Abstract
Current sequence databases now contain numerous whole genome sequences of pathogenic bacteria. However, many of the predicted genes lack any functional annotation. We describe an assumption-free approach, Rapid Virulence Annotation (RVA), for the high-throughput parallel screening of genomic libraries against four different taxa: insects, nematodes, amoeba, and mammalian macrophages. These hosts represent different aspects of both the vertebrate and invertebrate immune system. Here, we apply RVA to the emerging human pathogen Photorhabdus asymbiotica using “gain of toxicity” assays of recombinant Escherichia coli clones. We describe a wealth of potential virulence loci and attribute biological function to several putative genomic islands, which may then be further characterized using conventional molecular techniques. The application of RVA to other pathogen genomes promises to ascribe biological function to otherwise uncharacterized virulence genes.
Keywords: bacteria, pathogenomics, screening, toxins, entomopathogenic
The growing speed with which the genomes of bacteria can be sequenced is producing an ever expanding knowledge gap between sequence data and its functional annotation. In the case of bacterial pathogens, the identification of virulence factors has typically relied on genetic knock-out and demonstration that virulence is attenuated in a suitable animal model. This approach is not only time consuming and expensive but also, in the case of the use of vertebrate models, ethically debatable. Here, we demonstrate the power of using parallel screens for the identification of virulence genes using three invertebrate infection models and mammalian macrophages in tissue culture, collectively termed Rapid Virulence Annotation (RVA). RVA utility relies upon similarities between the immune responses of vertebrates and invertebrates. The innate immune systems of insects and mammals share many common features, both mechanistically and genetically (1, 2). In addition, much of the basic cellular machinery and pathways that bacterial pathogens can subvert during infection is also well conserved among eukaryotes, presenting common targets. This suggests that virulence mechanisms used by pathogens of mammals may work against invertebrates and vice versa. Indeed, we suggest many virulence strategies evolved initially to combat invertebrates have subsequently been redeployed against vertebrates (3). To test these predictions, we conducted an RVA-based screen on a recently emerging pathogen, Photorhabdus asymbiotica, which is a pathogen of both insects and man (4) and causes invasive soft tissue and disseminated bacteraemic infections (5, 6). Photorhabdus are members of the Enterobacteriaceae that live in association with soil dwelling entomopathogenic Heterorhabditid nematodes and that invade and kill insects. Infective juvenile nematodes seek out and penetrate the insect prey, regurgitating a small number of Photorhabdus cells that evade the immune system, kill the insect, and then bio-convert the tissues into more bacteria. The worm feeds off the bacteria and reproduces until the insect resource is exhausted, whereupon they repackage the bacteria and leave in search of new prey (7). The requirement to overcome the insect immune system and to keep the insect cadaver free of saprophytic organisms in the soil means that all Photorhabdus produce a range of bioactive molecules including immune inhibitors, toxins, and powerful antimicrobials. The fully annotated genome sequence of the insect-only pathogen Photorhabdus luminescens strain TT01 is available (8) and the genome sequence of the clinical isolate Photorhabdus asymbiotica ATCC43949 is almost completed (http://www.sanger.ac.uk/Projects/P_asymbiotica/). The availability of the two genomes allows us to correlate the RVA data with regions unique to the human pathogenic P. asymbiotica.
For RVA analysis we sheared the P. asymbiotica ATCC43949 genome and cloned it into recombinant Escherichia coli. The library, covering 91.4% of the estimated 5.0 Mb genome with an average insert size of 37 kb, was arrayed into 16 96-well plates. All 1,536 clones were sequenced at both ends and end-sequences were assembled onto the genomic scaffold. This cosmid library was screened for gain of toxicity (GOT) assays against the nematode Caenorhabditis elegans (nGOT), serving as an oral route model; the single-cell protozoa Acanthamoeba polyphaga (aGOT), used as a phagocytosis model; and two caterpillar models (iGOT), the Tobacco hornworm Manduca sexta and the Waxmoth Galleria mellonella, both of which represent the more complex insect immune systems. Numerous examples of the use of invertebrates as model hosts can be found in the literature (1, 9–12). Finally, we used the mouse BALB/c macrophage cell line J774–2 (mGOT) to represent the phagocytic component of the vertebrate immune system. The use of GOT studies in E. coli, in which the model host is challenged with individual clones, has the advantage over chromosomal mutagenesis of “unmasking” any virulence factors that would otherwise be hidden due to toxin redundancy or the presence of potent dominant toxins. To define the virulence-related regions (RVA regions), the end sequences of cosmids showing an effect were assembled onto a genomic scaffold and regions of minimum genetic overlap within these clusters were identified, often leading to the identification of specific candidate ORFs or operons. Cross-comparison of genomic regions identified in different assays also gives us an indication of the specificity of the virulence factors identified.
Results
The application of RVA to P. asymbiotica gave a high detection rate of candidate gene clusters, encoding virulence factors, which is a reflection of both the sensitivity of RVA method and also the high level of redundancy in encoded pathogenesis factors in Photorhabdus bacteria (7, 8, 13). The RVA regions were aligned against the current assembly of the P. asymbiotica genome (Fig. 1), each region representing a cluster of cosmids identified either in a single screen (with a minimum of two overlapping cosmids) or via a number of parallel screens in combination (Fig. 2). The complete RVA dataset and more detailed diagrammatic summaries of the 21 RVA regions identified are presented in Table 1 and in supporting information (SI) Materials and Methods, Figs. S1–S25, and Tables S1 and S2. Figs. S2–S22 show correlations between gene clusters and the effects on the mammalian and invertebrate model targets. For brevity, here we will only discuss specific examples of the different classes of virulence factors detected, illustrating the robust nature of RVA-based screening.
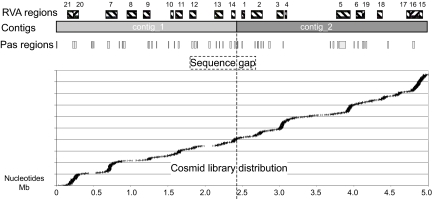
Fig. 1.
RVA functional genomics map for P. asymbiotica. Boxes on the top layer represent the RVA regions (see Figs. S2–S22). The two large contigs of the P. asymbiotica ongoing genome sequencing project are shown. “Pas” regions contain genes that do not identify homologues in P. luminescens TT01 (at 75% identity level). The lower panel represents the actual genomic locations of the cosmids from the library analyzed by RVA screen. The dotted line represents a sequencing gap, however, the order and orientation of the two large contigs is confirmed by the correct end sequence alignment of five cosmids across this region. Note that a sequence gap of unknown size also exists so the other ends of the contigs are not contiguous.
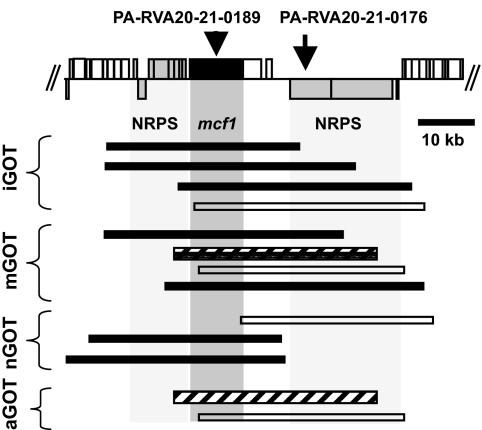
Fig. 2.
Biologically active cosmids in the RVA20 region. Boxes above and below the top central line represent ORFs on the forward and reverse strands. RVA positive cosmids are represented below as bars. Hatched bars represent cosmids containing only intact mcf1. Black bars represent cosmids containing intact mcf1 and at least one of the up- or downstream NRPS clusters. Open bars represent cosmids encoding the NRPS downstream of mcf1 but not the intact gene.
Table 1.
Regions of the P. asymbiotica ATCC43949 genome identified by RVA analysis
| RVA region | Accession number (locus tag range) | Assay | Gene clusters and predicted function |
|---|---|---|---|
| RVA1 | FM211043 (PA-RVA1–4467-4422) | iGOT nGOT | Type 3 secretion system genes and exoU effector |
| RVA2 | FM211044 (PA-RVA2–4341-4267) | iGOT mGOT | Hemolysin and PKS modules |
| RVA3 | FM211045 (PA-RVA3–4024-3964) | iGOT mGOT nGOT aGOT | Hemagglutinin-like gene and two NRPS clusters |
| RVA4 | FM211046 (PA-RVA4–3953-3929) | mGOT | xaxAB homologues and hemagglutinin-like gene |
| RVA5 | FM211047 (PA-RVA5–3244-3168) | iGOT mGOT nGOT aGOT | PVClumt, evp operon and hemagglutinin |
| RVA6 | FM211048 (PA-RVA6–3081-3019) | iGOT nGOT | Fimbrial operon and Pdl-GI-1 island |
| RVA7 | FM211049 (PA-RVA7–0626-0673) | iGOT mGOT nGOT aGOT | Invasin and unknown genes |
| RVA8 | FM211050 (PA-RVA8–2849-2974) | nGOT | Unknown |
| RVA9 | FM211051 (PA-RVA9–1890-1949) | iGOT mGOT aGOT | Prophage |
| RVA10 | FM211052 (PA-RVA10–2214-2246) | mGOT aGOT | Fimbrial operon |
| RVA11 | FM211053 (PA-RVA11–2280-2352) | iGOT mGOT nGOT aGOT | Hemagglutinin and type VI secretion system |
| RVA12 | FM211054 (PA-RVA12–2456-2515) | nGOT mGOT | Unknown |
| RVA13 | FM211055 (PA-RVA13–1315-1206) | iGOT nGOT mGOT | Pdl-GI-4 and ast enterotoxin homologue |
| RVA14 | FM211056 (PA-RVA14–1110-1068) | iGOT aGOT | Hemagglutinin-like and NRPS cluster |
| RVA15 | FM211057 (PA-RVA15_17–0915-0951) | iGOT aGOT | rtxA homologue, NRPS and evp-like operon |
| RVA16 | FM211057 (PA-RVA15_17–0951-0992) | iGOT mGOT nGOT aGOT | vgrG, rtxA homologues and NRPS |
| RVA17 | FM211057 (PA-RVA15_17–1004-1042) | iGOT nGOT mGOT | kdp operon and a perforin-like gene |
| RVA18 | FM211058 (PA-RVA18–1596-1632) | iGOT mGOT | NRPS and PKS-like gene clusters |
| RVA19 | FM211059 (PA-RVA19–1780-1813) | iGOT | Pdl-GI-2 and putative virulence factor mviN |
| RVA20 | FM211060 (PA-RVA20_21–0210-0176) | iGOT mGOT nGOT aGOT | Two NRPS clusters and mcf1 cytotoxin |
| RVA21 | FM211060 (PA-RVA20_21–0176-0072) | iGOT mGOT aGOT | PVCpnf and hemagglutinin-like gene |
Illustrations of this information can be seen in Figs. S2–S22. Full annotation of these regions can be found in the EMBL entries indicated by accession numbers.
Re-Identification of Known Virulence Factors.
The effectiveness of RVA is confirmed by the reidentification of previously known virulence factors, also providing an excellent “internal” positive control. Thus, as anticipated, RVA detected mcf1 (RVA20) that encodes a dominant insecticidal toxin (14) and also the phage-related toxin delivery system PVCpnf (RVA21) (15). Some cosmids containing part or the full PVCpnf cluster, detected in iGOT, aGOT, and mGOT assays, also contain a hemagglutinin-like gene. However, one iGOT cosmid (Fig. S22) carries a complete copy of PVCpnf along with a truncated nonribosomal peptide synthase (NRPS), indicating that PVCpnf alone exhibits toxicity to insects as previously demonstrated (15). Cosmids containing mcf1 were identified in all types of parallel screens, consistent with the ability of Mcf1 to cause apoptosis in both insect and mammalian tissues (16). Fig. 2 illustrates how a detailed examination of the cosmid locations relative to the genome can reveal the contribution of different gene clusters to toxicity. In several of the mcf1-containing cosmids, the further involvement of tightly linked upstream and downstream NRPS genes in the nGOT and iGOT screens could not be ruled out. Conversely, several cosmids were identified in the mGOT and aGOT assays, which contained only intact mcf1 and one cosmid from each assay that contained only the downstream NRPS cluster. Cosmid 3AG4, carrying only mcf1 (identified by aGOT), was tested against the other models resulting in toxic phenotypes, severe feeding delay in the nematodes, insect death at 24 h, and an exhibition of the typical “floppy” phenotype consistent with intoxication by Mcf1 (14).
Identification of Virulence Factor Homologues.
Further confidence in RVA comes from the detection of homologues of virulence factors from other organisms. A “type VI” virulence secretion system described in Vibrio cholerae V52 is proposed to provide a target for vaccines and therapeutic agents (17). The P. asymbiotica RVA screen identified a highly homologous gene cluster (RVA11) detected in the iGOT assay that is 46–90% similar at the predicted amino acid level (Figs. S12 and S23). Indeed, subsequent work has confirmed the importance of a homologous operon in Burkholderia mallei (18). Interestingly, cosmids toxic to macrophages (mGOT) also span an adjacent region that contains the putative vgrG toxin unique to the human pathogen P. asymbiotica and absent from P. luminescens, potentially representing a genomic island specialized for mammalian pathogenicity. Further, an RVA region showing activity in all assays (locus tag PA-RVA5–3205 to 3199, Fig. S6) is homologous (48–72% similar at the amino acid level) to the type-VI like evp operon involved in virulence in the fish pathogen Edwardsiella tarda (19, 20).
Functional Annotation of Secondary Metabolite Gene Clusters.
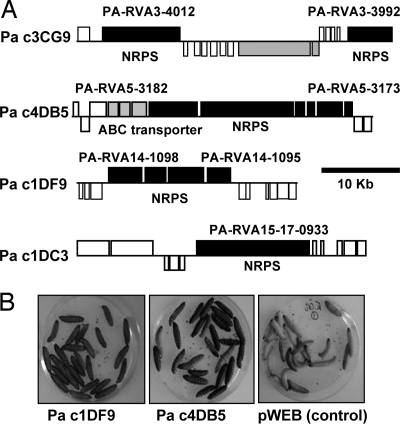
It is notoriously difficult to ascribe biological function to gene clusters responsible for the synthesis of secondary metabolites such as polyketide synthesis (PKS) and non-ribosomal peptide synthesis (NRPS) enzyme complexes. Like many pathogens, Photorhabdus dedicates a large amount of coding sequence to the manufacture of small molecules but their biological roles and structures remain unknown. RVA identified a number of regions predicted to encode NRPS/PKS-like complexes and Fig. 3A illustrates four examples of identified cosmids that exhibit a range of activities including the ability of the recombinant E. coli to kill G. mellonella upon injection (Fig. 3B). HPLC/MS analysis of preparations from cosmids 4DB5 (RVA5) and 1DF9 (RVA14) identified 265.4/468.4 and 586.4 as specific [M+H]+ masses for the respective molecules, which were not present in the E. coli control (Fig. S24). Two compounds were identified from cosmid 4DB5 (RVA5) encoding a gene cluster with high similarity to the yersiniabactin biosynthesis genes. The postulated sum formula C12H12ON2S2 is identical to the sum formula of ulbactin E, a short derivative of yersiniabactin, which has been described from a marine Alteromonas strain (21). The second compound C20H25O4N3S3 might represent a desmethyl-derivative of yersiniabactin (C21H27O4N3S3) that has not been described in the literature yet. The identified sum formula of C32H51O5N5, from 1DF9 cosmid (RVA14), fits well with the structure of a cyclic pentapeptide. Interestingly, an identical sum formula is found in sansalvamide A peptides (22–24), synthetic derivatives of the depsipeptide natural product sansalvamide A that were isolated from a marine fungus and showed potent anti-cancer activity (25). Isolation of all identified compounds for detailed structure elucidation is currently underway. Preliminary structural predictions of other secondary metabolites identified by RVA are presented in Table S1. One example is the predicted compound encoded by RVA 18 (Fig. S19) similar to a new antitumor antibiotic: glidobactin from Polyangium brachysporum (26). Two PKS clusters and one NRPS gene cluster were also encoded in mGOT RVA regions, suggesting that secondary metabolites produced from these operons could have potential roles in human pathogenicity. The rapid identification of new biological activities from these secondary metabolite synthetic genes suggests that RVA analysis can be a powerful method of drug discovery.
Fig. 3.
Functional NRPS gene clusters identified by RVA. (A) Gene maps of four cosmids encoding NRPS genes that were identified using RVA screens. Boxes above and below the lines represent ORFs on the forward and reverse strands respectively. (B) Two of these cosmids showed toxicity to G. mellonella compared with the control (pWEB vector).
Identification of Virulence Gene Clusters.
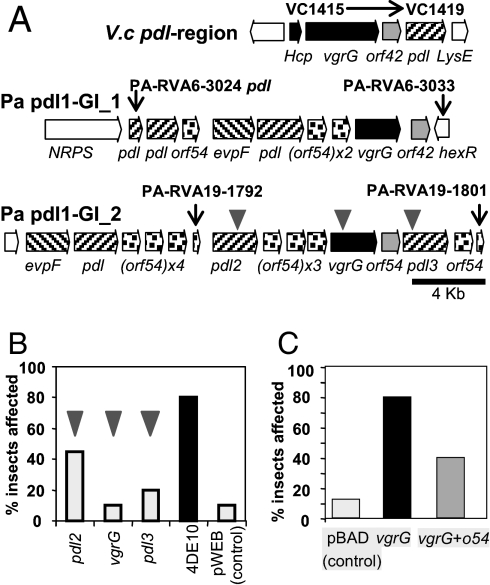
Our recent work on secretion of the orally active Toxin Complex d (Tcd) from P. luminescens indicates that a putative class III lipase pdl (27) is responsible for release of Tcd from the outer membrane of the bacterium and that this process is negatively regulated by the tightly linked gene orf54 (G. Yang and N.R.W., unpublished work). We detected several regions encoding pdl-o54 gene homologues in the P. asymbiotica genome that were toxic to either insects and/or nematodes. We therefore speculate that these represent virulence loci (Fig. 4A, and Figs. S7 and S20). We note that these regions have no effect on the mammalian macrophages again suggesting some specificity in their toxicity. Interestingly, pdl-islands are also present in the genomes of many other Gram-negative pathogens, including Vibrio cholerae, suggesting they also may be virulence determinants in other human pathogens. In characterizing RVA regions such as these, determination of the minimal region of overlap between “toxic” cosmids at any given locus may not provide sufficient clues to suggest the exact genes responsible. Thus, we carried out insertional mutagenesis to map the genes causing the phenotype by using an in vitro mutagenesis kit to construct insertion mutant libraries of toxic cosmids. Clones with insertions in different genes are then identified by sequencing out from the integrated transposon. The mutated cosmids are rescreened in the appropriate RVA assay to identify clones showing loss of function and therefore the ORF responsible. In the case of the Pa pdl1-GI_2 insecticidal region, we selected the 4DE10 toxic cosmid for this fine-scale mapping (Fig. 4B). This identified vgrG and two of the pdl-homologues as necessary for the toxic effect on G. mellonella. Subcloning and heterologous expression of vgrG in E. coli (Fig. 4C) confirmed the toxicity of this gene product and serves to illustrate how RVA allows us to move from a whole genome to an individual toxin gene with ease.
Fig. 4.
Putative novel virulence factors detected by RVA. (A) Comparison of a pdl operon in Vibrio cholerae O1 strain N16961 (Top) and two of the four pdl-o54 islands of P. asymbiotica ATCC43949. Homologous genes are coded by shading. Gray triangles represent transposon insertions in three individual genes for fine-scale mapping (see below). (B) Fine-scale mapping of the Pa pdl-GI_2 virulence island. The insertion mutants of the two pdl genes and the vgrG gene all show reduced toxicity when injected to G. mellonella, while the wild-type cosmid (4DE10) is fully toxic. Injection of control E. coli (pWEB cosmid) had negligible effect. (C) The vgrG toxin with or without the small flanking orf54 were cloned into the arabinose-inducible expression vector pBAD30. When induced, these clones were toxic by injection to G. mellonella. Interestingly, cloned orf54 reduced this effect suggesting its function is antagonistic to the toxic phenotype.
Importantly, the RVA approach is also powerful in identifying genes with functions that could not be predicted using conventional genetic approaches. Three cosmids were identified in the iGOT screen, 3CD1, 3DB8, and 1AA3 (RVA17, Fig. S18), causing a moribund phenotype when injected to M. sexta. Fine-scale mapping traced the toxic effect to kdpEDCBAF encoding the high affinity potassium pump (28) of P. asymbiotica (Fig. S25). Over-expression of the Kdp pump appears to allow the bacteria to persist and even grow in the hemocytes (I. Vlisidou and N.R.W., unpublished work). This striking fundamental discovery is likely to be relevant to all immune cell-pathogen interactions, which rely upon high potassium gradients for maturation of the phagolysosome (29). We propose that the assumption-free RVA screening technology is ideal for making such discoveries and expect many more from a range of pathogens.
Virulence Against Insect or Man?
It was anticipated that in the case of P. asymbiotica, a comparison between the invertebrate and macrophage (mGOT) RVA data could provide candidate genes responsible for facilitating human infection. Cosmids in RVA4 were identified by the mGOT screen but not in either of the invertebrate assays. This region does not encode genes unique to the human pathogenic P. asymbiotica and, therefore, has homologues in the insect-only pathogen P. luminescens TT01. RVA4 (Fig. S5) contains homologues of the proapoptotic xaxAB from Xenorhabdus (30) and a large hemolysin/hemagglutinin gene, either of which may represent potential virulence factors. Interestingly, the majority of mGOT regions are also shown to have effects in invertebrate hosts, supporting our hypothesis that genes evolved to combat invertebrates can be redeployed against mammalian hosts (3).
Discussion
Here, we have demonstrated the ability of RVA to identify virulence loci via the parallel screening of a recombinant DNA library in a non-pathogenic laboratory strain of E. coli against a range of different taxa. The RVA analysis of P. asymbiotica identified a range of different putative virulence factors including eight NRPS operons, two PKS operons, six hemolysin/hemagglutinin-like genes, three pili/fimbrial operons, seven putative specialized secretion systems (PVCpnf, T6SS, Evp-islands, T3SS and Pld-islands) and at least seven toxin-like genes (homologues of mcf1, xaxAB, vgrG, rtxA, xnt2, ast, and a VIP2 family gene). The identification of such a large number of virulence loci illustrates that RVA is useful as a means of generating preliminary biological annotation relating to pathogenic phenotypes. RVA annotation highlights operons or genes as potentially important in virulence that can then be investigated in greater detail either for accurate annotation or more specific applications (i.e., drug discovery or vaccine candidates). Therefore, fine-scale mapping of RVA regions, by transposon mutagenesis or cloning individual ORFs, is essential to confirm the exact genes responsible for toxicity in each cluster. The use of different invertebrate targets with less complex immune responses (insects, nematodes, and protozoa) allows the detection of less “potent” virulence factors that could otherwise be overlooked in whole animal mammalian models. It is interesting that most of the virulence factors identified in the mGOT assay were also toxic in the invertebrate screens confirming the general utility of invertebrates as model hosts for disease studies. Although it is likely that a small subset of host-specific virulence factors will not be detected by RVA, the screens are nevertheless sensitive enough to detect a very large number of more numerous general virulence factors.
The power of the RVA technique relies on several factors. First and highly important is the coverage and the depth of the genomic library used that covered >90% of the genome, with nearly 80% being covered by two or more clones. The analysis of the cosmid library distribution on the genome assembly confirmed that some regions are more represented than others and certain regions are absent. In future RVA studies it may be pertinent to “normalize” the library by selecting a subset of cosmids for the assays, reducing the level of redundancy in testing while still maximizing genome coverage. The second factor is the underlying genetic architecture of virulence factors. RVA's strength is in the identification of single-loci virulence factors and multilocus virulence factors that are tightly clustered within a bacterial genome. Molecular characterization of virulence factors to date suggests that the majority fall into these categories, further justifying our screening approach. However, there are several notable examples of virulence factors for which the underlying genetics basis is complex and involves multiple genes or regions of the genome, and these will naturally not be identified by RVA. This limitation can be overcome by using vectors that accommodate larger fragments such as BACs.
Frequent concerns regarding heterologous expression in E. coli include potential problems in differing G+C content between host and donor DNA and a failure of E. coli expression machinery either to recognize the donor gene expression signals or to provide the appropriate secretion machinery. Nevertheless, the successful application of RVA to Photorhabdus, with an average G+C content of 42%, significantly lower than the 50% of E. coli, indicates that this is not a current limitation to these biological screens. One reason why E. coli-based expression is more successful than expected may relate to the fact that pathogenicity islands that have been recently horizontally acquired may be more prone to gene expression in differing host backgrounds, and indeed this may actually be a requirement of such elements if they are to be positively selected in nature. Furthermore, the RVA technique does not only rely on correct secretion of the heterologously expressed virulence factors as whole cultures are used in the screens. We note that cytoplasmically accumulated toxins can be detected in these assays, as demonstrated by the insect toxicity of E. coli expressing but not secreting the Mcf1 toxin (14). The application of RVA to more diverse bacterial pathogens will ultimately define the limits of its utility, although in principle the host strain used need not be E. coli and libraries from Gram-positive pathogens could be screened in different host species (such as Lactococcus).
We believe that RVA represents an excellent way to provide basic information about general virulence determinants in poorly understood pathogens and to add functional relevance to the ongoing annotation of pathogen genome sequences. It also provides an alternative strategy for virulence gene detection across whole genomes, important for current efforts toward the reduction and replacement of animal testing.
Materials and Methods
Cosmid Library Construction and Statistical Analysis.
The P. asymbiotica ATCC43949 genomic library was constructed in the pWEB vector (Epicentre) in E. coli EP1305TM by MWG Biotech. The 1,536 resulting cosmids were arrayed into 16 96-well plates and end-sequenced before storage at −80 °C. Cosmid size and location were determined by BLASTN comparison of each end against the unfinished genome. Average cosmid length is 37,014 bp (SD = 4578.4) and 4,605,359 bp is covered representing 91.4% of the estimated 5.0 Mb genome. Average fold coverage is >7 fold and 79.75% of genome has twofold coverage or more. There are 17 gaps with a relatively large average gap size of 25,602bp (SD = 32,568) due to 2 large gaps while the remainder are smaller, typically <1000bp. Library plates were subcultured in LB medium supplemented with 100 μg/ml ampicillin and/or 50 μg/ml kanamycin. Transposon mutagenesis was done using the EZ-Tn5<Tet> kit (Epicentre).
Invertebrates Maintenance.
M. sexta were reared in controlled conditions (25 °C, 80% humidity) on artificial diet (31) and fifth instar larvae were used for iGOT. G. mellonella was purchased from Livefood Ltd for fine-scale mapping analysis. C. elegans were raised at 18 °C on NGM medium (32) using E. coli OP50 as feeding strain and transferred weekly to fresh plates. Adult worms were washed off plates with Phosphate Saline Buffer and the suspension adjusted to 10 nematodes per 10 μl drop. A. polyphaga was grown in PYG medium (33) at 25 °C, and subcultured weekly. Stationary phase (5 days) cultures were used for aGOT, adjusting the number of cells to 2 × 105 cells/ml using a haemocytometer.
Gain of Toxicity Assays (GOT).
100 μl of overnight cosmid cultures were injected into individual M. sexta larvae that were scored for death or severe delay in development daily during one week. Every cosmid showing an effect was retested on a further 10 insects allowing confirmation of the phenotype. For nGOT/aGOT, a 10 μl drop of overnight culture from each individual cosmid was grown on 25-well plates containing NGM agar with 50 μg/ml ampicillin (nGOT) or PYG agar with 50 μg/ml kanamycin (aGOT). The plates were inoculated with 10 μl of the adjusted suspensions of C. elegans or A. polyphaga respectively and incubated at 22 °C for 7 days. Toxicity was assessed both by eye and under an inverted microscope and avoidance or delay in feeding was scored using an arbitrary scale of 0 to 5 (5 being no consumption, 3 half-consumed and 0 completely consumed). For mGOT, the mouse BALB/c monocyte macrophage cell line J774–2 was seeded at 1 × 103 cells per ml into 96-well plates, in Dulbeccos Modified Eagles Medium (DMEM) supplemented with 10% fetal bovine serum, 5% non-essential amino acids, and 100 μg/ml ampicillin and incubated overnight (37 °C, 5% CO2). Cosmid cultures were harvested by centrifugation and cells subjected to lysozyme treatment followed by freeze-thaw cycles. Crude lysates were applied to the macrophage plates and incubated for 24 h. Subsequently, the mix was aspirated from the macrophages and replaced with phenol red-free DMEM, with 2% 100X solution penicillin/streptomycin and 50 μg/ml gentamicin and incubated under the same conditions for 2 h. Cell viability was ascertained using the XTT assay (34). Candidate cosmid clones were selected for their ability to reduce cell viability by 40% comparative to untreated cells.
HPLC/MS Analyses and Secondary Metabolite Structure Prediction.
Cosmids 4DB5 and 1DF9 were coexpressed in E. coli EC100 with pSUMtaA (35), which encodes a promiscuous phosphopantheteine transferase from Stigmatella aurantiaca DW4/3–1. After three days of growth in LB medium with 2% Amberlite XAD-16 adsorber resin (Fluka), the resin was harvested by centrifugation and extracted with 10 ml of MeOH. The organic extract was evaporated to dryness, dissolved in 1 ml of MeOH, centrifuged to get rid of solid material, and finally analyzed by HPLC/MS as described in ref. 36. Sum formulas were determined from the masses after HR-MS using a LTQ-Orbitrap Hybrid MS (Thermo) and databases searches were performed with the obtained predictions. The annotation of gene clusters putatively involved in secondary metabolite production was done by analyzing the protein sequences with a program written by Jacques Ravel (University of Maryland School of Medicine, Baltimore, MD) for overall domain organization and the NRPS predictor for adenylation specificity of the NRPS adenylation domains (37). In cases where no functional assignments could be made for full proteins or parts thereof, a BLASTP analysis was performed.
Supplementary Material
Acknowledgments.
We thank Sandra Barns for the maintenance of M. sexta and C. elegans, and Sharon Huws for A. polyphaga gift. This work was funded by the BBSRC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the EMBL database (accession nos. FM211043–FM211060).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711114105/DCSupplemental.
References
- 1.Khush RS, Lemaitre B. Genes that fight infection: What the Drosophila genome says about animal immunity. Trends Genet. 2000;16:442–449. doi: 10.1016/s0168-9525(00)02095-3. [DOI] [PubMed] [Google Scholar]
- 2.Sadd BM, Schmid-Hempel P. Insect immunity shows specificity in protection upon secondary pathogen exposure. Curr Biol. 2006;16:1206–1210. doi: 10.1016/j.cub.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 3.Waterfield NR, Wren BW, Ffrench-Constant RH. Invertebrates as a source of emerging human pathogens. Nat Rev Microbiol. 2004;2:833–841. doi: 10.1038/nrmicro1008. [DOI] [PubMed] [Google Scholar]
- 4.Gerrard J, Waterfield N, Vohra R, ffrench-Constant R. Human infection with Photorhabdus asymbiotica: An emerging bacterial pathogen. Microbes Infect. 2004;6:229–237. doi: 10.1016/j.micinf.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Farmer JJ, et al. Xenorhabdus luminescens (DNA hybridization group 5) from human clinical specimens. J Clin Microbiol. 1989;27:1594–1600. doi: 10.1128/jcm.27.7.1594-1600.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrard JG, et al. Nematode symbiont for Photorhabdus asymbiotica. Emerg Infect Dis. 2006;12:1562–1564. doi: 10.3201/eid1210.060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ffrench-Constant R, et al. Photorhabdus: Towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol Rev. 2003;26:433–456. doi: 10.1111/j.1574-6976.2003.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 8.Duchaud E, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 9.Sifri CD, Begun J, Ausubel FM. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Kaito C, et al. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol Microbiol. 2005;56:934–944. doi: 10.1111/j.1365-2958.2005.04596.x. [DOI] [PubMed] [Google Scholar]
- 11.Lauriano CM, et al. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci USA. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ffrench-Constant RH, et al. A genomic sample sequence of the entomopathogenic bacterium. Photorhabdus luminescens W14: Potential implications for virulence. Appl Environ Microbiol. 2000;66:3310–3329. doi: 10.1128/aem.66.8.3310-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daborn PJ, et al. A single. Photorhabdus gene makes caterpillars floppy (mcf) allows Esherichia coli to persist within and kill insects. Proc Natl Acad Sci USA. 2002;99:10742–10747. doi: 10.1073/pnas.102068099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang G, Dowling AJ, Gerike U, ffrench-Constant RH, Waterfield NR. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol. 2006;188:2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowling AJ, et al. The insecticidal toxin makes caterpillars floppy (Mcf) promotes apoptosis in mammalian cells. Cell Microbiol. 2004;6:345–353. doi: 10.1046/j.1462-5822.2003.00357.x. [DOI] [PubMed] [Google Scholar]
- 17.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schell MA, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 19.Rao PS, Yamada Y, Tan YP, Leung KY. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- 20.Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi K, et al. New alkaloids, ulbactins D and E, and their manufacture with Alteromonas. Jpn. Kokai Tokkyo Koho. 1998:6. CODEN: JKXXAF JP 10245377 A 19980914 Heisei. CAN 129:244217 AN 1998:600005 CAPLUS. [Google Scholar]
- 22.Gu WX, Liu SX, Silverman RB. Solid-phase, Pd-catalyzed silicon-aryl carbon bond formation. Synthesis of sansalvamide A peptide. Org Lett. 2002;4:4171–4174. doi: 10.1021/ol0269392. [DOI] [PubMed] [Google Scholar]
- 23.Carroll CL, et al. Synthesis and cytotoxicity of novel sansalvamide A derivatives. Org Lett. 2005;7:3481–3484. doi: 10.1021/ol051161g. [DOI] [PubMed] [Google Scholar]
- 24.Otrubova K, Lushington G, Velde DV, McGuire KL, McAlpine SR. Comprehensive study of sansalvamide a derivatives and their structure-activity relationships against drug-resistant colon cancer cell lines. J Med Chem. 2008;51:530–544. doi: 10.1021/jm070731a. [DOI] [PubMed] [Google Scholar]
- 25.Belofsky GN, Jensen PR, Fenical W. Sansalvamide: A new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett. 1999;40:2913–2916. [Google Scholar]
- 26.Schellenberg B, Bigler L, Dudler R. Identification of genes involved in the biosynthesis of the cytotoxic compound glidobactin from a soil bacterium. Environ Microbiol. 2007;9:1640–1650. doi: 10.1111/j.1462-2920.2007.01278.x. [DOI] [PubMed] [Google Scholar]
- 27.Waterfield NR, Daborn PJ, ffrench-Constant RH. Insect pathogenicity islands in the insect pathogenic bacterium Photorhabdus. Physiological Entomology. 2004;29:1–11. [Google Scholar]
- 28.Walderhaug MO, et al. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J Bacteriol. 1992;174:2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves EP, et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 30.Vigneux F, et al. The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J Biol Chem. 2007;282:9571–9580. doi: 10.1074/jbc.M604301200. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds SE, Nottingham SF, Stephens AE. Food and water economy and its relation to growth in 5th-instar larvae of the Tobacco Hornworm, Manduca-Sexta. J Insect Physiol. 1985;31:119–127. [Google Scholar]
- 32.Stiernagle T. Maintenance of C. elegans. [Accessed July 23, 2008];Wormbook. 2006 doi: 10.1895/wormbook.1.101.1. Available at http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 33.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scudiero DA, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- 35.Gaitatzis N, Hans A, Muller R, Beyer S. The mtaA gene of the myxothiazol biosynthetic gene cluster from Stigmatella aurantiaca DW4/3–1 encodes a phosphopantetheinyl transferase that activates polyketide synthases and polypeptide synthetases. J Biochem. 2001;129:119–124. doi: 10.1093/oxfordjournals.jbchem.a002821. [DOI] [PubMed] [Google Scholar]
- 36.Brachmann AO, Forst S, Furgani GM, Fodor A, Bode HB. Xenofuranones A and B: Phenylpyruvate dimers from Xenorhabdus szentirmaii. J Nat Prod. 2006;69:1830–1832. doi: 10.1021/np060409n. [DOI] [PubMed] [Google Scholar]
- 37.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.