Abstract
Enzyme replacement therapy for lysosomal storage diseases is currently based on endocytosis of lysosomal enzymes via the mannose or mannose 6-phosphate receptors. We are developing a technology for endocytosis of lysosomal enzymes that depends on generic, chemically conjugated reagents. These reagents are aptamers (single-stranded nucleic acid molecules) selected to bind to the extracellular domain of the mouse transferrin receptor. After selection, an RNA aptamer and a DNA aptamer were modified with biotin and linked to dye-labeled streptavidin for detection by confocal microscopy. Aptamer–streptavidin conjugates showed saturable uptake into mouse fibroblasts (Ltk− cells), which could be inhibited by an excess of free aptamer but not by tRNA, calf thymus DNA, or transferrin. The RNA aptamer–streptavidin conjugate was mouse-specific, as human cells (293T) did not take it up unless first transfected with the mouse transferrin receptor. Some streptavidin separated from the recycling pathway of transferrin and colocalized with lysosomes. After characterization in the model system, the DNA aptamer was conjugated to a lysosomal enzyme, α-l-iduronidase, from which mannose 6-phosphate had been removed. The aptamer had been modified by attachment of terminal glycerol for oxidation by periodate and reaction of the resulting aldehyde with amino groups on the protein. Dephospho-α-l-iduronidase-aptamer conjugate was taken up in saturable manner by α-l-iduronidase-deficient mouse fibroblasts, with half-maximal uptake estimated as 1.6 nM. Endocytosed enzyme–aptamer conjugate corrected glycosaminoglycan accumulation, indicating that it reached lysosomes and was functional in those organelles. Both uptake and correction were inhibited by unconjugated aptamer, confirming the role of the aptamer in receptor-mediated endocytosis.
Keywords: lysosomal storage disease, transferrin receptor
Receptor-mediated endocytosis of lysosomal enzymes underlies enzyme replacement therapy (ERT) for lysosomal storage diseases (1, 2). The first lysosomal enzyme to be developed as a pharmaceutical was β-glucosidase for ERT of Gaucher disease (3). Endocytosis of this enzyme depends on terminal mannose residues, which are recognized by the mannose receptor on cells of macrophage lineage (4). Five more enzymes have been recently approved for ERT: α-l-iduronidase, iduronate sulfatase, and N-acetylgalactosamine 4-sulfatase (arylsulfatase B) for mucopolysaccharidoses (MPS) I, II, and VI, respectively (5–7); α-galactosidase for Fabry disease (8); and α-glucosidase for Pompe disease (9). Others are in clinical trial or in preclinical studies. Endocytosis and therapeutic effectiveness of these lysosomal enzymes depend on recognition of mannose 6-phosphate residues on the enzyme by the widely distributed mannose 6-phosphate/IGFII receptor (10).
Useful as these endocytosis systems may be, challenges remain for introducing lysosomal enzymes into deficient cells of human patients or animal models. Some recombinant lysosomal enzymes may have insufficient mannose 6-phosphate residues for efficient uptake (11, 12). Another problem is that, because of the blood–brain barrier (13), current ERT provides lysosomal enzymes to somatic organs but not to the brain; that is a serious shortcoming, given that most lysosomal storage disorders have a neurologic component. Even some somatic tissues may be hard to reach. To address these problems, there has been substantial interest in developing alternative technologies for targeted delivery of lysosomal enzymes into cells and tissues. These include modification of the carbohydrate residues (14), preparation of fusion proteins of enzymes with antibodies to specific receptors such as the Fc receptor (15) or the insulin receptor (16), or attachment of enzyme to nanocarriers targeted by antibodies to the adhesion molecule ICAM (17).
We have envisioned the benefit of chemical modification of lysosomal enzymes with generic reagents that would mediate entry into cells or tissues of choice. The reagents to modify the enzymes would be aptamers (single-stranded nucleic acids) selected against an endocytosis receptor. Aptamers are selected from very large random libraries by a procedure known as SELEX (systematic evolution of ligands by exponential enrichment)—an iterative process of enriching the mixture in molecules with high binding affinity and selectivity against the desired target (18, 19). Aptamers themselves are considered promising candidates for many therapeutic applications (20) and have also been used to facilitate targeted uptake of other therapeutic molecules. For example, aptamers directed against prostate-specific membrane antigen have been linked to drug-laden nanoparticles (21), small interfering RNA (22), and a ribosomal toxin (23) to destroy prostate cancer cells.
Our target molecule for aptamer selection was the extracellular domain of the mouse transferrin receptor (TfR). This receptor, ubiquitous in mammalian cells, is essential for the delivery of iron to cells via binding and endocytosis of transferrin-Fe+3, release of iron in endosomes, and dissociation of apotransferrin at the cell surface (24, 25). Although other receptors that participate in endocytosis could have been used as targets for aptamer selection, we chose the transferrin receptor because it has been implicated in transcytosis across brain capillary endothelial cells (26–28). The latter is important for our ultimate objective of introducing therapeutic enzymes across the blood–brain barrier. In this study, we chose an RNA and a DNA aptamer binding to the extracellular domain of mouse TfR (TfR-ECD) and demonstrated their ability to deliver streptavidin to Ltk− cells. We then showed the ability of the DNA aptamer to deliver a lysosomal enzyme into deficient mouse fibroblasts and correct the defective glycosaminoglycan (GAG) degradation in these cells.
Results
Characterization of RNA and DNA Aptamers.
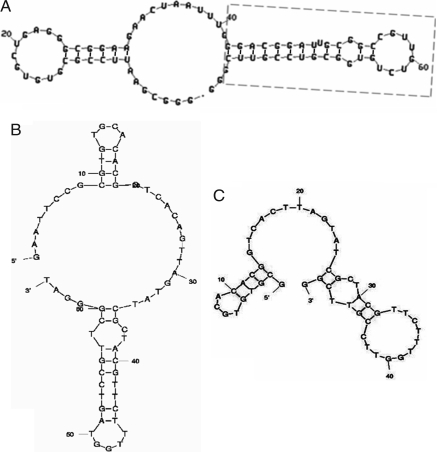
RNA aptamers targeted to TfR-ECD were selected from a library of ≈1014 molecules containing 49 nucleotides in random sequence, as described in supporting information (SI) Materials and Methods. After nine rounds of selection (two by filter binding and seven by affinity spin columns) and cloning, aptamers were obtained from individual clones. Of 69 aptamers analyzed by filter binding assay, 30 showed significant binding and were sequenced. Five aptamers had sequence similarity (Fig. S1), and the remainder were unrelated. Based on the predicted secondary structure (29) of the five aptamers that shared part of the primary sequence, a truncated version (Fig. S1, boxed) was generated and synthesized. The folded structures of full-length aptamer “FB4” and of its truncated form are shown in Fig. 1A.
Fig. 1.
Folded structures of aptamers used in this study. (A) Full-length RNA aptamer FB4, with the box outlining the truncated form; additional details are provided in Fig. S1. (B) Full-length DNA aptamer GS24. (C) Truncated GS24, reduced to 50 nucleotides.
DNA aptamers targeted to TfR-ECD were selected from a library of ≈1014 single-stranded DNA molecules containing 36 nucleotides in random sequence. After five rounds of selection by affinity spin columns and a sixth round by gel shift, aptamers were obtained from individual clones. The binding of each aptamer was analyzed by gel shift because filter binding gave a very high background. Of 40 aptamers analyzed, 10 showed significant binding. One aptamer, “GS24,” was truncated to 50 nucleotides based on its folding structure (Fig. 1 B and C).
The truncated forms of the RNA aptamer FB4 and the DNA aptamer GS24 were used throughout unless the full-length form is specifically indicated.
Endocytosis of Streptavidin–Aptamer Conjugates by Cultured Cells.
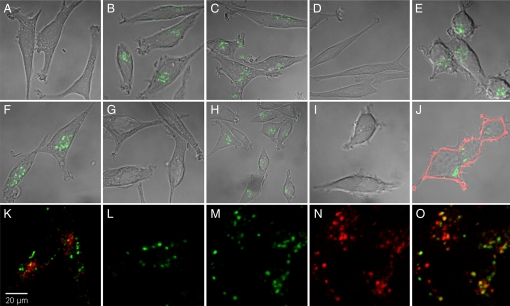
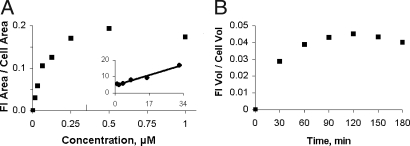
Aptamers were conjugated to streptavidin and used to test endocytosis into Ltk− cells. The results, obtained by confocal microscopy, are shown in Fig. 2. The streptavidin had been labeled with Cy5, which is displayed in green. Whereas there was no endocytosis of free streptavidin (Fig. 2A), the streptavidin conjugated to the RNA aptamer (FB4-streptavidin) was readily taken up and was seen after a 30-min incubation in a punctate pattern that occupied an area close to the nucleus (Fig. 2B) and at longer times extended to the tips of the cell (data not shown). Endocytosis of FB4-streptavidin was not inhibited by tRNA (Fig. 2C) or poly(rA) (data not shown) but was inhibited by free full-length FB4 (Fig. 2D), indicating a requirement for the aptamer structure. It was inhibited neither by 34 μM transferrin, a concentration found in serum of C57BL/6 mice (30) (Fig. 2E), nor by 10 mM methyl-β-cyclodextrin, an inhibitor of raft/caveolar uptake (31) (data not shown). Similar uptake of streptavidin conjugated to the full-length DNA aptamer GS24 was observed (Fig. 2F); it was inhibited by free full-length GS24 (Fig. 2G) but not by transferrin (Fig. 2H). Quantitation of fluorescence showed that endocytosis of the FB4-streptavidin was saturable and half-maximal at ≈80 nM (Fig. 3A). Accumulation of this conjugate reached a plateau at 90 min (Fig. 3B), suggesting that the rate of streptavidin degradation or exit from the cell had become equal to its rate of accumulation or, alternatively, that all transferrin receptor had been internalized during the prolonged exposure to the conjugate and was no longer available for endocytosis.
Fig. 2.
Endocytosis of streptavidin–aptamer conjugates by cultured Ltk− cells. Streptavidin was labeled with Cy5, shown in green, and confocal microscopy was used for its detection in the cells. The cells were incubated at 37°C for 30 min, with free streptavidin (A), RNA aptamer conjugate, FB4-streptavidin (B), FB4-streptavidin with 34 μM tRNA (C), FB4-streptavidin with 50-fold free, full-length FB4 (D), or FB4-streptavidin with human transferrin-Fe3+ (E). The cells were incubated with the DNA aptamer conjugate, full-length GS24-streptavidin, with no additions (F), with 50-fold free full-length GS24-streptavidin (G), or with human transferrin-Fe3+ (H). (I) Human (293T) cells did not internalize FB4-streptavidin unless transfected with mouse TfR-HA (J); antibody to the HA tag (pink) marks the presence of mouse TfR on the cell surface. Cells were not permeabilized before staining. (K) Human transferrin (red) and FB4-streptavidin after a 30-min incubation; (L) the same, with a 30-min chase. (M–O) Partial colocalization of internalized streptavidin after a 1-h incubation and a 2-h chase (M) with lysosomes that had been prelabeled by incubation of the cells with dextran-Texas red for 3 h (N). (O) Merged image of M and N. Magnification, indicated in K, is the same for all panels.
Fig. 3.
Endocytosis of FB4-streptavidin conjugate as a function of concentration and time, measured by confocal microscopy. (A) FB4-streptavidin internalized as function of concentration, determined as the fraction of the cell area that was fluorescent. The Inset shows a double reciprocal plot of the data. (B) FB4-streptavidin internalized as a function of time, measured as the fraction of the cell volume that was fluorescent. The ratio was obtained by summing the fluorescence in each optical slice of a z-stack and estimating the cell volume on the assumption that the cells are ellipsoidal. Fl, fluorescence
Human 293T cells were found not to take up FB4-streptavidin (Fig. 2I) unless first transfected with the mouse transferrin receptor (Fig. 2J). The mouse transferrin receptor had been fused to a hemagglutinin (HA) tag to label the plasma membrane of transfected cells, and it can be seen as a pink outline of the cell. Fig. 2J shows that a representative 293T cell transfected with the mouse transferrin receptor had taken up FB4-streptavidin.
The fate of endocytosed FB4-streptavidin did not follow that of transferrin. After 30 min of incubation in medium containing both transferrin and FB4-streptavidin, the transferrin was located in a small area inside the cell (red), whereas streptavidin was located further out (Fig. 2K). After 30 min of chase in medium devoid of transferrin or conjugate, no transferrin was visible within the cells whereas the streptavidin was still present (Fig. 2L). The eventual destination of the labeled streptavidin appeared to be the lysosome, as shown by the partial colocalization of streptavidin with dextran-Texas red (used to label lysosomes) after a 1-h incubation with FB4-streptavidin and a 2-h chase (Fig. 2 M–O).
Endocytosis and Functionality of Aptamer Conjugate of a Lysosomal Enzyme in Deficient Cells.
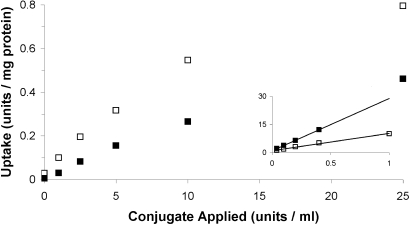
Because of its greater stability, only the DNA aptamer (GS24) was used for the enzyme studies. The aptamer conjugate of dephosphorylated α-l-iduronidase (GS24-dePIdu) was taken up by α-l-iduronidase-deficient (Idua−/−) mouse fibroblasts in a saturable manner, with half-maximal uptake at ≈1.6 nM (Fig. 4). It was competitively inhibited by the presence of aptamer, with a Ki of ≈40 nM. Because there was some phosphorylated α-l-iduronidase in the dephosphorylated preparation (≈2%), these studies were performed in the presence of 10 mM mannose 6-phosphate.
Fig. 4.
Aptamer-dependent endocytosis of the GS24-dePIdu conjugate by Idua−/− mouse cells. Activity internalized is plotted as a function of concentration of aptamer–enzyme conjugate applied, with (■) or without (□) free GS24 (100 nM). Mannose 6-phosphate (10 mM) was included in all reactions. The double reciprocal plot in the Inset shows competitive inhibition by the aptamer.
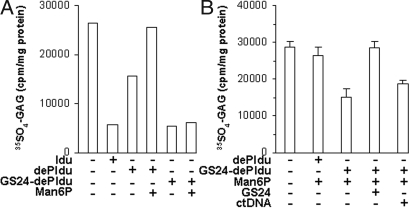
The enzyme–aptamer conjugate accelerated the degradation of GAG within the mouse Idua−/− cells into which it had been taken up (Fig. 5). Initially, the fibroblasts were incubated for 17 h in medium containing 35SO4, and the labeled GAG was examined at the end of that period. The accumulation of 35SO4-GAG in the Idua−/− cells was greatly elevated over normal (data not shown). It could be reduced (corrected) when GS24-dePIdu was present during the incubation (Fig. 5A). Correction was half-maximal at 130 pM enzyme–aptamer conjugate and was not affected by the presence of mannose 6-phosphate. However, we were unable to demonstrate inhibition of correction by free aptamer using this experimental design (data not shown). We considered it likely that the free aptamer might be degraded faster than the enzyme–aptamer conjugate during the prolonged incubation, allowing endocytosis of the conjugate and resulting in correction. We therefore modified the experimental design to allow the cells to be in the presence of conjugate and free aptamer for a shorter time.
Fig. 5.
Aptamer-dependent correction of glycosaminoglycan metabolism in Idua−/− mouse cells by the GS24-dePIdu conjugate. (A) Reduction of 35SO4 GAG in cells incubated for 22 h in radiolabeled medium with 50 units/ml enzyme or enzyme–aptamer conjugate, with or without 10 mM mannose 6-phosphate. (B) Reduction in 35SO4-GAG in cells incubated for 22 h with radiolabeled medium, then allowed to take up 20 units/ml GS24-dePIdu for 1 h with or without inhibitors and chased for 22 h. The bars indicate the mean and SD of triplicate reactions. ctDNA, calf thymus DNA.
To demonstrate inhibition by free aptamer, the fibroblasts were first preincubated with medium containing 35SO4 for 22 h to label sulfated GAG and were then allowed to take up the aptamer–enzyme conjugate for 1 h in the presence of mannose 6-phosphate, with or without potential inhibitors. Correction was demonstrated by the drop in intracellular radiolabeled GAG in cells that had received GS24-dePIdu (Fig. 5B). The correction was inhibited by the presence of 10 μM free aptamer in the medium but not by an equivalent amount of calf thymus DNA.
Discussion
Two major lines of evidence show that aptamer directed against the transferrin receptor is able to mediate the endocytosis of protein conjugates and their transport to lysosomes. The first was obtained with RNA and DNA aptamers conjugated to labeled streptavidin. Uptake of aptamer–streptavidin was readily visualized by confocal microscopy; it was saturable, inhibited by free aptamer, and specifically dependent on the presence of the mouse transferrin receptor. With a sufficiently long incubation time, part of the labeled streptavidin was found in the same vesicles as endocytosed dextran. The second line of evidence was obtained with a DNA aptamer conjugate of a lysosomal enzyme. Uptake of the conjugate was saturable and was inhibited by free aptamer but not by mannose 6-phosphate. Most importantly, the aptamer–enzyme conjugate could function in correction—i.e., normalization of defective GAG catabolism in deficient cells—in a manner inhibited by free aptamer, but not by mannose 6-phosphate or double-stranded calf thymus DNA. Because correction requires not only uptake but also enzymatic function within lysosomes, this shows that at least some of the aptamer-conjugated lysosomal enzyme reached its lysosomal destination.
Because transferrin is a major protein in plasma and the eventual goal of our studies is to use aptamers in vivo for therapeutic purposes, we had to find aptamers that would not compete with transferrin. Both the RNA aptamer and the DNA aptamer used in these studies fulfilled that requirement: internalization of streptavidin–aptamer conjugates was not inhibited by transferrin-Fe+3 at a concentration similar to that found in mouse serum (30). This fortuitous result can be understood by examining the properties of the TfR-ECD made by two expression systems (see Materials and Methods). One bound transferrin with low affinity (Kd in the μM range), suggesting a conformational defect in the transferrin binding region. That preparation was used for all nine rounds of selection of RNA aptamers and for the first round of selection of DNA aptamers. A second preparation of TfR-ECD had much tighter binding of transferrin (Kd in the nM range), similar to the binding of human TfR-ECD to transferrin (32). Because the aptamers bound equally well to both preparations of TfR-ECD (data not shown), their binding sites had to be distinct from the transferrin binding site.
But even though the aptamer conjugates were not bound to the transferrin site, we believe that, like transferrin, they were brought into the cells through coated pits. Although that has not been shown directly, the alternate endocytic pathway by lipid rafts and caveolae was excluded because uptake of the FB4-streptavidin conjugate was not inhibited by methyl-β-cyclodextrin, a cholesterol-sequestering agent that inhibits the raft–caveolar pathway (31). Once inside the cell, the conjugate followed a path different from that of transferrin. Separation of the two ligands was seen after 30 min of endocytosis, the transferrin being found in a compact area close to the nucleus with the streptavidin dispersed to other parts of the cell. When the endocytosis was followed by a 30-min chase, the transferrin, but not the streptavidin, was gone from the cells. The loss of the endocytosed transferrin is in keeping with the well known recycling pathway by which endocytosed transferrin loses iron in an acidic endosome compartment and is returned to the cell surface, where it dissociates from TfR (33). By contrast, streptavidin was found partially colocalized with dextran-labeled lysosomes after prolonged incubation. On the assumption that TfR accompanied the streptavidin conjugate during vesicular transport, this suggests that some TfR was not recycled but was routed to lysosomes. Routing of TfR to lysosomes has been reported in certain circumstances (34, 35).
The aptamer-mediated endocytosis of dephospho-α-l-iduronidase conjugate by mouse fibroblasts was half-maximal at 1.6 nM, close to the half-maximal concentration (0.7 nM) obtained for mannose 6-phosphate-mediated endocytosis of α-l-iduronidase by human fibroblasts (36). However, half-maximal correction of GAG accumulation in mouse fibroblasts by the aptamer conjugate occurred at 130 pM, a value 200 times higher than the 0.7 pM value reported for correction of GAG accumulation in human fibroblasts by α-l-iduronidase (36). The reason for the difference is not known but could result from the difference in cells and/or the efficiency of targeting to lysosomes. It should be noted, however, that uptake is not the limiting factor for either the aptamer-mediated or mannose 6-phosphate-mediated system.
Our results represent proof-of-principle that proteins conjugated to aptamers that bind to the extracellular domain of the transferrin receptor can undergo receptor-mediated endocytosis and reach lysosomes by a non-carbohydrate-mediated mechanism. If the protein is a lysosomal enzyme such as α-l-iduronidase, it can function in lysosomes and degrade substrate accumulated in deficient cells. We predict that the principle can be applied to other enzymes and other receptors that participate in endocytosis. However, we have no evidence that the DNA aptamer we selected by binding could participate in transcytosis, a process more complicated than endocytosis in that it requires not only binding and translocation through the cell, but also release from the receptor and exocytosis (37). It may be that cell-based selection will be required to find aptamers that can mediate transcytosis.
Materials and Methods
Reagents.
Full-length mouse and human TfR cDNA clones were kindly provided by Nancy Andrews (Duke University, Durham, NC) and Pamela Bjorkman (California Institute of Technology, Pasadena), respectively. α-l-Iduronidase (BioMarin Pharmaceuticals) and dephosphorylated α-l-iduronidase were provided by Merry Passage (Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA). Other reagents were purchased from commercial sources, as listed in SI Materials and Methods.
Cell Culture.
Methods are described in SI Materials and Methods.
Selection of RNA and DNA Aptamers.
Details of the selection, using the SELEX procedure, are provided in SI Materials and Methods.
Preparation of the Extracellular Domain of TfR.
We made two constructs for overexpressing TfR-ECD, the extracellular domain of TfR, using a different insect cell system for each. In the first system, a clone was generated for transfection in the S2 (Drosophila melanogaster Schneider 2) cell system by PCR amplification of the cDNA encoding mouse TfR-ECD, followed by restriction with BglII and XhoI and ligation into pMT/BiP/V5. A stable cell line was generated by cotransfection with a plasmid carrying the hygromycin resistance gene as selection marker. The induction was carried out with 500 μM CuSO4 for 2 days at a density of 6 × 106 cells per milliliter. The TfR-ECD (amino acid 90 to amino acid 757) thus produced by S2 cells had both hexa-His and V5 epitope tags fused to its C terminus. In the second system, we used High Five cells (Trichopulsia ni) of the baculovirus system to produce TfR-ECD fused to a hexa-His tag at the N terminus, with an intervening factor Xa protease cleavage site. This construct was generated by replacing the portion of human TfR cDNA in a modified pAcGP67A expression vector (32) with the mouse TfR-ECD (amino acid 90 to amino acid 763) cDNA at EcoRI and BglII sites. The generation of recombinant virus using this construct and the production of baculovirus-infected High Five cells were carried out at the Caltech Protein Expression Center. The TfR-ECD proteins were purified from the respective insect cell supernatants using Ni columns. Although both preparations of TfR-ECD bound mouse holotransferrin [made from apotransferrin as described (38)], the first bound only weakly, with a Kd of ≈9 μM, whereas the second bound much more tightly, with a Kd of ≈8 nM, as determined by plasmon resonance.
Expression of Full-Length Mouse TfR Fusion Proteins in Human Cells.
Full-length mouse TfR was generated by PCR amplification of its cDNA using pBluescript II KS plus cDNA. The PCR product was then restricted with KpnI and XhoI and ligated with T4 ligase to pcDNA3 expression vector (Invitrogen) that had been digested with the same nucleases. To create the construct of TfR with HA at the C terminus, DNA encoding the HA was cloned into pcDNA3-mTfR at the XhoI and ApaI sites. A transiently transfected 293T cell line was generated by using Lipofectamine 2000 with the appropriate construct.
Endocytosis of Streptavidin–Aptamer Conjugates.
5′ biotinylated RNA aptamer FB4 and 5′ biotinylated DNA aptamer full-length GS24 were conjugated to Cy-5-labeled streptavidin by mixing with an approximately stoichiometric amount of tetrameric streptavidin in PBS. A slight excess (10%) of streptavidin was used to ensure that all of the aptamer was bound. Ltk− cells were plated on cover slips in six-well dishes and grown to subconfluence. Standard conditions for incubation with RNA aptamer conjugate consisted of a 30-min incubation in DMEM containing 1% BSA, 20 μM tRNA, 80 units of RNasin, and 2 μM FB4-streptavidin, in a total volume of 200 μl, at 37°C in 5% CO2. To avoid the presence of NH4+, a known inhibitor of endocytosis (39), glutamax was added to glutamine-free MEM immediately before use. Incubation with DNA aptamer conjugate was performed under similar conditions with 2 μM full-length GS24-streptavidin, but without RNasin and with 0.2 mg/ml calf thymus DNA instead of tRNA. At the end of the reaction, the cells were fixed with 4% paraformaldehyde and 4% sucrose in PBS. Deviations from these conditions for specific experiments are described in Results. For 293T cells transfected with HA-tagged mouse TfR, the fixation procedure was followed by staining with anti-HA antibody clone 12CA5 and a secondary donkey anti-mouse IgG Alexa Fluor 488 without permeabilizing the cells. Images were acquired by laser scanning electron microscopy (Pascal; Carl Zeiss Microimaging) and processed by Axiovision software (Zeiss) and Photoshop (Adobe Systems). Fluorescence was quantitated by using the LSM5 Pascal software program.
To determine whether FB4-streptavidin conjugate could be used to deliver streptavidin to lysosomes, Ltk− cells were preincubated at 37°C with 0.5 mg/ml dextran-Texas red for 3 h to label lysosomes. The cells were then incubated with the conjugate for 1 h, washed, and further incubated for 2 h. Cells were then fixed and analyzed by confocal microscopy.
To compare the fate of aptamer protein conjugate with that of transferrin, Ltk− cells were incubated for 30 min at 37°C with 2 μM FB4-streptavidin and 313 nM human transferrin-Alexa Fluor 488. The cells were then washed and incubated for an additional 30 min or longer. They were fixed and analyzed by confocal microscopy.
Enzyme Activity.
α-l-Iduronidase activity was determined essentially as described (36). A unit is defined as the activity catalyzing the hydrolysis of 1 nmol of 4-methylumbelliferyl-α-l-iduronide per hour. Protein was determined by the bicinchoninic acid (BCA) method by using the manufacturer's recommendation. For determining the specific activity of aptamer–enzyme conjugates, which were available in limited amount, the protein was determined by the micro-BCA method as recommended by the manufacturer.
Preparation of Aptamer–Enzyme Conjugates.
For conjugating to enzyme, the aptamer used was DNA aptamer GS24, which had been extended at the 3′ end by a linker region of 12 CH2 groups and glycerol. GS24 terminated with glycerol (without linker) was also used. The DNA aptamer was allowed to react with 100 mM NaIO4 in 30 mM Na acetate buffer (pH 5.6) at ambient temperature for 1.5 h. After removal of periodate by two passes through G25 Microspin columns, the aptamer was added to either α-l-iduronidase or its dephosphorylated form in a 10:1 molar ratio of aptamer to enzyme. This was followed by the immediate addition of 0.8 M NaCNBH3 in 10 mM NaOH, to a final concentration of 40 mM. The mixture was kept at ambient temperature for 3 h, then at 4°C overnight. Conjugation was followed by electrophoresis on a 2% agarose gel, which separates the conjugate from the faster-moving aptamer, and DNA was visualized with ethidium bromide.
Purification of aptamer–enzyme conjugate was carried out by using the Biologic System (Bio-Rad) on a Superdex 200 sizing column equilibrated with 100 mM Na phosphate/150 mM NaCl (pH 5.8). To achieve better separation, two sizing columns were sometimes used in series. The fractions were analyzed by electrophoresis on a 2% agarose gel. Specific activity determination after purification showed that the GS24 conjugate of α-l-iduronidase had retained the specific activity of the enzyme, whereas the conjugate of the dephosphorylated α-l-iduronidase had lost approximately half. The reason for this difference is not clear.
To estimate the number of DNA aptamers conjugated to enzyme, the aptamer GS24 was modified by addition of biotin at the 5′ end. The amount of biotin incorporated into the conjugates was determined with the FluoroReporter Biotin Quantitation kit. At the 10:1 ratio of aptamer to enzyme used for conjugation, there was an average of three aptamers per enzyme in the major fraction and up to 10 in minor fractions. Fractions were pooled for experiments.
An in situ enzyme assay that distinguishes between unreacted enzyme and enzyme–aptamer conjugate proved particularly useful in early conjugation experiments. After electrophoresis on agarose gel and visualization of the aptamer with ethidium bromide, a strip of Whatman #3 filter paper soaked in buffer (0.4 M Na formate buffer, pH 3.5/0.15 M NaCl/0.9 mg/ml BSA) was applied to the gel for 10 min to acidify it; then a strip of Whatman #1 filter paper soaked in substrate solution (250 μM 4-MU-α-l-iduronide in 0.4 M formate buffer, pH 3.5/0.15 M NaCl) was applied for 10 min. The reaction was alkalinized by a 10-min exposure to NH3 vapor in a desiccator, and the fluorescent product was visualized at 365 nm with a hand-held UV light.
Endocytosis of Enzyme–Aptamer Conjugate and Correction of GAG Accumulation.
Endocytosis of aptamer–enzyme conjugate was studied in mouse Idua−/− fibroblasts, which have no endogenous α-l-iduronidase. The fibroblasts were grown to confluence in six-well dishes. Growth medium was replaced by 1 ml of α-MEM containing 15% FBS, aptamer–enzyme conjugate at various concentrations, 10 mM mannose 6-phosphate, and aptamer as indicated. After incubation for 1 h at 37°C, the medium was removed and the cells were washed with PBS, and detached from the plate by trypsinization. The cells were collected by centrifugation, washed again with PBS, and disrupted by freeze-thawing two times for assay of α-l-iduronidase activity.
Correction of the abnormal catabolism of GAG in Idua−/− cells was determined by measurement of intracellular radiolabeled GAG present after accumulation or after chase (40). Cells, grown to confluence in 100-mm Petri dishes, were incubated for ≈22 h with 5 ml of medium containing 4 × 106 cpm/ml 35SO4; the radiolabeled GAG was determined as radioactive cellular material insoluble in boiling 80% ethanol. The medium used for these experiments was α-MEM with 15% FBS, but with penicillin/streptomycin removed to reduce the amount of sulfate. In the first method, enzyme–aptamer conjugate and inhibitors were present during the 22-h incubation of the cells, after which the cells were detached from the plates by trypsinization and their content of radiolabeled GAG was determined. In the second method, the cells were incubated with 5 ml of 35SO4-labeled medium for 22 h; the radioactive medium was removed and the cells were incubated for 1 h in 5 ml of medium containing enzyme–aptamer conjugate, with or without inhibitors, to permit endocytosis. After a change of medium, incubation was continued for an additional 22 h; cells were detached from the plate by trypsinization, and the radiolabeled GAG was measured.
Supplementary Material
Acknowledgments.
We thank Dr. Kazuhiro Ohmi for initiating primary cultures of mouse fibroblasts, Dr. Nancy Andrews for a clone of mouse TfR, Dr. Pamela Bjorkman for a clone of human TfR, and Merry Passage for recombinant α-l-iduronidase and dephosphorylated α-l-iduronidase. This work was supported in part by National Institutes of Health Grants GM21199 (to C.-h.B.C.), NS048124 (to E.F.N.), and NS053808 (to E.F.N.) and by a grant from the Children's Medical Research Foundation (to E.F.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808360105/DCSupplemental.
References
- 1.Desnick RJ, Schuchman EH. Enzyme replacement and enhancement therapies: Lessons from lysosomal disorders. Nat Rev Genet. 2002;3:954–966. doi: 10.1038/nrg963. [DOI] [PubMed] [Google Scholar]
- 2.Neufeld EF. In: Lysosomal Disorders of the Brain. Platt F, Walkley S, editors. Oxford: Oxford Univ Press; 2004. pp. 319–330. [Google Scholar]
- 3.Barton NW, et al. Replacement therapy for inherited enzyme deficiency—macrophage-targeted glucocerebrosidase for Gaucher's disease. N Engl J Med. 1991;324:1464–1470. doi: 10.1056/NEJM199105233242104. [DOI] [PubMed] [Google Scholar]
- 4.Furbish FS, Steer CJ, Krett NL, Barranger JA. Uptake and distribution of placental glucocerebrosidase in rat hepatic cells and effects of sequential deglycosylation. Biochim Biophys Acta. 1981;673:425–434. doi: 10.1016/0304-4165(81)90474-8. [DOI] [PubMed] [Google Scholar]
- 5.Wraith JE, et al. Enzyme replacement therapy for mucopolysaccharidosis I: A randomized, double-blinded, placebo-controlled, multinational study of recombinant human alpha-L-iduronidase (laronidase) J Pediatr. 2004;144:581–588. doi: 10.1016/j.jpeds.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 6.Muenzer J, et al. A phase II/III clinical study of enzyme replacement therapy with idursulfase in mucopolysaccharidosis II (Hunter syndrome) Genet Med. 2006;8:465–473. doi: 10.1097/01.gim.0000232477.37660.fb. [DOI] [PubMed] [Google Scholar]
- 7.Harmatz P, et al. Enzyme replacement therapy for mucopolysaccharidosis VI: A phase 3, randomized, double-blind, placebo-controlled, multinational study of recombinant human N-acetylgalactosamine 4-sulfatase (recombinant human arylsulfatase B or rhASB) and follow-on, open-label extension study. J Pediatr. 2006;148:533–539. doi: 10.1016/j.jpeds.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Wilcox WR, et al. Long-term safety and efficacy of enzyme replacement therapy for Fabry disease. Am J Hum Genet. 2004;75:65–74. doi: 10.1086/422366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishnani PS, et al. Recombinant human acid α-glucosidase: Major clinical benefits in infantile-onset Pompe disease. Neurology. 2007;68:99–109. doi: 10.1212/01.wnl.0000251268.41188.04. [DOI] [PubMed] [Google Scholar]
- 10.Dahms NM, Lobel P, Kornfeld S. Mannose 6-phosphate receptors and lysosomal enzyme targeting. J Biol Chem. 1989;264:12115–12118. [PubMed] [Google Scholar]
- 11.Zhao KW, Neufeld EF. Purification and characterization of recombinant human α-N-acetylglucosaminidase secreted by Chinese hamster ovary cells. Protein Expression Purif. 2000;19:202–211. doi: 10.1006/prep.2000.1230. [DOI] [PubMed] [Google Scholar]
- 12.Weber B, Hopwood JJ, Yogalingam G. Expression and characterization of human recombinant and alpha-N-acetylglucosaminidase. Protein Expression Purif. 2001;21:251–259. doi: 10.1006/prep.2000.1361. [DOI] [PubMed] [Google Scholar]
- 13.Rubin LL, Staddon JM. The cell biology of the blood-brain barrier. Annu Rev Neurosci. 1999;22:11–28. doi: 10.1146/annurev.neuro.22.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Grubb JH, et al. Chemically modified beta-glucuronidase crosses blood–brain barrier and clears neuronal storage in murine mucopolysaccharidosis VII. Proc Natl Acad Sci USA. 2008;105:2616–2621. doi: 10.1073/pnas.0712147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubb JH, et al. Infused Fc-tagged beta-glucuronidase crosses the placenta and produces clearance of storage in utero in mucopolysaccharidosis VII mice. Proc Natl Acad Sci USA. 2008;105:8375–8380. doi: 10.1073/pnas.0803715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boado RJ, Zhang Y, Xia CF, Wang Y, Pardridge WM. Genetic engineering of a lysosomal enzyme fusion protein for targeted delivery across the human blood-brain barrier. Biotechnol Bioeng. 2008;99:475–484. doi: 10.1002/bit.21602. [DOI] [PubMed] [Google Scholar]
- 17.Muro S, Schuchman EH, Muzykantov VR. Lysosomal enzyme delivery by ICAM-1-targeted nanocarriers bypassing glycosylation- and clathrin-dependent endocytosis. Mol Ther. 2006;13:135–141. doi: 10.1016/j.ymthe.2005.07.687. [DOI] [PubMed] [Google Scholar]
- 18.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Farokhzad OC, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNamara JO, II, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 23.Chu TC, et al. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 24.Klausner RD, et al. Receptor-mediated endocytosis of transferrin in K562 cells. J Biol Chem. 1983;258:4715–4724. [PubMed] [Google Scholar]
- 25.Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jefferies WA, et al. Transferrin receptor on endothelium of brain capillaries. Nature. 1984;312:162–163. doi: 10.1038/312162a0. [DOI] [PubMed] [Google Scholar]
- 27.van Gelder W, Cleton-Soeteman MI, Huijskes-Heins MI, van Run PR, van Eijk HG. Transcytosis of 6.6-nm gold-labeled transferrin: An ultrastructural study in cultured porcine blood-brain barrier endothelial cells. Brain Res. 1997;746:105–116. doi: 10.1016/s0006-8993(96)01179-1. [DOI] [PubMed] [Google Scholar]
- 28.Lee HJ, Engelhardt B, Lesley J, Bickel U, Pardridge WM. Targeting rat anti-mouse transferrin receptor monoclonal antibodies through blood-brain barrier in mouse. J Pharmacol Exp Ther. 2000;292:1048–1052. [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernstein SE. Hereditary hypotransferrinemia with hemosiderosis, a murine disorder resembling human atransferrinemia. J Lab Clin Med. 1987;110:690–705. [PubMed] [Google Scholar]
- 31.Parton RG, Richards AA. Lipid rafts and caveolae as portals for endocytosis: New insights and common mechanisms. Traffic. 2003;4:724–738. doi: 10.1034/j.1600-0854.2003.00128.x. [DOI] [PubMed] [Google Scholar]
- 32.Lebron JA, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 33.Hanover JA, Beguinot L, Willingham MC, Pastan IH. Transit of receptors for epidermal growth factor and transferrin through clathrin-coated pits. Analysis of the kinetics of receptor entry. J Biol Chem. 1985;260:15938–15945. [PubMed] [Google Scholar]
- 34.Hopkins CR. Intracellular routing of transferrin and transferrin receptors in epidermoid carcinoma A431 cells. Cell. 1983;35:321–330. doi: 10.1016/0092-8674(83)90235-0. [DOI] [PubMed] [Google Scholar]
- 35.Lepelletier Y, et al. Prevention of mantle lymphoma tumor establishment by routing transferrin receptor toward lysosomal compartments. Cancer Res. 2007;67:1145–1154. doi: 10.1158/0008-5472.CAN-06-1962. [DOI] [PubMed] [Google Scholar]
- 36.Kakkis ED, Matynia A, Jonas AJ, Neufeld EF. Overexpression of the human lysosomal enzyme α-L-iduronidase in Chinese hamster ovary cells. Protein Expression Purif. 1994;5:225–232. doi: 10.1006/prep.1994.1035. [DOI] [PubMed] [Google Scholar]
- 37.Tuma PL, Hubbard AL. Transcytosis: Crossing cellular barriers. Physiol Rev. 2003;83:871–932. doi: 10.1152/physrev.00001.2003. [DOI] [PubMed] [Google Scholar]
- 38.Hamilton DH, Turcot I, Stintzi A, Raymond KN. Large cooperativity in the removal of iron from transferrin at physiological temperature and chloride ion concentration. J Biol Inorg Chem. 2004;9:936–944. doi: 10.1007/s00775-004-0592-6. [DOI] [PubMed] [Google Scholar]
- 39.Sando GN, Titus-Dillon P, Hall CW, Neufeld EF. Inhibition of receptor-mediated uptake of a lysosomal enzyme into fibroblasts by chloroquine, procaine and ammonia. Exp Cell Res. 1979;119:359–364. doi: 10.1016/0014-4827(79)90364-1. [DOI] [PubMed] [Google Scholar]
- 40.Fratantoni JC, Hall CW, Neufeld EF. The defect in Hurler's and Hunter's syndromes: Faulty degradation of mucopolysaccharide. Proc Natl Acad Sci USA. 1968;60:699–706. doi: 10.1073/pnas.60.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.