Abstract
The basic helix-loop-helix transcription factor Twist1 plays an essential role in mesenchymal cell populations during embryonic development and in pathological disease. Remodeling of the cardiac outflow track (OFT) into the functionally separate aortic arch and pulmonary trunk is dependent upon the dynamic, coordinated contribution of multiple mesenchymal cell populations. Here, we report that Twist1−/− mice exhibit OFTs that contain amorphic cellular nodules within their OFT endocardial cushions. The nodular mesenchyme expresses the related bHLH factors Hand1 and Hand2, but reduced levels of the normal cushion marker Periostin. Lineage mapping confirms that nodule cells are exclusively of cardiac neural crest origin (cNCC), and are not ectopic cardiomyocytes or smooth muscle cells. These studies also reveal a delay in cNCC colonization of the OFT cushions. Furthermore, these mapping studies uncover nodules in the pharyngeal arches, and identify Twist1−/− neural crest cell defects within the dorsal neural tube, which exhibits an expanded domain of Wnt1-Cre-lineage marked cells. Together, these data support a model where Twist1 is required both for proper cNCC delamination, and for emigration from the dorsal neural tube and along cNCC migration pathways. Within the Twist1−/− neural crest cell populations that do emigrate to the OFT, a Hand-expressing subpopulation displays defective maturation, tracking, and, presumably, cell-cell adhesion, further compromising cNCC morphogenesis.
Keywords: Twist1, heart development, neural crest, Outflow track, mouse
Introduction
Development of the OFT from a simple tube to the highly structured junction between the ventricles and great arteries entails an array of tightly orchestrated morphological modifications. Reflective of its inherent developmental complexity and indispensability for proper cardiac function, abnormalities of the OFT and the tissues that govern its morphogenesis account for roughly one third of congenital heart defects (Clark, 1996; Ferencz et al., 1985; Rothenberg et al., 2003). During OFT development, mesenchymal cells derived from disparate sources accumulate within the OFT cushions, the progenitors of the OFT septum and valves. Integral to our understanding of the etiology of cardiac OFT defects is a clarification of the molecular mechanisms which enable these mesenchymal cell populations to emigrate into the lumen of the cardiac jelly of the OFT cushions, and which govern their differentiation upon arrival.
The OFT originates as a cardiomyocyte-lined vascular channel through which blood exits the primitive ventricle and fills the aortic sac (AS). First, the OFT cushions, which initially form as a deposit of extracellular matrix termed cardiac jelly, intervenes the conotruncal myocardium and the endocardium. The cardiac jelly is then progressively invaded by mesenchymal cells, some of which are cxardiac neural crest cells (cNCCs) (Snider et al., 2007; Stoller and Epstein, 2005), and the remainder of which are endocardial cells that have undergone epithelial-to-mesenchymal transition (EMT). The OFT cushions ultimately divide the OFT into two separate vessels, the aorta and the pulmonary trunk, and contribute to the subvalvular component of the interventricular septum (IVS). This process of septation establishes the division between the systemic and pulmonary circulation (Conway et al., 1997; Jiang et al., 2000).
The cNCC are essential for the patterning of the pharyngeal arch arteries (PAAs) as they are remodeled to form the aortic arch and contribute directly to the smooth muscle component of these tissues (Creazzo et al., 1998; Hutson and Kirby, 2003). Furthermore, proper cNCC contribution to and behavior within the OFT cushions is required for their morphogenesis into the progenitors of the conotruncal septum (Jiang et al., 2000; Qayyum et al., 2001). Additionally, the cNCCs ventral to the pharyngeal pouch form the OFT septum, a wedge that divides the aortic sac into the proximal origin of the aorta and pulmonary trunk (Waldo et al., 1998). Thus, septation and alignment of the developing cardiac OFT are contingent upon the contribution of cNCCs.
A diverse array of transcription factors is employed to mediate the local morphogenetic cues and guide the developing OFT through its intermediate stages (Firulli and Thattaliyath, 2002). Studies indicate that basic helix–loop–helix (bHLH) transcription factors contribute to a number of these processes (Firulli, 2003). bHLH factors conform to an evolutionarily conserved secondary structure, comprised of a DNA-binding and dimerization motif containing a short stretch of basic amino acids, followed by a pair of amphipathic α-helices intervened by a loop of varying length (for review, see Massari and Murre, 2000). These α-helices mediate protein-protein interactions between bHLH factors, enabling them to bind DNA, via their basic domains, at canonical consensus sites termed E-boxes (CANNTG) and to modulate transcription as either homo- or heterodimers.
Members of the Twist sub-class of bHLH transcription factors have been shown to exhibit broad dimer-partner choices. An evolutionarily conserved phosphoregulatory circuit modulates, in part, formation of these transcriptional complexes (Firulli et al., 2003; Firulli et al., 2005; Firulli et al., 2007). Twist1 displays a broad mesodermal domain of expression (Fuchtbauer, 1995; Glackin et al., 1994; Stoetzel et al., 1995), is essential for the development of multiple organ systems, and has been correlated with EMT during tumor cell metastasis (Yang et al., 2004). Within the developing mammalian heart, Twist1 is expressed in the endocardium of the AV canal, where it has been proposed to induce EMT (Ma et al., 2005).
The Twist1-related factors Hand1 and Hand2 are also required for heart development. Knockout studies show that Hand2 is necessary for the formation of the presumptive right ventricle and PAAs (Srivastava et al., 1997). Conditional ablation of Hand1 within the myocardial lineage confirmed its contribution to the development of the left ventricle, and revealed a role for Hand1 during AV valvulogenesis (McFadden et al., 2005).
Here, we demonstrate that Twist1 is expressed within components of the OFT and its progenitors in a manner partially overlapping the expression domains of its putative dimerization partners. We further demonstrate that cNCC derivatives are abnormal in Twist1−/− mutants, and that loss of Twist1 function causes delayed and diminished cNCC emigration to, and aberrant cNCC morphology within the developing OFT. Importantly, we note that these defects are restricted to the subpopulation of cNCCs, which express the potential Twist1 dimer partners Hand1 and Hand2. Finally, we correlate these cNCC defects with an expansion of Wnt1-marked cells within the dorsal neural tube of Twist1−/− embryos that suggests a trapping of NCC cells resulting in thickened neural tubes as compared to wild-type. As a whole, these data provide insight into the transcriptional regulation of cNCC development and novel understanding of the cellular processes underlying NCC and OFT morphogenesis.
Materials and Methods
Mouse strains
The targeting and PCR-based genotyping strategies for the Twist1tm1Bhr, Hand1tm1Eno and Hand2tm1Eno null alleles have been previously described (Chen and Behringer, 1995; Firulli et al., 1998; Srivastava et al., 1997). For cell lineage trace analyses, either the Wnt1-Cre (Danielian et al., 1998) or R26R allele (Soriano, 1999) were introduced into the Twist1 mutant background to generate <Cretg(+);Twist1+/−> and <R26R+/−;Twist1+/−> offspring, respectively. These mice were subsequently intercrossed to generate embryos for analysis.
Histology
Embryos (E9.5 – E11.5) were fixed in 4% paraformaldehyde, dehydrated through a methanol gradient and embedded in paraffin. Embryos were sectioned at 7µm unless otherwise noted. Nuclear Fast Red (Vector Laboratories) staining was performed as per manufacturer’s instructions. Hematoxylin and Eosin (H&E) stain was performed exactly as described (Conway et. al. 2000). Propidium iodide (PI) staining was performed using 50µg/mL PI in 2X SSC. A minimum of 3 viable embryos (assayed via the presence of a heart beat) per genotype were used for these and all subsequent analyses.
In situ hybridization
Digoxygenin labeled section in situ hybridizations were carried out using established protocols on 10µm paraffin sections (Chen et al., 2004; Nagy, 2003) using T7, T3 or SP6 polymerases (Promega) and DIG-Labeling Mix (Roche). Sense and antisense digoxygenin-labeled riboprobes were transcribed for Hand1 and Hand2, PlexinA2 (provided by Jonathan Epstein), Twist1, Periostin, Msx2, Wnt1, Pax3, Shh (provided by Simon Conway).
X-gal staining
X-gal staining of embryos for β-galactosidase activity was performed essentially as previously described (Firulli et al., 1998).
BrdU, TUNEL, and immunohistochemistry
Pregnant mothers were injected intraperitonially with 100mg/g body weight of BrdU (Zymed Laboratories, Inc.) 1 hour prior to embryo dissection and processing as described above. BrdU immunodetection was carried out exactly as described (Conway et al., 2000). TUNEL assays were performed using Apoptag Cell Death Detection Kit (Chemicon) as per manufacturer’s instruction. αSMA and Periostin antibody immunodetection was carried out exactly as described (Rios et al., 2005).
Cell counts and statistics
Cell counts were preformed manually from 7µm sections of Twist1 mutants and WT littermates immediately caudal to the otic placodes. Sections were nuclear stained with either propidium iodide or hematoxylin. The exact dorsoventral midline was demarcated in these NTs, and cells in both the dorsal and ventral domains were counted. To account for variance in total cell number due to embryo stage, the number of cells in each Twist1 mutant NT was normalized against a somite-matched WT littermate. Statistical significance was confirmed via one tailed student’s t-Test assuming equal variances.
Results
Twist1 is required for normal mesenchymal cell contribution to and behavior within the PAs and the OFT cushions
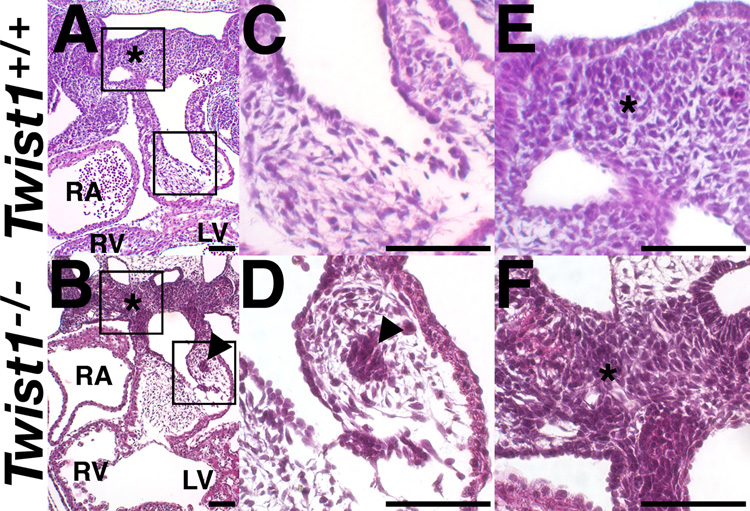
To assess a potential requirement for Twist1 in heart development, we performed histological analyses of Twist1−/− mutants at E11.5. Twist1−/− mice die at E11.5 and exhibit exencephaly and hypoplastic limb buds (Chen and Behringer, 1995). However, although cardiac expression of Twist1 has recently been reported (Ma et al., 2005), no cardiac phenotypes have been described in these mutants. Examination of H&E stained transverse sections revealed that portions of the mesenchyme within the Twist1−/− mutant OFT cushion are abnormally condensed into nodules of cells (compare Fig. 1C, D). These nodules are confined to the cNCC populated truncal cushions and are absent from the EMT-derived conal cushions. Myocardium in Twist1−/− mice is not significantly different from that of wild-type littermates (Fig. 1).
Figure 1. Defects of the OFT cushions and PAs, but not AV cushions in Twist1−/− mutant embryos.
(A–F) H&E stained transverse sections of somite-matched E11.5 embryos. (C, F) High power magnification of the insets in A and B. Arrowheads denote mesenchymal nodules. Asterisks denote pharyngeal mesenchyme. LV, left ventricle; RA, right atrium; RV, right ventricle. Scale bars = 100µm.
We further examined the caudal PA mesenchyme. Proximal to the OFT, the PA mesenchyme appears abnormally condensed when compared with wild-type embryos (Fig. 1E, F, asterisk). PA mesenchyme within Twist1−/− embryos appears more dense and irregular than wild-type tissues and has a grossly similar appearance to the nodule clusters that appear in the OFT.
Additionally, we examined the mesenchyme of the AV cushions, which is derived solely from an endocardial lineage. Gross comparison of AV cushions of wild-type and Twist1−/− mice show no significant differences in structure (data not shown). Given that the novel defects observed in Twist1−/− embryos are more pronounced in the truncal OFT cushions and PAs; not the AV cushions, this suggests that endocardium does not require Twist1 for EMT and normal AV cushion formation, and implicates normal behavior of the mesenchyme present in the PAs that populates the OFT as likely to be Twist1-dependent. We noted no observable myocardial or endocardial phenotypes within the hearts of Twist1−/− embryos (data not shown). We therefore initiated molecular characterization of these mesenchymal cell populations in wild-type and in Twist1−/− mice.
Twist1, Hand1 and Hand2 are expressed within tissues that contribute to the OFT in partially overlapping patterns
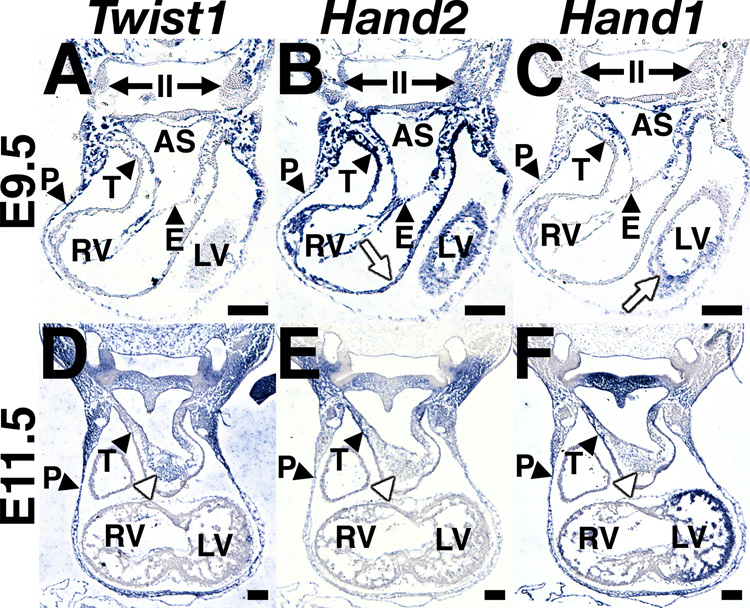
Given its established role in NCC function and EMT, we investigated the potential for Twist1-mediated regulation of OFT cushion development. We sought to create a detailed profile of Twist1 expression relative to that of Hand1 and Hand2 within the developing OFT cushions and its progenitor cell populations. We performed DIG-labeled in situ hybridization for Twist1, Hand1 and Hand2 upon adjacent sections of wild-type embryos at E9.5 and E11.5 (Fig. 2A–F). We observed that Twist1, Hand1 and Hand2 are coexpressed in the body wall overlying the pericardial cavity at all three developmental stages, while both Twist1 and Hand2, but not Hand1 are expressed within the endocardium. As previously reported, Hand1 and Hand2 are expressed in myocardial cell populations (white arrows, Fig. 2B, C), most strongly at the truncus Twist1 expression is not detectable within myocardium (Fig. 2A, D).
Figure 2. Twist1 and Hand gene expression is partially overlapping in the OFT.
(A–F) DIG-labeled in situ hybridization upon serial transverse sections of E9.5 (A–C) and E11.5 (D–F) embryos showing Twist1 (A, D), Hand2 (B, E) and Hand1 (C, F) expression. II, second pharyngeal arch; AS, aortic sac; E, endocardium; LV, left ventricle; P, pericardium; RA, right atrium; RV right ventricle; T, truncus; white arrows, myocardium; white arrowhead, OFT cushion mesenchyme. Scale bars = 100µm.
Twist1 displays a broad expression domain, including the mesenchymal cells occupying all pharyngeal arches (PAs) as well as the dorsal aorta (Fig. 2A). In the PAs more proximal to the OFT, staining is weaker, reflecting a reduction either in Twist1 expression levels, or in number of Twist1 expressing cells. At E9.5, it is undetectable at the distal lip of the truncus, which, at this stage, would include myocardium derived from the second heart field, endothelium, and cNCC-derived mesenchyme migrating into the lumen of the OFT cushions (Fig. 2A). Hand2 is also expressed in the ventral PA mesenchyme, overlapping with the domains of low-level Twist1 expression (Fig. 2A, B). Additionally, Hand1 and Hand2 are expressed strongly within the distal lip of the truncus (Fig. 2B, C), cells in which Twist1 expression is absent (Fig. 2A). However, at E11.5 Twist1 expression is strongly detected uniformly throughout the mesenchymal cells of the OFT cushions (Fig, 2D, white arrowhead), ostensibly representing a mixed populace of both cNCC and endocardial-derived cells. These cells do not express Hand2 at detectable levels (Fig. 2E), but some of these cells do express Hand1 (Fig. 2F). Given that Twist1−/− OFTs display both normal and abnormally condensed mesenchymal cell populations, and both Hand1 and Hand2 are tightly spatiotemporally regulated in these affected cells, we assessed a potential aberrant expression of these two genes within the OFTs of Twist1−/− mice.
Abnormal mesenchyme within the OFT cushions of Twist1−/− mice expresses Hand1 and Hand2
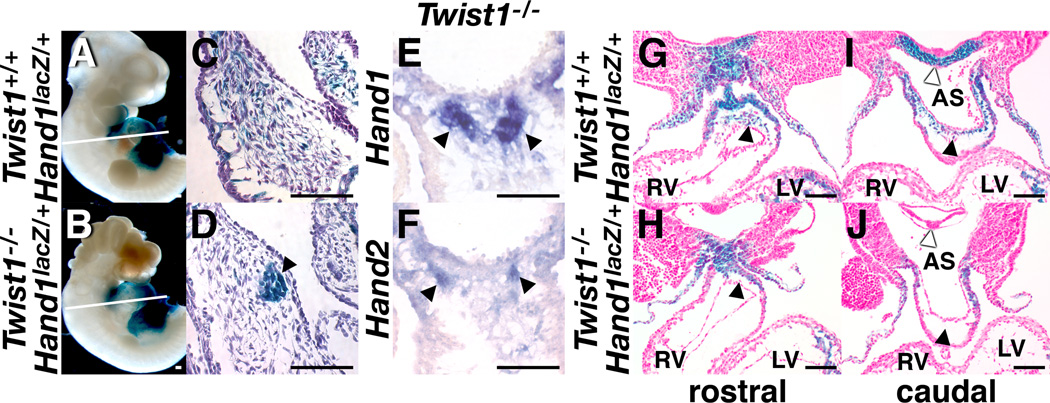
As the majority of the mesenchymal cells within the PAs that emigrate into the OFT and differentiate into smooth muscle are cNCC derivatives, we predict that Twist1 is necessary for proper cNCC development within the OFT. Hand1 and Hand2 are post-migratory markers of cNCCs (Srivastava et. al. 1997) and interestingly, are also expressed within the cNCCs in a pattern partially overlapping that of Twist1 (Fig. 2). mRNA expression analysis shows that nodular cells within the OFT cushions of Twist1−/− mice express both Hand1 and Hand2, whereas the non-nodular mesenchyme expresses less of each Hand-factor (Fig. 3E, F). To validate this observation, we took advantage of our Hand1lacZ mice, crossing this allele onto the Twist1−/− background. Given that Twist1 and Hand factors can form transcriptional complexes and that in the developing limb genetic interactions between Twist1 and Hand2 are evident (Firulli et al., 2003; Firulli et al., 2005; Firulli et al., 2007), we carefully looked for changes in the Twist1−/− OFT phenotype when presented on a Hand1 haploinsufficient background. In all embryos examined (8 total), no phenotypic differences were observed (compare Fig 1D with Fig. 3D, E). Thus, we conclude that relative gene dosages of Hand1 and Twist1 are not critical for OFT development. Analysis of Hand1lacZ expression shows that the nodular cells within the Twist1−/− mice express Hand1 whereas, in wild-type mice, these Hand1lacZ expressing cells are evenly intermixed with a population of Hand1lacZ negative mesenchymal cells, validating in situ analysis (Fig. 3C–F). In situ hybridization of the cNCC marker PlexinA2 confirms that at this level of the OFT the majority of the cells are migratory cNCC (data not shown). Together, these data show that the nodular mesenchymal cells within the OFT of the Twist1−/− embryos robustly express both the post-migratory NCC markers Hand1 and Hand2. Coupled with the observation that the unaffected mesenchyme within the OFT of these mutants expresses Hand1 and Hand2 at lower levels or, more likely, within fewer cells, these data indicate that Hand1 and Hand2 expressing cells are selectively affected by a loss of Twist1.
Figure 3. Defective Hand-factor-expressing mesenchyme in Twist1−/− mutants.
Whole mount (A, B) and transverse sections (C, D) of X-gal-stained, somite-matched Twist1+/+;Hand1lacZ and Twist1−/−;Hand1lacZ embryos at E11.5 showing lacZ expression localized to the mesenchymal nodules (black arrowheads) of the Twist1−/− OFT. White lines in A and B denote plane of section. (E, F) DIG-labeled in situ hybridization upon serial transverse sections of somite-matched E11.5 embryos showing Hand1 (G) and Hand2 (H) expression in the mesenchymal nodules (black arrowheads) of the Twist1−/− OFT. (G–J) transverse sections of X-gal-stained Twist1+/+;Hand1lacZ and Twist1−/−;Hand1lacZ embryos at E10.5 showing an absence of lacZ expressing cells in the OFT (black arrowheads) and pharyngeal mesenchyme (white arrowheads) of Twist1−/− mutants. AS, aortic sac; LV, left ventricle; RV, right ventricle.
We next sought to determine whether these mesenchymal morphology defects were associated with cell migration defects. We therefore examined the behavior of the Hand1lacZ-expressing mesenchymal subpopulation at E10.5. At this stage, we observed a noticeable decrease in the number of Hand1lacZ-expressing cells that have moved into the OFT (black arrowheads, Fig. 3G, H) and ventral pharynx (white arrowheads, Fig. 3I, J), suggesting a delay in cNCC migration into these structures. Taken together, these data support that a novel cardiac defect is present in Twist1−/− mice that effects spatiotemporal mesenchymal-cell migration. Moreover, the defective mesenchymal cell population is marked by the expression of Hand1 and Hand2.
Defective mesenchyme in the Twist1−/− OFT is not composed cardiac or smooth muscle and exhibits a lack of mesenchymal maturation
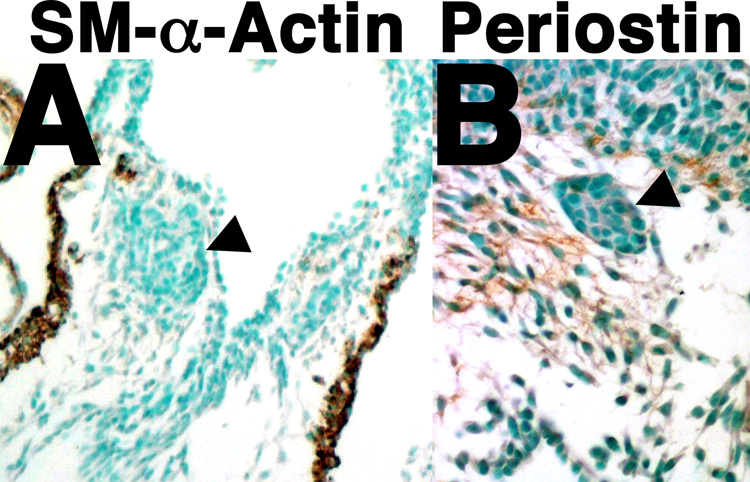
cNCC-derived ectomesenchyme localizes to the PAs and OFT cushions, a portion of which ultimately differentiates and contributes to the vascular smooth muscle of the mature aortic arch (Creazzo et al., 1998; Firulli and Conway, 2004). As histological analyses indicate that mesenchymal components of both the PAs and OFT cushions are abnormal in Twist1−/− mutant embryos, we employed immunohistochemistry to look at the smooth and early cardiac muscle marker SM-α-Actin (Smart et al., 2002), and the mesenchymal maturation marker and a putative downstream target of Twist1, Periostin (Lindsley et al., 2007) to assess whether the defective mesenchyme within the Twist1−/− OFTs contain ectopic cardiac muscle and/or are able to mature and to ultimately differentiate. Results show that although SM-α-Actin is robustly expressed within the cardiomyocytes of the Twist1−/− OFT and more weakly within the ostensibly normal OFT cushion mesenchyme, it is undetectable within the abnormal Twist1−/− mesenchymal condensations (arrowhead, Fig. 4A) indicating that nodules are not likely to be composed of ectopic cardiomyocytes or differentiated smooth muscle. Periostin, which is broadly expressed within the OFT cushions during cushion maturation, is reduced within the OFT nodules (arrowhead, Fig. 4B) supporting the idea that these nodular cells are molecularly abnormal. Therefore, these data suggest that the nodular cells within the Twist1−/− OFT are not ectopic cardiomyocytes, but, rather, immature cNCCs.
Figure 4. Defects of mesenchymal cell maturation in Twist1−/− mutant embryos.
Immunohistochemical staining on transverse sections of E11.5 Twist1−/− embryos show an absence of SM-α-actin (A) and periostin (B) protein within the abnormal OFT mesenchymal nodules.
Wnt1-Cre lineage mapping indicates that cNCC cells have defective migration
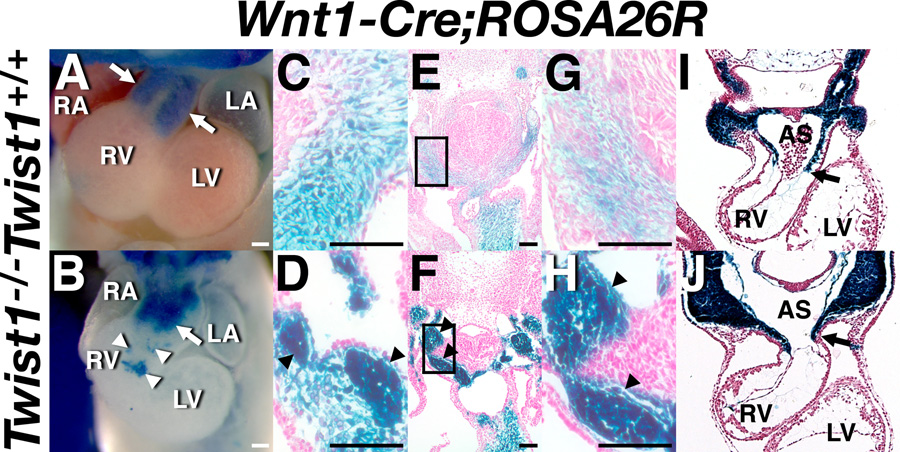
Wnt1-Cre-mediated recombination of the R26R allele marks all NCCs from the point at which they delaminate from the neural tube and initiate migration throughout the embryo (Chai et al., 2000). Taking advantage of real-time expression of Hand1lacZ within the pathologically defective mesenchymal cells of the Twist1−/− OFT cushions allows us to examine these Hand1lacZ expressing cells in the context of all cNCCs within the OFT. Wnt1-Cre-mediated R26R expression in wild-type and Twist1−/− mice was first examined at E11.5 (Fig. 5A–H). In the wild-type embryo, the OFT cushions have segregated into the conotruncal ridges that will ultimately fuse to form the septum dividing the aorta and pulmonary trunk (arrows in Fig. 5A). In the Twist1−/− mutant, these ridges are poorly defined (arrow in Fig. 5B). The aberrant morphology of this cushion is consistent with other models of cNCC dysfunction (Epstein et al., 2000; Kaartinen et al., 2004), although the early embryonic lethality of Twist1−/− mutants precludes characterization of potential persistent truncus arteriosis (PTA) or double outlet right ventricle (DORV) defects. Additionally, satellite clusters of cNCCs are evident proximal to the RV (white arrowheads in Fig. 5B) and within the pericardium (arrowheads in Fig. 5B) in Twist1−/− but not wild-type embryos, suggesting that cNCC migration paths are altered in the absence of Twist1. These path-finding defects are consistent with previous studies of Twist-deficient cranial NCCs (Soo et al., 2002). Further histological analyses of these embryos reveal that, within the OFT cushions, both the abnormally condensed and phenotypically normal mesenchyme are of cNCC origin (arrowheads in Fig. 5C, D). Given that the large majority of Hand1LacZ positive cells contribute to the nodules within the OFT cushions, whereas the phenotypically normal cushion mesenchyme is largely Hand1LacZ negative but still of cNCC origin, it appears likely that Hand factor-expression is defining a subpopulation of cNCC that are sensitive to Twist1 loss-of-function.
Figure 5. Defective cNCC mesenchyme in Twist1−/− mutants.
Whole mount (A, B) and transverse sections (C–J) of X-gal-stained, somite-matched Twist1+/+;Wnt1-Cre(+);R26R (C, E, G, I) and Twist1−/−;Wnt1-Cre(+);R26R (D, F ,H, J) embryos at E11.5 (A-H) and E9.5 (I, J). White arrows in A and B denote the OFT cushions. White arrowheads in A and B denote mis-emigrating cNCCs. LacZ expression is apparent in the mesenchymal nodules of the Twist1−/− OFT and PAs as well as the NCC-derived truncal endocardial cushions (black arrowheads in D, F, H). AS, aortic sac; LA, left atrium; LV, left ventricle; RA, right atrium; RV right ventricle; Scale bars = 100µm.
We further verified NCC migration defects, examining Wnt1-Cre;R26R-expressing cNCCs at E9.5 (Fig. 5I, J). At this stage, clusters of lacZ-positive cells were evident within the pericardium and endocardium of the Twist1−/− mutants (data not shown), confirming cNCC path-finding defects. Additionally, cNCC emigration was delayed in entering the OFT cardiac jelly (compare position of arrows, Fig. 5I, J). Together, these data support the hypothesis that Hand-factor expression delineates a specific subpopulation of cNCCs, which are defective in their maturation, emigration, path-finding, and, presumably, adhesion, in the absence of Twist1 expression.
Additional histological analyses of cNCC lineage mapping in both wild-type and Twist1−/− mice confirms that cNCCs are also present as nodular condensations within the PAs (Fig. 5E–H) and that these nodular cNCCs appear to emigrate as such into the OFTs of Twist1−/− mice. This suggested to us that these condensations might appear more proximal to the neural tube. In order to further explore the etiology of these Twist1-mediated OFT defects, we next examined cNCCs proximal to the neural tube to pinpoint the source of the nodule formation and defective migration behavior.
Loss of Twist1 results in an expansion of NCC ventrally along the neural tube
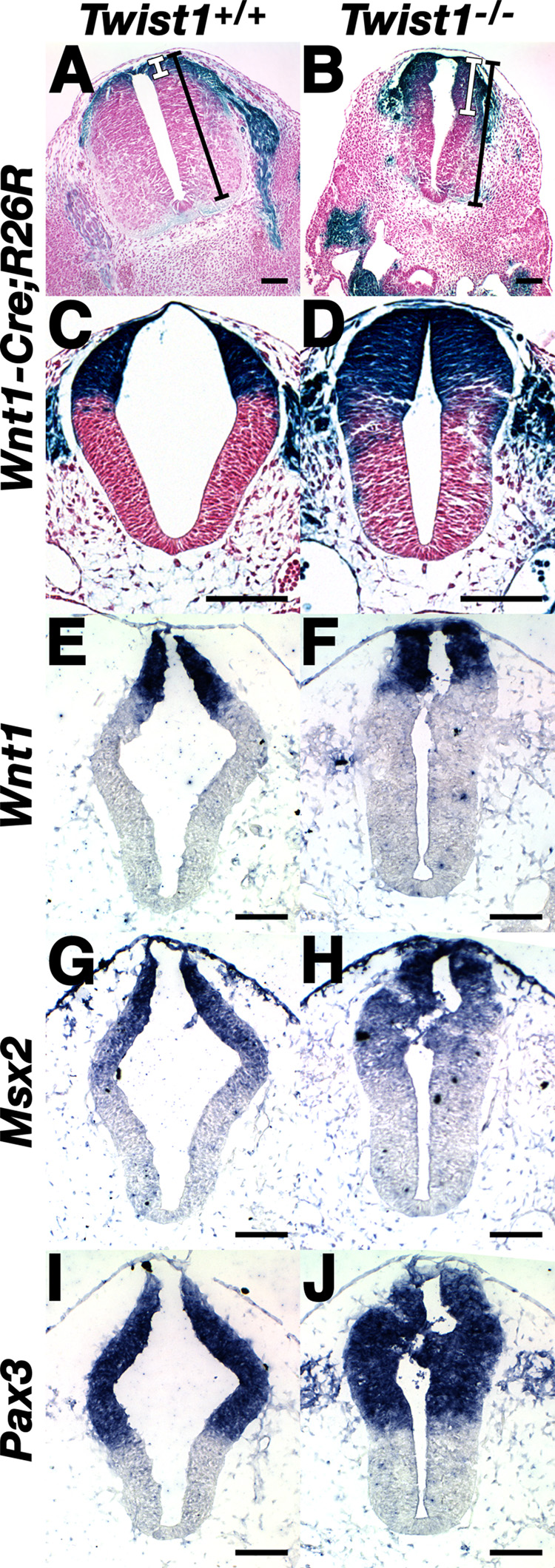
In order to better define the origins of observed cNCC defects in Twist1−/− embryos, we examined E9.5–E11.5 wild-type and Twist1−/− neural tubes marked via the Wnt1-Cre;R26R alleles (Fig. 6). As expected, at E11.5 expression of Wnt1-Cre marks the dorsal lip of the neural tube corresponding to the domain from which the cNCCs have undergone EMT and emigrated towards the PAs (Fig. 6A). Surprisingly, analysis of transverse sections shows a clear ventral expansion of lacZ-positive cells in the Twist1−/− neural tubes (compare white bars in Fig. 6A, B). This expansion is less apparent in the more caudal neural tube (data not shown), consistent with the increasingly pronounced defects within the more rostral neural tube of Twist1−/− mutants (Chen and Behringer, 1995). At E9.5, dorsoventral expansion is less apparent; however, the Wnt1-Cre;R26R domain is larger owing to an obviously thicker neural tube in the Twist1−/− mutant (compare Fig. 6C, D). These results support the idea that cNCC delamination from the neural tube may be inhibited/delayed in Twist1−/− mutants, and as embryonic development proceeds, cNCC lineage mapping renders this altered delamination more apparent.
Figure 6. Expansion of the Wnt1-Cre-expressing lineage in the rostral neural tube Twist1−/− mutants in the absence of DV patterning defects.
(A, B) Transverse sections of X-gal-stained, somite-matched Twist1+/+;Wnt1-Cre(+);R26R and Twist1−/−;Wnt1-Cre(+);R26R embryos at E11.5 (A, B) and E9.5 (C, D) showing expansion of lacZ expressing domain (white bars) in the dorsal neural tube (compare to total dorsoventral width of neural tube, black bars). E9.5 dorsal Wnt1-Cre expansion is less apparent in contrast to increased thickness of the neural tube (compare C & D). (E–J) DIG-labeled in situ hybridization upon transverse sections of somite-matched E9.5 embryos showing Wnt1, Msx2, and Pax3 expression in wild-type (E,G, I) in Twist1−/− mutants (F, H, J). Scale bars = 100µm.
To determine whether this expansion of the Wnt1 lineage represents a dorsalization of the Twist1−/− neural tube, we examined mRNA expression of Wnt1, Wnt3a (Ikeya et al., 1997), Shh (Munsterberg et al., 1995), Pax3 (Conway et al., 1997) and Msx2 (Kwang et al., 2002), which delimit specific dorsal/ventral domains of the developing neural tube. Although expression of these markers appears unaffected in regards to dorsal-ventral neural tube patterning within the Twist1−/−, expression of these markers is, as a whole, more robust, reflecting the increase in neural tube thickness in Twist1−/− embryos when compared to wild-type controls (Fig. 6E–J and data not shown).
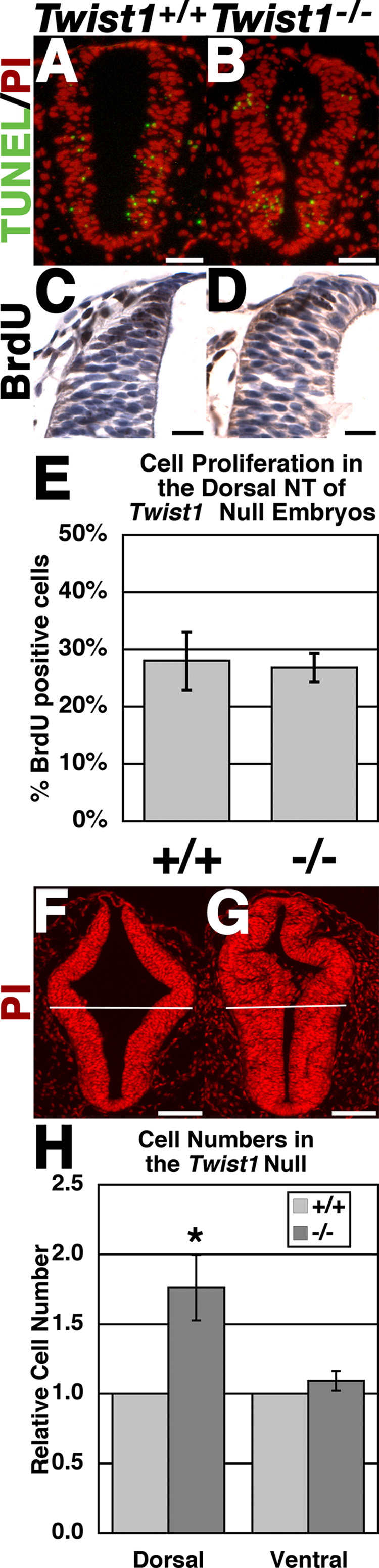
To determine if the observed neural tube thickening was a result of decreased cell death or increased cell proliferation, we performed both TUNEL and BrdU and analyses upon Twist1−/− mutants at E9.5 (Fig. 7). Extensive mesenchymal apoptosis has been reported in Twist1−/− embryos within the cranial mesenchyme, first branchial arch and the somite at E9.5; however, apoptosis in the dorsal neural tube was not reported (Ota et al., 2004). We observe no significant differences in the number of apoptotic cells within the neural tubes of wild-type and Twist1−/− embryos (Fig. 7A, B). Similarly, the number of BrdU positive nuclei is not significantly different between wild-type and Twist1−/− embryos, indicating that increased proliferation is not likely involved in the expansion of Twist1−/− neural tubes (Fig. 7C–E). To validate that the total number of cells within the neural tube is altered in the absence of Twist1, transverse sections of wild-type and Twist1−/− embryos at E10.0 were nuclear stained (Fig. 7F, G). The neural tubes were divided along the dorsoventral midline (white lines, Fig. 7F, G), and the number of nuclei manually counted. Quantification of total cell number with the neural tube shows a nearly 2-fold increase in the number of cells within the Twist1−/− dorsal neural tube, whereas ventral cell counts are similar between wild-type and Twist1−/− embryos (Fig. 7H). The increased cell number in the absence of decreased cell death or increased proliferation within the dorsal neural tubes of E9.5 Twist1−/− mice, in conjunction with the expanded domain of Wnt1-Cre marked cells within the neural tube observed at later developmental stages, is consistent with the hypothesis that NCC become trapped within the neural tube of Twist1−/− mice, which results in poor delamination, altered cell adhesion, and delayed cNCC emigration.
Figure 7. Cell death and cell proliferation is unaltered but cell number is increased within the dorsal neural tube of Twist1−/− mutant embryos.
(A, B) TUNEL green and propidium iodide (PI; red) staining of somite-matched E9.5 wild-type (A) and Twist1−/− (B) neural tubes. (C-E) α-BrdU immunohistochemical staining on transverse sections of somite-matched E9.5 wild-type (C) and Twist1+/+ embryos (D) quantified and represented as a percentage of the total number of cells within the dorsal neural tube (E). (F-H) 10µM sections of somite-matched wild-type (F) and Twist1−/− (G) neural tubes axially matched immediately caudal to the otic placodes stained with propidium iodide (PI; red). Neural tubes were bisected along the dorsoventral midline (white lines). Total cell counts were made of dorsal vs. ventral neural tubes of each respective genotype and represented graphically (H). Scale bars (A, B) = 100µm, (C, D) = 20µm.
Discussion
Twist1 is necessary for cNCC migration, maturation and morphology
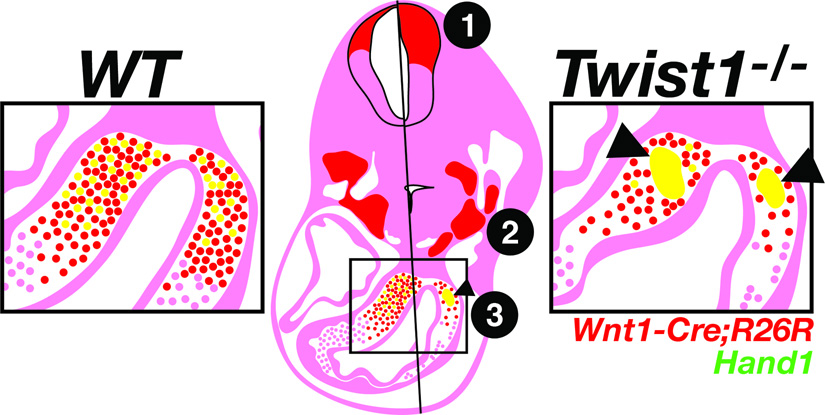
We have shown that mice null for Twist1, in addition to exencephaly and hypoplastic limbs, also exhibit cardiac OFT defects that result from defects in cNCC emigration, maturation, and, presumably, cell adhesion. These phenotypes can be traced back to the cNCC origins at the dorsal neural tube where an expansion of Wnt1-Cre expressing cells is observed, suggesting that the cNCCs may be partially trapped from emigrating out of the neural tube, and those that do escape exhibit adhesion, emigration and maturation defects. These phenotypes are summarized in Figure 8.
Figure 8. Schematic representation of the NCC phenotypes associated with a loss of Twist1.
Phenotypes are depicted for the Twist1−/− (left) compared to the wild-type (right) at E11.5. 1) Denotes expansion of the Wnt1-Cre lineage (red) in the dorsal neural tube. 2) Denotes abnormal condensation of the mesenchyme within the pharyngeal arches. 3) Denotes aggregation of the Hand1-expressing subpopulation of cNCCs (yellow) in the OFT cushions.
Morphologically affected cells within the OFTs of Twist1−/− mutants selectively express Hand1 and Hand2 at E11.5. Given the dynamic expression profiles of these two genes described in Figure 2, we may interpret these results to indicate that Hand-factors are broadly expressed in cNCCs occupying the PAs and dorsal lip of the OFT at E9.5. As development proceeds, the domains of Hand1 and Hand2 are restricted, and persist solely within a subset of these cells in the OFT at E11.5. It is these cells, which appear to be most affected by a loss of Twist1 at this stage. Alternatively, Hand-factor expression may be restricted to early-migrating cNCCs, the progression of which appears delayed in Twist1−/− mutants. As these cNCCs are presumably intermixed with ostensibly unaffected late-migrating cNCCs within the E11.5 OFT, this would explain the difference in phenotype evident between Hand-factor expressing and non-expressing cells. Differentiation between these two possibilities would necessitate detailed lineage trace analyses employing NCC-specific Hand1- or Hand2-Cre alleles. In either case, these observations raise the intriguing possibility that Hand-factors ultimately mark a distinct subpopulation of cNCCs, which may contribute to specific cardiac tissues as the OFT matures.
Mesenchymal cell invasion of OFT cushions in Twist1−/− mutants is delayed. As Twist1−/− cNCCs within the PAs is abnormal and condensed, the observed delay of mesenchymal cell invasion of the cushions may reflect altered cNCC adhesive properties impeding cell emigration. The presence of Wnt1-Cre marked cells in ectopic locations within the heart further shows that Twist1−/− cNCC cells have compromised ability to accurately track to their intended destinations. These migration phenotypes are consistent with the defects in cranial NCC migration that were previously observed in relation to the exencephalic phenotype (Chen and Behringer, 1995; O'Rourke and Tam, 2002).
Twist1−/− defective mesenchymal cells within the OFT fails to upregulate a marker of maturation Periostin and are not ectopic cardiomyocytes. Previous studies have reported that cranial NCCs fail to differentiate to osteogenic and myogenic lineages in Twist1−/− mutants (Soo et al., 2002). Our data suggest that cNCCs also require Twist1 function to progress through their developmental programs.
In contrast, the AV cushions, which are derived solely from endocardium that has undergone EMT, appear normal in Twist1−/− mutants, and express at least two expected mesenchymal markers, Periostin and Tbx20 in a manner indistinguishable from wild-type mice (data not shown). This further supports the conclusion that Twist1−/− mutant cardiac defects are cNCC in origin and that, although Twist1 has been shown to be a key player in mediating EMT, its absence is not required for the EMT of endocardial cells. Perhaps functional redundancy or synergy with Hand2 or another Twist family member that is expressed in the endocardium is at play. Hand1 seems unlikely to play such a role as it is not expressed at detectable levels in endocardium (Fig. 2C, F) and its haploinsufficiency does not noticeably alter the Twist1−/− cNCC phenotype. We have, however, previously shown genetic and functional interactions between Twist1 and Hand2 during limb development and perturbation of this antagonistic relationship can cause SCS (Firulli et al., 2005). It would be interesting to look for genetic interactions between Twist1 and Hand2; however Hand2 null mice die at E9.5 (Srivastava et al., 1997), precluding analysis without employing a conditional Hand2 allele.
cNCCs fail to segregate normally in Twist1−/− embryos. Previous chimeric analyses demonstrate that Twist1−/− cells within the head mesenchyme fail to segregate normally with wild-type mesenchymal cells (Chen and Behringer, 1995). It has been proposed that these abnormal cells are cranial NCCs that have failed to assume a mesenchymal character despite their ability to emigrate from the neural tube. These data echo the abnormal cell interactions we observe in the PA mesenchyme and OFT. At the level of the neural tube corresponding to the presumptive cNCCs, it is unclear whether the cNCCs delaminate as nodules. However, within the head, we observe the cranial NCCs delaminating from the dorsal lip as nodules directly proximal to the neural tube (data not shown). We feel it is likely that the delaminating cNCCs exhibit similar behavior. As a whole, these observations raise the intriguing possibility that the potential adhesion, emigration, tracking, and maturation defects of Twist1−/− cNCCs can be attributed to abnormal delamination from the dorsal neural tube, although we cannot discount the possibility that these cNCCs have been mis-specified to a neuronal cell fate. Importantly, Twist1 expression is not detected within NC stem cells or the dorsal neural tube (Fuchtbauer, 1995; Glackin et al., 1994; Stoetzel et al., 1995, our data; Wolf et al., 1991), but it has been proposed that ECM plays a significant role in cell recognition, initiation and regulation of NCC migration (Coles et al., 2007; Lofberg et al., 1985). Given that Twist1 has been reported to regulate a number of ECM genes in both development and cancer, this supports the hypothesis that the initial effects of the Twist1−/− NCC phenotype are non-cell autonomous.
Twist1 function has been implicated in establishing a boundary between cranial NCCs and mesoderm during formation of the coronal suture of the skull vault (Merrill et al., 2006). The data presented here are consistent with a role for Twist1 function in boundary formation. Normal OFT cushion mesenchyme comprises an integrated population of cNCC and endocardially-derived cells. Integration of these distinct cell populations into a unified structure presumably entails modulation of cell adhesion programs. Our findings suggest that Twist1 is critical for cNCC emigration and, most likely, in regulation of cell adhesion, influencing the manner in which cNCCs interact with the cell populations they encounter, such as the endocardial-derived OFT cushion cells which have undergone EMT and moved into the cardiac jelly.
Loss of Twist1 function causes an expansion of the Wnt1-Cre lineage in the neural tube in the absence of defective dorsoventral patterning
We demonstrate that the cNCC defects characteristic of Twist1−/− mutants are associated with a pronounced expansion of the Wnt1-expressing lineage within the dorsal neural tube. The expansion of the dorsal neural tube may cause the cNCCs migration and morphogenetic phenotypes seen in these mutants. However, it is possible that this dorsal neural tube phenotype does not cause the observed cNCC phenotypes, but rather, both phenotypes reflective of a common, non-autonomous requirement of Twist1 to regulate a process such as cell-cell adhesion. Alternatively, we cannot rule out that the cNCC and dorsal neural tube phenotypes are unrelated. The relationship between the expansion of the dorsal neural tube and cNCC phenotypes could be clarified by spatiotemporally ablating Twist1 function employing the recently described conditional allele bred to Cre alleles specific for the neural crest, such as Wnt1-Cre (Danielian et al., 1998) or P3Pro-Cre (Li et al., 2000), or the lateral mesenchyme, such as T-cre (Perantoni et al., 2005).
Following the Twist1−/− cNCC defects back to the neural tube, we observed an expansion of Wnt1-Cre marked cells that is not associated with increased cell proliferation or decreased cell death. This increase in cell number without change in proliferation or death suggests that delaminating NCCs become trapped in the neural tube. Similar trapping of NCCs without changes in dorsoventral patterning is observed in Wnt1/Wnt3a double knockout mice (Ikeya et al., 1997), which is interesting as Twist1 expression is known to be regulated by Wnt signaling (Howe et al., 2003; Reinhold et al., 2006). Previous studies have implicated Twist1 function in the regulation of dorsoventral patterning in the brain (Soo et al., 2002). Twist1−/− mutants display a loss of dorsal markers in the forebrain and expanded or ectopic ventral marker expression in the midbrain and hindbrain (Soo et al., 2002). These observations are reflective of the exencephaly characteristic of Twist1−/− mutants (Chen and Behringer, 1995). In situ hybridization marker analyses within the neural tube shows increased expression of these markers reflective of the thickened-distorted neural tubes. As the expansion of dorsal neural tube cells as well as NCC lineage-marked cells correlates with decreased cNCC emigration into the OFT, and Twist1 expression is not detectable within the neural tube, we conclude that cNCC trapping in the neural tube is the result of a non-cell autonomous mechanism, most likely the regulation of microenvironment ECM. Twist1 is robustly expressed in the surrounding non-NCC mesenchymal cell populations proximal to the dorsal neural tube (Fuchtbauer, 1995; Glackin et al., 1994; Stoetzel et al., 1995, our data; Wolf et al., 1991), and these cells are likely the indirect source of the putative trapping defect. Moreover, Twist1 is known to regulate the expression of ECM genes, such as N-cadherin, in coordination with its role in EMT (Yang et al., 2004). Interfering antibodies to N-CAM and N-cadherin within the dorsal neural tube have been shown to result in grossly distorted neural tubes, ectopic migration, and abnormal adhesion (Bronner-Fraser et al., 1992) Interestingly, the effects of these antibodies are temporally dependent. Given that NCCs leave the neural tube in waves during early embryogenesis (Artinger and Bronner-Fraser, 1992; Loring and Erickson, 1987; Serbedzija et al., 1994), we hypothesize that Twist1 is necessary for early NCC emigration owing to its regulation of ECM components and cell adhesion molecules. Thus, in the absence of Twist1, cNCCs are temporally delayed in entering the OFT and cannot track or adhere properly. Later waves of NCC emigration, such as those, which populate more caudal structures, one would predict, would be less sensitive to a Twist1 early function. Phenotypic observations of Twist1−/− mice support this idea. Additionally, in chick-quail transplant experiments, it has been shown that transplanted quail NCCs from non-cNCC locations cannot normally contribute to the OFT. In addition to later stage phenotypes such as PTA, these abnormal chimeric OFTs display mesenchymal nodules (Kirby, 1989). This classic study put forth the idea that local environment is critical for normal cNCC behavior and strongly suggests that loss of Twist1 in the local environment surrounding the dorsal neural tube disrupts NCC positional information necessary for normal function.
Acknowledgements
We would like to thank Ellen Gunn and members of the Conway lab for technical assistance. We thank Henry Sucov for generously providing Wnt1-Cre transgenic mice. We would also like to thank the Herman B Wells Center Cardiac Developmental Biology Group for helpful input during group discussions. Infrastructural support at the Herman B Wells Center is in part supported by the generosity of the Riley Children’s Foundation. This work is supported by the AHA and NIH (ABF) and NIH (SJC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Artinger KB, Bronner-Fraser M. Partial restriction in the developmental potential of late emigrating avian neural crest cells. Dev Biol. 1992;149:149–157. doi: 10.1016/0012-1606(92)90271-h. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, et al. Effects of antibodies against N-cadherin and N-CAM on the cranial neural crest and neural tube. Dev Biol. 1992;153:291–301. doi: 10.1016/0012-1606(92)90114-v. [DOI] [PubMed] [Google Scholar]
- Chai Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chen CM, et al. Tumor formation in p53 mutant ovaries transplanted into wild-type female hosts. Oncogene. 2004;23:7722–7725. doi: 10.1038/sj.onc.1208037. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Clark EB. Pathogenetic mechanisms of congenital cardiovascular malformations revisited. Semin Perinatol. 1996;20:465–472. doi: 10.1016/s0146-0005(96)80062-0. [DOI] [PubMed] [Google Scholar]
- Coles EG, et al. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev Biol. 2007;312:533–544. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, et al. Decreased neural crest stem cell expansion is responsible for the conotruncal heart defects within the splotch (Sp(2H))/Pax3 mouse mutant. Cardiovasc Res. 2000;47:314–328. doi: 10.1016/s0008-6363(00)00098-5. [DOI] [PubMed] [Google Scholar]
- Conway SJ, et al. Pax3 is required for cardiac neural crest migration in the mouse: evidence from the splotch (Sp2H) mutant. Development. 1997;124:505–514. doi: 10.1242/dev.124.2.505. [DOI] [PubMed] [Google Scholar]
- Creazzo TL, et al. Role of cardiac neural crest cells in cardiovascular development. Annu Rev Physiol. 1998;60:267–286. doi: 10.1146/annurev.physiol.60.1.267. [DOI] [PubMed] [Google Scholar]
- Danielian PS, et al. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Epstein JA, et al. Migration of cardiac neural crest cells in Splotch embryos. Development. 2000;127:1869–1878. doi: 10.1242/dev.127.9.1869. [DOI] [PubMed] [Google Scholar]
- Ferencz C, et al. Congenital heart disease: prevalence at livebirth. The Baltimore-Washington Infant Study. Am J Epidemiol. 1985;121:31–36. doi: 10.1093/oxfordjournals.aje.a113979. [DOI] [PubMed] [Google Scholar]
- Firulli AB. A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix-loop-helix transcription factors. Gene. 2003;312:27–40. doi: 10.1016/s0378-1119(03)00669-3. [DOI] [PubMed] [Google Scholar]
- Firulli AB, Conway SJ. Combinatorial transcriptional interaction within the cardiac neural crest: a pair of HANDs in heart formation. Birth Defects Res C Embryo Today. 2004;72:151–161. doi: 10.1002/bdrc.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB, et al. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Firulli AB, Thattaliyath BD. Transcription factors in cardiogenesis: the combinations that unlock the mysteries of the heart. Int Rev Cytol. 2002;214:1–62. doi: 10.1016/s0074-7696(02)14002-2. [DOI] [PubMed] [Google Scholar]
- Firulli BA, et al. PKA, PKC, and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue-specific transcriptional regulation. Mol Cell. 2003;12:1225–1237. doi: 10.1016/s1097-2765(03)00425-8. [DOI] [PubMed] [Google Scholar]
- Firulli BA, et al. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, et al. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282:27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchtbauer EM. Expression of M-twist during postimplantation development of the mouse. Dev Dyn. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- Glackin CA, et al. Transcripts encoding the basic-helix-loop-helix factor TWIST, are expressed in mouse embryos, cell lines and adult tissues. Mol Cell Diff. 1994;2:309–328. [Google Scholar]
- Howe LR, et al. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Research. 2003;63:1906–1913. [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Neural crest and cardiovascular development: a 20-year perspective. Birth Defects Res C Embryo Today. 2003;69:2–13. doi: 10.1002/bdrc.10002. [DOI] [PubMed] [Google Scholar]
- Ikeya M, et al. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jiang X, et al. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, et al. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- Kirby ML. Plasticity and predetermination of mesencephalic and trunk neural crest transplanted into the region of the cardiac neural crest. 1989 doi: 10.1016/0012-1606(89)90112-7. [DOI] [PubMed] [Google Scholar]
- Kwang SJ, et al. Msx2 is an immediate downstream effector of Pax3 in the development of the murine cardiac neural crest. Development. 2002;129:527–538. doi: 10.1242/dev.129.2.527. [DOI] [PubMed] [Google Scholar]
- Li J, et al. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Lindsley A, et al. Identification and characterization of a novel Schwann and outflow tract endocardial cushion lineage-restricted periostin enhancer. Dev Biol. 2007;307:340–355. doi: 10.1016/j.ydbio.2007.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofberg J, et al. Stimulation of initial neural crest cell migration in the axolotl embryo by tissue grafts and extracellular matrix transplanted on microcarriers. Dev Biol. 1985;107:442–459. doi: 10.1016/0012-1606(85)90326-4. [DOI] [PubMed] [Google Scholar]
- Loring JF, Erickson CA. Neural crest cell migratory pathways in the trunk of the chick embryo. Dev Biol. 1987;121:220–236. doi: 10.1016/0012-1606(87)90154-0. [DOI] [PubMed] [Google Scholar]
- Ma L, et al. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, et al. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Merrill AE, et al. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet. 2006;15:1319–1328. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- Munsterberg AE, et al. Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 1995;9:2911–2922. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the mouse embryo : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- O'Rourke MP, Tam PP. Twist functions in mouse development. Int J Dev Biol. 2002;46:401–413. [PubMed] [Google Scholar]
- Ota MS, et al. Twist is required for patterning the cranial nerves and maintaining the viability of mesodermal cells. Dev Dyn. 2004;230:216–228. doi: 10.1002/dvdy.20047. [DOI] [PubMed] [Google Scholar]
- Perantoni AO, et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- Qayyum SR, et al. Septation and valvar formation in the outflow tract of the embryonic chick heart. Anat Rec. 2001;264:273–283. doi: 10.1002/ar.1162. [DOI] [PubMed] [Google Scholar]
- Reinhold MI, et al. The Wnt-inducible transcription factor Twist1 inhibits chondrogenesis. Journal of Biological Chemistry. 2006;281:1381–1388. doi: 10.1074/jbc.M504875200. [DOI] [PubMed] [Google Scholar]
- Rios H, et al. periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Mol Cell Biol. 2005;25:11131–11144. doi: 10.1128/MCB.25.24.11131-11144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg F, et al. Sculpting the cardiac outflow tract. Birth Defects Res C Embryo Today. 2003;69:38–45. doi: 10.1002/bdrc.10007. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, et al. Developmental potential of trunk neural crest cells in the mouse. Development. 1994;120:1709–1718. doi: 10.1242/dev.120.7.1709. [DOI] [PubMed] [Google Scholar]
- Smart N, et al. A differential screen for putative targets of the bHLH transcription factor Hand1 in cardiac morphogenesis. Gene Expr Patterns. 2002;2:61–67. doi: 10.1016/s0925-4773(02)00380-5. [DOI] [PubMed] [Google Scholar]
- Snider P, et al. Cardiovascular development and the colonizing cardiac neural crest lineage. ScientificWorldJournal. 2007;7:1090–1113. doi: 10.1100/tsw.2007.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo K, et al. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev Biol. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srivastava D, et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, et al. Dorso-ventral and rostro-caudal sequential expression of M-twist in the postimplantation murine embryo. Mech Dev. 1995;51:251–263. doi: 10.1016/0925-4773(95)00369-x. [DOI] [PubMed] [Google Scholar]
- Stoller JZ, Epstein JA. Cardiac neural crest. Semin Cell Dev Biol. 2005;16:704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Waldo K, et al. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev Biol. 1998;196:129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- Wolf C, et al. The M-twist gene of Mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus X-twi and the Drosophila twist genes. Dev Biol. 1991;143:363–373. doi: 10.1016/0012-1606(91)90086-i. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]