 peptide. These T cells produced higher levels of IL-4 and reduced levels of IFN-γ in response to OVA323–339 peptide (Fig. 2C), indicating that conventional IRF-4−/−CD4+ T cells produced IL-4 at high levels in response to TCR stimulation. In contrast, total CD4+ cells from IRF-4−/− mice produced IL-4, IL-2, and IFN-γ at reduced levels in response to α-GalCer (Fig. 2B). The proportion of CD4+ NK T cells (DX5+ cells) in IRF-4−/− mice was lower than BALB/c mice (Fig. S2), partly explaining the reduced IL-4 production of CD4+ cells from IRF-4−/− mice in response to α-GalCer (Fig. 2B). Purified CD4+DX5+ cells from IRF-4−/− mice, however, produced equivalent or higher levels of IL-4 in response to α-GalCer than those from BALB/c mice (Fig. S2), suggesting that IRF-4−/−CD4+DX5+ NK T cells are able to produce sufficient levels of IL-4 in response to α-GalCer.
Fig. 2.
Conventional CD4+ T cells from IRF-4−/− mice produce IL-4 at levels higher than wild-type CD4+ T cells. (A) Splenic CD4+ T cells (1 × 105) from BALB/c (open circles) or IRF-4 −/− (closed circles) mice were cultured with mitomycin C-treated T-depleted spleen cells (5 × 105) pulsed with SEB or on plates coated with anti-TCR Ab for 48 h at the indicated concentrations. The cytokine levels in the supernatant were determined by ELISA. (B) Splenic CD4+ T cells (2 × 105) from BALB/c (open circles) or IRF-4−/− (filled circles) mice were cultured in the presence of mitomycin C-treated T-depleted spleen cells (5 × 105) and SEA or α-GalCer at the indicated concentrations for 48 h. (C) Naïve CD4+ T cells (1 × 105 CD62L+CD4 + T cells) from DO11.10 (open circles) or IRF-4−/− DO11.10 (closed circles) mice were stimulated with mitomycin C-treated T-depleted spleen cells (4 × 105) in the presence of OVA323–339 peptide (0–10 μg/ml) for 48 h. Representative results of three independent experiments are shown.
IL-4 Production by Naïve vs. Effector/Memory IRF-4−/−CD4+ T Cells.
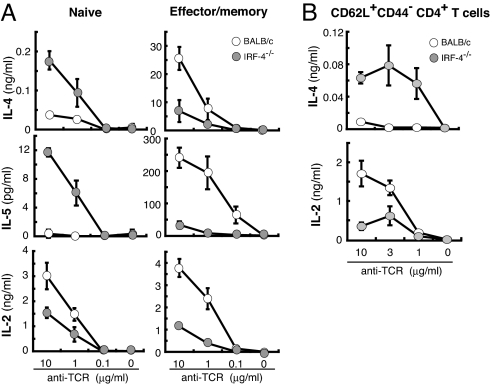
Peripheral CD4+ T cells contain both naïve and effector/memory-type T cells that can be distinguished by their cell surface phenotype such as CD62L. Naïve lymphocytes have higher levels of CD62L expression than effector and effector memory T cells (26). The proportion of CD62L+ cells in CD4+ T cells was not significantly different between BALB/c and IRF-4−/− mice (data not shown). To determine which cell type produces higher levels of Th2 cytokines in IRF-4−/− mice, we prepared naïve (CD62L+) and effector/memory (CD62L−) CD4+ T cells and examined their cytokine production in response to anti-TCR mAb (Fig. 3A). Naïve IRF-4−/−CD4+ T cells produced IL-4 and IL-5 at levels higher than BALB/c naïve CD4+ T cells and IL-2 at reduced levels. Effector/memory CD4+ T cells from IRF-4−/− BALB/c mice, however, produced IL-4, IL-5, and IL-2 at levels lower than BALB/c CD4+ T cells. CD62L+CD4+ T cell population may contain central memory CD4+ T cells (CD62L+CD44+) in addition to naïve CD4+ T cells (CD62L+CD44−). To confirm that naïve CD4+ T cells produce higher levels of IL-4, we purified CD62L+CD44−CD4+ T cells by sorting and cultured in the presence of anti-TCR mAb (Fig. 3B). CD62L+CD44−CD4+ T cells from IRF-4−/− mice produced IL-4 at levels higher than those BALB/c mice and IL-2 at reduced levels. Thus, we concluded that naïve IRF-4−/−CD4+ T cells rather than effector/memory IRF-4−/−CD4+ T cells were responsible for the high levels of Th2 cytokines produced. We also determined the expression of Th2 cytokine mRNAs by naïve CD4+ T cells (Fig. S3). The expression levels of cytokine mRNA were detected basically in parallel to their protein production. Also, up-regulation of GATA3 mRNA expression was observed in naïve IRF-4−/−CD4+ T cells and not in BALB/c CD4+ T cells. Taken together, these results suggest that IRF-4 plays an inhibitory role in the expression of Th2 cytokines in naïve T cells. This inhibitory effect, however, was not seen in effector/memory-type CD4+ T cells.
Fig. 3.
Th2 cytokine production by naïve vs. effector/memory CD4+ T cells is differentially regulated in IRF-4−/− mice. (A) Naïve (CD62L+) or effector/memory (CD62L−) CD4+ T cells (1 × 105 per well) from BALB/c (open circles) or IRF-4−/− (filled circles) mice were purified by sorting and were cultured with plates coated with anti-TCR mAb for 48 h. The percentage of CD62L+ cells and CD62L− cells within BALB/c and IRF-4−/−CD4+ T cells was 97–99% and >99%, respectively. Cytokine levels in the supernatant were determined by ELISA. Representative results of three independent experiments are shown. (B) CD62L+CD44−CD4+ T cells (2 × 105) from BALB/c (open circles) or IRF-4−/− (filled circles) mice were purified by sorting and were cultured on plates coated with anti-TCR mAb for 48 h. The purity of CD62L+CD44−CD4+ T cells was >92%.
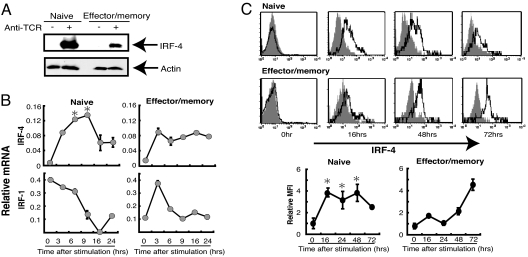
To examine the mechanisms underlying the differential effect of IRF-4 in naïve and effector/memory T cells, we compared the expression levels of IRF-4 in BALB/c CD4+ T cells after stimulation with anti-TCR mAb. Naïve (CD62L+) CD4+ T cells expressed IRF-4 protein at a level significantly higher than effector/memory (CD62L−) CD4+ T cells (Fig. 4A). We examined the kinetics of IRF-4 expression after T cell activation both at the RNA and protein levels (Fig. 4 B and C). The expression of IRF-4 was induced after activation with TCR stimulation in both naïve and effector/memory CD4+ T cells. Compared with effector/memory CD4+ T cells, however, naïve CD4+ T cells showed higher levels of IRF-4 mRNA expression at early hours after TCR stimulation. Similarly, intracellular staining of IRF-4 protein indicated that the expression of IRF-4 was induced at higher levels in naïve CD4+ cells than effector/memory CD4+ cells during the early time point after activation (Fig. 4C). In contrast, the expression of IRF-1 was not induced in naïve CD4+ T cells and was only transiently increased in effector/memory CD4+ T cells (Fig. 4B). These results indicate that the expression pattern of IRF-4 after T cell activation is distinct in naïve and effector/memory CD4+ cells.
Fig. 4.
Induction of IRF-4 expression in naïve and effector/memory CD4+ T cells after TCR stimulation. (A) CD62L+ and CD62L−CD4+ T cells were prepared from BALB/c mice and were cultured on plates coated with or without anti-TCR mAb (1 μg/ml for A and B, 10 μg/ml for C). After culture for 16 h, cells were lysed, separated on 12.5% SDS/PAGE, blotted, and probed with anti-IRF-4 Ab. The membrane was stripped and reprobed with anti-actin Ab. (B) After culture for 0–24 h, total RNA was prepared, and the levels of IRF mRNA were determined by real-time PCR. The mRNA expression was expressed as the ratio of DNA to G3PDH. *, P < 0.05, Mann–Whitney U test between naïve and effector/memory. (C) After culture for 0–72 h, cells were fixed, permeabilized, and stained with anti-IRF-4 Ab, biotin-anti-goat IgG, and streptavidin–phycoerythrin. Gray shadow represents staining of the cells at each time point with biotinylated anti-goat IgG and streptavidin–phycoerythrin only without anti-IRF-4 Ab. Relative mean fluorescent intensity (MFI) of IRF-4 staining was expressed as the ratio of MFI between the staining with and without anti-IRF-4 Ab. Representative results of three independent experiments are shown. *, P < 0.05 between naïve and effector/memory.
Regulation of Th2 Cytokine Production by IRF-4.
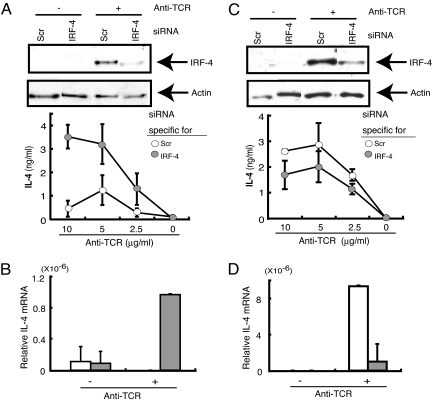
To determine whether the ability of CD4+ T cells to express Th2 cytokines is directly regulated by IRF-4, we reconstituted the IRF-4 gene in IRF-4−/−CD4+ T cells by transfection. CD4+ T cells reconstituted with IRF-4 produced reduced levels of IL-4 and IL-5 in response to anti-TCR mAb (Fig. S4). We next used the siRNA technique to inhibit expression of IRF-4 in CD4+ T cells. Naïve CD4+ T cells were prepared by depletion of CD44high cells from total CD4+ T cells. After transfer of IRF-4 siRNA by electroporation, cells were stimulated with anti-TCR mAb. Naïve CD4+ T cells expressing greatly decreased levels of IRF-4 produced IL-4 at levels much higher than control CD4+ T cells in response to TCR stimulation at both the RNA and protein levels (Fig. 5 A and B). We also evaluated the effect of inhibiting IRF-4 expression in effector/memory CD4+ T cells because these cells normally produce IL-4 at levels higher than IRF-4−/− effector/memory CD4+ T cells. Effector/memory CD4+ T cells were prepared by depletion of CD62L+ cells from total CD4+ T cells, transfected with IRF-4 siRNA, and stimulated with anti-TCR mAb. The production of IL-4 by effector/memory CD4+ T cells was significantly inhibited by IRF-4-specific siRNA at both protein and RNA levels (Fig. 5 C and D). These results suggest that IRF-4 expressed in CD4+ T cells differentially regulates Th2 cytokine production in naïve and effector/memory CD4+ T cells. IRF-4 is inhibitory to IL-4 production in naïve CD4+ T cells and is stimulatory in effector/memory CD4+ T cells.
Fig. 5.
The levels of IRF-4 expression in CD4+ T cells affect the levels of Th2 cytokine production. Naïve CD4+ T cells (CD44low) (A and B) and effector/memory CD4+ T cells (CD62L−) (C and D) from BALB/c mice were transfected with IRF-4-specific (IRF-4) or control (Scr) siRNA (100 pmol). Cells were stimulated with plate-bound anti-TCR mAb for 48 h (A), 60 h (C), or 4 h (B and D). The expression of IRF-4 protein was determined by Western blot analysis by using cells stimulated with anti-TCR mAb at the concentrations of 0 (−) and 2.5 (+) μg/ml (A and C). The blot was stripped and reprobed with anti-actin Ab. The IL-4 levels in the supernatant were determined by ELISA (A and C). After culture for 4 h on plates coated with anti-TCR mAb (3 μg/ml), RNA was prepared, and the levels of IL-4 were determined by real-time PCR (B and D).
Discussion
We have shown a complete lack of Th2-type immune responses in vivo in IRF-4−/− mice of BALB/c genetic background. IRF-4−/− mice were unable to expel N. brasiliensis in vivo because of the lack of Th2-type effector cells. These results are consist with our previous reports using IRF-4 KO mice on a B6 background, in which we and others showed in vitro that Th2 responses of IRF4−/−CD4+ B6 T cells were impaired (19–21). Rengarajan et al. (19) described that IRF-4 physically interacts with NFATc2 and enhances transcriptional activation of the IL-4 promoter. In human studies, Hu et al. (27) reported that stable expression of IRF-4 in the Jurkat human T cell line led to production of Th2 cytokines and that IRF-4 binds the IL-4 promoter element triggering the transcription of IL-4. These studies used immortalized cells, which may represent effector/memory cell types, to evaluate the effect of IRF-4 on IL-4 production, and they are consistent with our observation that IRF-4−/−CD4+ T cells are defective in Th2 development and that IL-4 production by effector/memory IRF-4−/−CD4+ T cells is much lower than BALB/c CD4+ T cells. This work, however, has added a finding that IRF-4 has an inhibitory effect on the production of Th2 cytokines by naïve CD4+ T cells in response to TCR stimulation. These results are seemingly paradoxical but are consistent with the results of IRF-4 knockdown experiments using siRNA in naïve vs. effector/memory type CD4+ T cells; the inhibition of IRF-4 expression in naïve CD4+ T cells conferred the ability to produce IL-4 on naïve CD4+ T cells, whereas inhibition of IRF-4 expression in effector/memory CD4+ T cells down-regulated IL-4 expression.
Studies on the function of IRF-4 indicated that it could function as either a transcriptional activator or repressor depending on the context of the DNA-binding sequences and/or protein-interacting partners (14, 19, 28–31). IRF-4 contains an amino-terminal DNA-binding domain and a carboxyl-terminal IRF association domain that can interact with a variety of proteins, including IRFs. IRF-4 interacts with Ets family member PU.1, transcription factor E47, Stat6, Bcl-6 or NFATc2 to synergistically enhance transcriptional activity of a variety of genes including the Ig light chain gene in B cells and IL-4 in T cells (14, 19, 28, 29). However, IRF-4 has been shown to repress gene activation induced by other IRFs and does not appear to require association with other proteins (30, 31). We have shown that IRF-4 inhibits IRF-1-dependent transcription of genes, including TRAIL, and that the IRF association domain of IRF-4 was dispensable for this inhibition, suggesting that IRF-4 acts as a natural antagonist of IRF-1 and inhibits its transactivation (31). There are at least three IRF-binding sites located in the IL-4 promoter region, some of which correspond with sequences that negatively regulate IL-4 promoter activity (32, 33). Elser et al. (33) showed that IRF-1 and IRF-2 bind to these sites and inhibit IL-4 promoter activity and that naïve IRF-1−/−CD4+ T cells produce IL-4 at levels higher than wild type. Because IRF-1 and IRF-2 are induced by IFN-γ, these IRFs were believed to mediate the inhibitory role of IFN-γ on expression of the IL-4 gene. Our study shows that naïve IRF-4−/−CD4+ T cells produce higher levels of IL-4 upon TCR stimulation. Therefore, both IRF-4 and IRF-1 appear to be required for optimum inhibition of IL-4 production in naïve CD4+ T cells. Unlike IRF-1 and IRF-2, which are constitutively expressed in T cells, IRF-4 expression is induced by antigen receptor-mediated stimuli (Fig. 4B) (13). Thus, IRF-1 and IRF-4 may cooperate temporally to regulate the induction of IL-4 in naïve CD4+ T cells tightly. Taken together, these studies suggest that IRF-4 plays a dual function in the production of IL-4 in CD4+ T cells; IRF-4 promotes IL-4 production in effector/memory CD4+ T cells in association with NFATc2 and inhibits IL-4 transcription in naïve CD4+ T cells. The precise mechanisms underlying the inhibition of IL-4 transcription by IRF-4 remain to be determined.
The question, then, is how the same transcription factor can have two opposing functions in naïve vs. effector/memory CD4+ T cells. One possibility lies in the differential expression levels of IRF-4 in naïve and effector/memory-type CD4+ T cells. IRF-4 expression is induced after activation of T cells through TCR signaling and reaches higher levels in naïve CD4+ T cells than in effector/memory CD4+ T cells. The early and strong induction of IRF-4 may be required for silencing of IL-4 expression. Second, IRF-4 might affect the signaling of TCR or threshold of T cell activation, which ultimately dictate the differential production of cytokines. This possibility may also explain the lack of Th2 differentiation in IRF-4 KO mice because the strength of TCR signaling appear to affect the generation of Th1/Th2 cells (34). We have reported that IRF-4 regulates the TLR signaling pathway, including the activation of MAP kinases and NF-κB in macrophages (17), and it is possible that IRF-4 affects TCR signaling in T cells. Third, IRF-4 might interact with partners distinct from NFAT, which dictates the repressive function of IRF-4 on the IL-4 promoter in naïve CD4+ T cells. However, IL-4 expression is controlled not only by the IL-4 promoter, but by several other regulatory elements, including enhancers, silencers, and locus control regions, which become targets of transcription factors and chromatin-remodeling factors (3, 4). There are IRF-binding sites within the silencer region (35). Therefore, IRF-4 may affect not only promoter activity of the target genes, but also the chromatin environment of the target genes in regulating gene expression (36). Our study indicates that IRF-4 is involved in regulating the expression of the majority of Th2 cytokines, including IL-4 and IL-5, supporting the view that IRF-4 is involved in the coordinated expression of Th2 cytokines through influences on chromatin structure. Control of Th2 cytokine expression is brought about by the coordinated actions of IRF-4 and a variety of transcriptional elements and chromatin structure influences.
Taken together, our findings suggested that IRF-4 plays dual roles in Th2 cytokine production in CD4+ T cells. IRF-4 is a negative regulator of Th2 cytokines during the early activation phase of naïve CD4+ T cells, whereas it promotes Th2 development and Th2 cytokine production in effector/memory CD4+ T cells. Th2 T cells are critical for protection against extracellular parasites and for the pathogenesis of allergic immune responses. Our work reveals critical dual roles of IRF-4 in the regulation of Th2 function.
Materials and Methods
Mice and N. brasiliensis.
IRF-4 KO mice were initially mated to C57BL/6 mice as described (17, 21). IRF-4 KO mice of BALB/c background (IRF-4−/− mice) were generated by backcrossing IRF-4 KO mice onto BALB/c for 10 generations and were maintained by intercrossing. The genotype was determined by PCR as described in ref. 21. DO11.10 mice, provided by M. Kubo (Riken, Yokohama, Japan), were crossed with IRF-4−/− mice to generate IRF-4−/− DO11.10 mice. These mice were maintained by intercrossing in the Laboratory Animal Center for Animal Research at Nagasaki University. C57BL/6 (B6) and BALB/c mice were purchased from Japan SLC Inc. Mice were used at 6–8 weeks old. All animal experiments were conducted with approval from the Nagasaki University Institutional Animal Care Committee. N. brasiliensis was provided by K. Ishiwata (Jikei University School of Medicine, Tokyo, Japan). Third-stage larvae (L3) of N. brasiliensis were injected into mice s.c. (500 per mouse) at the base of tail as described in ref. 37. Worm burden was analyzed after longitudinal dissection of the small intestine. Blood smear was stained by using Diff-Quik (Sysmex), and eosinophils were counted under a microscope. N. brasiliensis antigens were prepared by freezing and thawing.
Infection with L. major is discussed in SI Methods.
Cell Culture, ELISA, and Flow Cytometry.
Mesenteric lymph node cells (1 × 105) were cultured in the presence of N. brasiliensis antigen. In other T cell responses, lymphocytes were collected from a mixture of lymph nodes (mesenteric and inguinal) and spleen. CD4+ T cells (>98%) were prepared by using anti-CD4 IMag (BD Biosciences). CD62L+CD4+ T cells were purified by sorting (purity, 97≈99%) by using FACSAria (BD Biosciences) (Fig. 3A) or separated by using the CD4+CD62L+ isolation kit (purity, 70–80%) (Miltenyi Biotec) (Fig. 4, 5). CD62L−CD4+ T cells were purified by sorting (purity, >99%, Fig. 3A) or were prepared by treating the negative fraction of CD4+CD62L+ isolation kit with complement at 37°C for 30 min to deplete the remaining CD62L+ cells (purity, 93–97%, Figs. 4 and 5). T cells were stimulated with plate-bound anti-TCRβ mAb (H57) or mitomycin C-treated T-depleted spleen cells pulsed with α-GalCer (KRN7000, Kirin Brewery), SEA (Toxin Technology), or SEB. Supernatant was collected 48 h after stimulation, and the levels of cytokines in the supernatants were determined by sandwich ELISA. IL-2, IL-4, and IFN-γ levels were determined as described (17, 21). IL-5 levels were determined by an ELISA kit (R&D Systems). We did not find any significant differences in the cytokine profiles between CD62L+CD4+ or CD62L−CD4+ T cells that were purified by sorting and those purified by CD4+CD62L+ isolation kit (data not shown).
For intracellular staining of IRF-4, CD62L+CD4+ T cells and CD62L−CD4+ T cells were purified by using a CD4+CD62L+ isolation kit, stimulated with plate-coated anti-TCR mAb, fixed, and permeabilized by using Cytofix/Cytoperm kits (BD PharMingen), and were stained with anti-IRF-4 Ab (Santa-Cruz Biotechnology), biotin–anti-goat IgG Ab, and phycoerythrin–streptavidin. Cells were analyzed by using FACScan (BD Biosciences) (Fig. 4C).
Real-Time PCR.
RNA was prepared from cells by using Isogen (Nippon Gene). Total RNA (500 ng) was reverse-transcribed to cDNA by using random hexamers, and real-time PCR was performed as described in ref. 17. The mRNA expression was determined as the ratio of each DNA to glucose-3-phosphate dehydrogenase. The sequence of primers for GATA3 and Tbet was described in ref. 21. The sequences of other primer pairs are shown in Table S1.
Western Blotting.
Cells were washed and resuspended in sample buffer. The lysate was size-fractionated on 12.5% SDS/PAGE and transferred to a PVDF membrane. The blot was blocked with 5% milk TBS–Tween, washed twice with PBS–Tween, and incubated with anti-IRF-4 Ab (Santa Cruz Biotechnology). The membrane was incubated with horseradish peroxidase–anti-goat Ig Ab (MBL), washed, and visualized by using ECL reagent (Amersham Pharmacia). The same blot was stripped and reprobed with anti-actin Ab (Sigma).
Transfection and RNA Interference Assay.
Full-length mouse IRF-4 cDNA was cloned into pcDNA3 (Invitrogen). CD4+ T cells were prepared by using CD4+ T cell isolation kit (Miltenyi Biotech) and AutoMACS, and were transfected with the plasmid DNA (30 μg) by using a Nucleofector apparatus (Amaxa) in a 2.0-mm electroporation cuvette according to the protocol X-01. Three hours later, cells (5 × 105) were washed and seeded in 96-well flat-bottom plates coated with anti-TCR Ab and cultured for 48 h.
Double-stranded RNA for IRF-4 and control (22) were purchased from Takara Bio. CD4+ T cells were prepared by using the CD4+ T cells isolation kit (Miltenyi Biotec). Naïve and effector/memory CD4+ T cells were prepared from these cells by negative selection by using biotin–anti-CD44 mAb plus streptavidin–microbeads and FITC–anti-CD62L plus anti-FITC microbeads, respectively, followed by AutoMACS. Cells were transfected with FITC-labeled dsRNA (100 pmol) in a 2.0-mm electroporation cuvette according to the protocol X-01 by using a Nucleofector apparatus, and were cultured on plates coated with anti-TCR mAb for 4–48 h. The proportion of cells that incorporated the dsRNA was ≈97% as assessed by flow cytometry.
Statistics.
Significance levels were determined by the Mann–Whitney U test for unpaired observations. Results were considered significant when P < 0.05.
Supplementary Material
Acknowledgments.
We thank M. Ueda, K. Kimura, T. Ikeda, and M. Yoshida for excellent technical assistance. We also thank Dr. K. Nagai for discussion and help, Dr. K. Ishiwata for N. brasiliensis, and Kirin Brewery for α-GalCer. This work was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture, Japan, by the 21st Century Center of Excellence program, and by the president's discretionary fund of Nagasaki University, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803171105/DCSupplemental.
References
- 1.Mosmann T-R, Coffman R-L. Th1 and Th2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 2.Weaver C-T, Hatton R-D, Mangan P-R, Harrington L-E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 3.Ansel K-M, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and IL4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 4.Lee G-R, Kim S-T, Spilianakis C-G, Fields P-E, Flavell RA. T helper cell differentiation: Regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.O'Garra A. Cytokines induce the development of functionally heterogeneous T helper cell subsets. Immunity. 1998;8:275–283. doi: 10.1016/s1074-7613(00)80533-6. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz J, et al. Induction of interleukin 4 (IL-4) expression in T helper (Th) cells is not dependent on IL-4 from non-Th cells. J Exp Med. 1994;179:1349–1353. doi: 10.1084/jem.179.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noben-Trauth N, Hu-Li J, Paul WE. Conventional, naïve CD4+ T cells provide an initial source of IL-4 during Th2 differentiation. J Immunol. 2000;165:3620–3625. doi: 10.4049/jimmunol.165.7.3620. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh C-S, Macatonia S-E, O'Garra A, Murphy K-M. T cell genetic background determines default T helper phenotype development in vitro. J Exp Med. 1995;181:713–721. doi: 10.1084/jem.181.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guler M-L, Jacobson N-G, Gubler U, Murphy K-M. T cell genetic background determines maintenance of IL-12 signaling: Effects on BALB/c and B10. D2 T helper cell type 1 phenotype development. J Immunol. 1997;159:1767–1774. [PubMed] [Google Scholar]
- 10.Bix M, Wang Z-E, Thiel B, Schork N-J, Locksley R-M. Genetic regulation of commitment to interleukin 4 production by a CD4+ T cell-intrinsic mechanism. J Exp Med. 1998;188:2289–2299. doi: 10.1084/jem.188.12.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Launois P, et al. IL-4 rapidly produced by Vβ4Vα8 CD4+ T cells instructs Th2 development and susceptibility to. Leishmania major in BALB/c mice. Immunity. 1997;6:541–549. doi: 10.1016/s1074-7613(00)80342-8. [DOI] [PubMed] [Google Scholar]
- 12.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 13.Matsuyama T, et al. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenbeis C-F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 15.Marecki S, Atchison M-L, Fenton M-J. Differential expression and distinct functions of IFN regulatory factor 4 and IFN consensus sequence-binding protein in macrophages. J Immunol. 1999;163:2713–2722. [PubMed] [Google Scholar]
- 16.Suzuki S, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8α− dendritic cell development. Proc Natl Acad Sci USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honma K, et al. Interferon regulatory factor 4 negatively regulates the production of proinflammatory cytokines by macrophages in response to LPS. Proc Natl Acad Sci USA. 2005;102:16001–16006. doi: 10.1073/pnas.0504226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittrucker H-W, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. [PubMed] [Google Scholar]
- 19.Rengarajan J, et al. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med. 2002;195:1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohoff M, et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci USA. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tominaga N, et al. Development of Th1 and not Th2 immune responses in mice lacking IFN-regulatory factor-4. Int Immunol. 2003;15:1–10. doi: 10.1093/intimm/dxg001. [DOI] [PubMed] [Google Scholar]
- 22.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 23.Finkelman F-D, et al. Interleukin-4- and interleukin-13-mediated host protection against intestinal nematode parasites. Immunol Rev. 2004;201:139–155. doi: 10.1111/j.0105-2896.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao A, et al. Dependence of IL-4, IL-13, and nematode-induced alterations in murine small intestinal smooth muscle contractility on Stat6 and enteric nerves. J Immunol. 2003;171:948–954. doi: 10.4049/jimmunol.171.2.948. [DOI] [PubMed] [Google Scholar]
- 25.Kawano T, et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 26.Seder R-A, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 27.Hu C-M, Jang S-Y, Fanzo J-C, Pernis AB. Modulation of T cell cytokine production by interferon regulatory factor-4. J Biol Chem. 2002;277:49238–49246. doi: 10.1074/jbc.M205895200. [DOI] [PubMed] [Google Scholar]
- 28.Gupta S, Jiang M, Pernis A-B. IFN-α activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J Immunol. 1999;163:3834–3841. [PubMed] [Google Scholar]
- 29.Nagulapalli S, Atchison M-L. Transcription factor Pip can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol Cell Biol. 1998;18:4639–4650. doi: 10.1128/mcb.18.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamagata T, et al. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/mcb.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida K, et al. Active repression of IFN regulatory factor-1-mediated transactivation by IFN regulatory factor-4. Int Immunol. 2005;17:1463–1471. doi: 10.1093/intimm/dxh324. [DOI] [PubMed] [Google Scholar]
- 32.Li-Weber M, Eder A, Krafft-Czepa H, Krammer P-H. T cell-specific negative regulation of transcription of the human cytokine IL-4. J Immunol. 1992;148:1913–1918. [PubMed] [Google Scholar]
- 33.Elser B, et al. IFN-γ represses IL-4 expression via IRF-1 and IRF-2. Immunity. 2002;17:703–712. doi: 10.1016/s1074-7613(02)00471-5. [DOI] [PubMed] [Google Scholar]
- 34.Constant S, Pfeiffer C, Woodard A, Pasqualini T, Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naïve CD4+ T cells. J Exp Med. 1995;182:1591–1596. doi: 10.1084/jem.182.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubo M, et al. T cell subset-specific expression of the IL-4 gene is regulated by a silencer element and STAT6. EMBO J. 1997;16:4007–4020. doi: 10.1093/emboj/16.13.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozato K, Tailor P, Kubota T. The interferon regulatory factor family in host defense: Mechanism of action. J Biol Chem. 2007;282:20065–20069. doi: 10.1074/jbc.R700003200. [DOI] [PubMed] [Google Scholar]
- 37.Ishiwata K, Nakao H, Nakamura-Uchiyama F, Nawa Y. Immune-mediated damage is not essential for the expulsion of. Nippostrongyrus brasiliensis adult worms from the small intestine of mice. Parasite Immunol. 2002;24:381–386. doi: 10.1046/j.1365-3024.2002.00472.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.