Abstract
Ran, the small, predominantly nuclear GTPase, has been implicated in the regulation of a variety of cellular processes including cell cycle progression, nuclear-cytoplasmic trafficking of RNA and protein, nuclear structure, and DNA synthesis. It is not known whether Ran functions directly in each process or whether many of its roles may be secondary to a direct role in only one, for example, nuclear protein import. To identify biochemical links between Ran and its functional target(s), we have generated and examined the properties of a putative Ran effector mutation, T42A-Ran. T42A-Ran binds guanine nucleotides as well as wild-type Ran and responds as well as wild-type Ran to GTP or GDP exchange stimulated by the Ran-specific guanine nucleotide exchange factor, RCC1. T42A-Ran·GDP also retains the ability to bind p10/NTF2, a component of the nuclear import pathway. In contrast to wild-type Ran, T42A-Ran·GTP binds very weakly or not detectably to three proposed Ran effectors, Ran-binding protein 1 (RanBP1), Ran-binding protein 2 (RanBP2, a nucleoporin), and karyopherin β (a component of the nuclear protein import pathway), and is not stimulated to hydrolyze bound GTP by Ran GTPase-activating protein, RanGAP1. Also in contrast to wild-type Ran, T42A-Ran does not stimulate nuclear protein import in a digitonin permeabilized cell assay and also inhibits wild-type Ran function in this system. However, the T42A mutation does not block the docking of karyophilic substrates at the nuclear pore. These properties of T42A-Ran are consistent with its classification as an effector mutant and define the exposed region of Ran containing the mutation as a probable effector loop.
INTRODUCTION
Like other proteins of the Ras family, Ran acts as a molecular switch through a GTPase cycle (Rush et al., 1996; Sazer, 1996). Ran binds GTP, catalyzes its slow hydrolysis to GDP, and slowly exchanges the bound GDP for free nucleotide, which in vivo is predominantly GTP. GTP hydrolysis and guanine nucleotide exchange rates are each increased approximately 100,000-fold by accessory proteins (RanGAP1 and RCC1, respectively) (Klebe et al., 1995). Human Ran GTPase-activating protein, RanGAP11, has been purified to homogeneity, and human, mouse, budding yeast (Saccharomyces cerevisiae), and fission yeast (Schizosaccharomyces pombe) RanGAP1 genes have been cloned. Ran-specific guanine nucleotide exchange factor, first identified genetically and named RCC1 (regulator of chromosome condensation-1), has also been purified to homogeneity from human cells, and human, Xenopus, S. cerevisiae, and S. pombe genes encoding it have been cloned (for review, see Avis and Clarke, 1996; Dasso, 1993; Rush et al., 1996; Sazer, 1996). Given the low intrinsic rates of GTP hydrolysis and GDP release by Ran, both the Ran·GTP/Ran·GDP ratio and the rate of Ran·GTP turnover in vivo are likely to be determined largely by activities of RanGAP1, RCC1, or other accessory proteins (Nehrbass and Blobel, 1996; Rush et al., 1996; Sazer, 1996).
Mutations in Ran, RanGAP1, or RCC1 are associated with defects in nuclear protein import, the synthesis, processing, and export of nuclear RNA, cell cycle progression, DNA synthesis, the restoration of nuclear structure after mitosis, and the maintenance of interphase nuclear structure (for review, see Dasso, 1993; Elliott et al., 1994; Moore and Blobel, 1994a; Melchior and Gerace, 1995; Tartakoff and Schneiter, 1995; Avis and Clarke, 1996; Rush et al., 1996; Sazer, 1996). These findings raise two biochemical questions. What mechanism links the Ran GTPase cycle to its downstream target(s)? Which of these targets are directly regulated by the Ran GTPase cycle, and which are affected secondarily?
Two distinct mechanisms can couple GTPase switches to their downstream targets. The first is exemplified by the role of true Ras proteins in intracellular signaling and the second by the role of Rab proteins in vesicular sorting. In the Ras paradigm, the GTP-bound form of the GTPase interacts with an effector molecule to activate the latter and stimulate a process. The amount of stimulation depends on the amount of GTPase·GTP complex. In the Rab paradigm, stimulation requires GTP hydrolysis, suggesting GTPase interaction with at least two different effectors, one specific for GTPase·GTP and the other for GTPase·GDP. The amount of stimulation depends on the turnover of the GTPase·GTP complex.
Nuclear protein import is the best characterized of the cellular processes affected by the Ran GTPase cycle. Import of proteins containing polybasic nuclear localization signals occurs in two steps (Newmeyer and Forbes, 1988; Richardson et al., 1988; Moore and Blobel, 1994a; Melchior and Gerace, 1995). The first is energy independent and involves docking of the karyophilic protein to the nuclear pore complex. The second is energy dependent and involves transport of the protein through the pore. When mammalian cells are treated with digitonin, their plasma membranes become permeable and many endogenous cytoplasmic and nuclear macromolecules are lost, while nuclei and most cytoskeletal structures remain intact. Nuclear protein import halts in such permeabilized cells, but can be restored by addition of cytosolic proteins and an energy source (Adam et al., 1990). Docking requires karyopherin α (importin α), which is the receptor for the nuclear localization signal, and karyopherin β (importin β), a factor that binds to both karyopherin α and the nuclear pore. Addition of Ran (Melchior et al., 1993; Moore and Blobel, 1993), a small Ran-interacting protein named p10 [or nuclear transport factor (NTF) 2] (Moore and Blobel, 1994b; Paschal and Gerace, 1995), and GTP is necessary and sufficient for the import of proteins already docked at the nuclear pore. Studies using permeabilized cells and other systems have shown that Ran trapped in either its GTP- or GDP-bound forms does not support import and, in addition, inhibits import stimulated by wild-type Ran (Melchior et al., 1993; Moore and Blobel, 1993; Tachibana et al., 1994; Corbett et al., 1995; Palacios et al., 1996; Schlenstedt et al., 1995a). These data suggest that Ran functions here by a Rab-like mechanism.
Four putative Ran effector proteins, RanBP1, RanBP2, karyopherin β, and p10, have been identified and roles for all of them have been demonstrated in nuclear import. RanBP2/Nup358 is a nuclear pore protein that binds to Ran·GTP but not to Ran·GDP (Wu et al., 1995; Yokoyama et al., 1995). It is located on the cytosolic face of the pore, at positions similar to those where Ran charged with nonhydrolyzable GTP analogues accumulate in permeabilized cell nuclear import assays, and to the positions where Ran·GTP binds when incubated with purified nuclear envelopes (Melchior et al., 1995; Wu et al., 1995). RanBP2/Nup358 is the only known Ran·GTP-binding protein in the nuclear envelope (Melchior et al., 1995), and antibodies directed against it inhibit nuclear protein import (Yokoyama et al., 1995). Ran·GTP also binds to karyopherin β (Rexach and Blobel ,1995; Lounsbury et al., 1996b) and to RanBP1 (see below), while Ran·GDP binds to p10 (Nehrbass and Blobel, 1996). Ran·GDP does not bind either karyopherin β or RanBP1, nor does karyopherin β bind RanBP1. However, the three proteins together form a stable ternary complex, detected experimentally by the ability of RanBP1 to stimulate the interaction between karyopherin β and Ran·GDP (Chi et al., 1996, 1997). RanBP1 also increases the affinity of the interaction between Ran·GTP and karyopherin β (Lounsbury et al., 1996b; Chi et al., 1996, 1997). These findings have led to models in which the interaction of Ran with nucleoporins, RanBP1, karyopherin β, and p10 drives karyophilic proteins through the nuclear pore (Chi et al., 1996, 1997; Koepp and Silver, 1996; Lounsbury et al., 1996b; Nehrbass and Blobel, 1996). Many of the other processes disrupted by defects in the Ran GTPase cycle depend on the timely delivery of proteins to the cell nucleus; therefore, effects on these processes could be secondary to disruption of nuclear protein import.
Additional studies of the fourth putative Ran effector, RanBP1, however, suggest that it may mediate effects of the Ran GTPase cycle that are independent of the cycle’s role in nuclear protein import. RanBP1 is a small (203-aa residues in mice and humans), acidic, predominantly cytosolic protein. It binds to Ran·GTP. Although it does not possess GAP or guanine nucleotide exchange factor activities, it interacts with RanGAP1 in a yeast double-hybrid assay and stimulates RanGAP activity in vitro (Coutavas et al., 1993; Lounsbury et al., 1994; Beddow et al., 1995; Bischoff et al., 1995; Ren et al., 1995). As noted above, it also stimulates Ran-karyopherin β interactions. RanBP1 is about one-fifth as abundant as Ran (2 × 106 copies of RanBP1 and 107 copies of Ran per mammalian cell), and it might serve as a Ran coregulator. These biochemical studies have not identified a clear effector role for RanBP1, but a mutant Ran·GTP protein lacking its six carboxyl-terminal amino acids loses the ability to bind tightly to RanBP1, retains the ability to bind karyopherin β and reconstitute nuclear protein import in digitonin-permeabilized cells, but loses the ability to perturb cell cycle progression in transfected 293/Tag cells (Ren et al., 1995; Lounsbury et al., 1996b; Chi et al., 1997). These findings suggest that RanBP1 may be an effector that links the Ran GTPase cycle to cellular targets independent of and in addition to nuclear protein import.
To better define the mechanisms of Ran’s interactions with its putative effectors and regulators, we constructed a missense mutant of Ran homologous to the mutations of RAS residue 35 that disrupt interactions of the latter protein with its GAP and effector proteins (Bourne et al., 1991; Vojtek et al., 1993) and characterized the interactions of the Ran missense mutant protein with RanGAP1, RCC1, and the putative Ran effector proteins. We also examined the ability of this mutant to support and/or inhibit nuclear protein import in digitonin-permeabilized cells.
MATERIALS AND METHODS
Glutathione S-transferase (GST) Fusion Proteins
Polymerase chain reaction- (PCR) generated fragments of mouse RanBP1 and mouse RanGAP1 cDNAs with upstream EcoRI sites and downstream XhoI sites added to the respective PCR primers were cloned in-frame downstream of the GST coding domain of pGEX5–1 (Pharmacia LKB Biotechnology, Piscataway, NJ). GST-RanBP1 A (full-length RanBP1, aa 1–203), GST-RanBP1 B (aa 1–159), and GST-RanBP1 D (aa 69–203) were generated by cloning these portions of the coding region of a mouse RanBP1 cDNA (Coutavas et al., 1993). Two GST-RanGAP1 clones were generated from a mouse RanGAP1 cDNA (Ren et al., 1995): full-length (aa 1–589) and c-del (aa 1- 358). GST and GST fusion proteins were expressed by induction of transformed bacterial cultures with 0.1 mM isopropylthio-β-d-galactoside and purified batchwise using glutathione-agarose (Sigma Chemical Co., St. Louis, MO). Proteins were eluted with 5 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0) and 0.1 mM phenylmethylsulfonyl fluoride.
Site-directed Mutagenesis of Ran
nucleotide sequence encoding Ala at codon 42 of Ran was introduced into human Ran cDNA cloned in M13 mp18 (Coutavas et al., 1993; Ren et al., 1995) using an oligonucleotide with an A to G transition mutation at the 10th position (5′-TATGTAGCCGCCTTGGGTGTT-3′) and the reagents and protocol of the Amersham Life Sciences “Sculptor” in vitro mutagenesis system. The resulting mutation was confirmed by sequence analysis.
Recombinant Ran Proteins
Wild-type and mutant Ran coding regions were cloned into pET9c (Novagen, Madison, WI), and bacteria transformed with these constructs were grown, induced, and lysed as described previously (Ren et al., 1995). To purify wild-type and T42A-Ran proteins, crude cell lysate (40 ml) from 4 l of culture was subjected to ammonium sulfate fractionation. The precipitate from a 30 to 55% ammonium sulfate fraction was resuspended in 10 ml of 10 mM Tris (pH 8.0)/1 mM dithiothreitol (DTT)/1 mM GTP, and incubated for 5 min on ice. The sample was adjusted to a volume of 400 ml in 10 mM Tris (pH 8.0)/1 mM DTT/1 mM MgCl2 and concentrated to a volume of 50 ml using positive pressure in an Amicon concentrator cell with a YM-10 membrane. The material was clarified by centrifugation, applied to a MonoQ HR10/10 column (Pharmacia, Pistcataway, NJ) and eluted with a linear gradient of 0–500 mM NaCl. Ran proteins usually eluted at a concentration of approximately 250 mM NaCl. Column fractions were assayed for Ran by SDS-PAGE. Ran-containing fractions were pooled and applied to a Superdex HR75 26/60 gel filtration column (Pharmacia) in 10 mM HEPES (pH 7.5)/160 mM potassium acetate/1 mM DTT/5 mM magnesium acetate and eluted at 3.00 ml/min. Both wild-type and T42A-Ran proteins eluted from this column as single peaks with the mobility of 25-kDa globular proteins. Peak fractions were pooled and concentrated with Amicon YM-10 Centricon units spun at 3000 × g for 30 to 60 min at 4°C. The proteins, greater than 90% pure by SDS-PAGE, were stored in aliquots (2–3 mg/ml) at −80°C. Protein concentrations were determined with the Bradford method using the Bio-Rad protein assay kit. To charge proteins, GTP or GDP was added to a final concentration of 1 mM and EDTA to a final concentration of 5 mM. The mixture was incubated for 20 min at room temperature (RT). MgCl2 was then added to a final concentration of 20 mM to stabilize Ran·nucleotide complexes.
Constructs of Ran proteins with N-terminal histidine-tagged (His*Tag, Novagen) fusions (23 aa) were made by cloning the wild-type, T42A, and GTPase defective (dm) (G19V, Q69L) forms of Ran into the pET19b vector, with a cloning strategy similar to that described for GST fusion proteins but with PCR primers generating NdeI or BamHI cleavable sites. His*Tag fusion proteins were expressed by induction of transformed bacterial cultures with 1.0 mM isopropylthio-β-d-galactoside and purified in a batchwise manner using nickel resin and the reagents and protocol of the Novagen pET System.
Other Recombinant Proteins
Proteins expressed from pET21B constructs encoding His*Tag fusions of the first or fourth Ran-binding domains (RanBD1, aa 1152–1321; RanBD4, aa 2892–3060) of RanBP2/Nup358 (Wu et al., 1995; Yokoyama et al., 1995) were a gift from J. Wu (Laboratory of Cell Biology, The Rockefeller University, New York, NY). His*Tag fusions of human karyopherin α and rat karyopherin β were purified as described by Schwoebel and Moore (manuscript in preparation), as were untagged Xenopus RCC1 and untagged human p10 and karyopherin β. E. coli expression vectors were obtained from the following investigators: His*Tag rat karyopherin β (lacking its amino terminal 12 residues) from A. Radu (The Rockefeller University) (Moroianu et al., 1995), full-length human karyopherin β from D. Görlich (University of Heidelberg, Heidelberg, Germany), His*Tag human karyopherin α (clone hSRP1α) from A. Lamond (European Molecular Biology Laboratories, Heidelberg, Germany) (Weis et al., 1995), human p10 (“pp15” expression clone) from U. Grundmann (Behringwerke, Marburg, Germany) (Lehmeier and Amann, 1992; Grundmann et al., 1988), and Xenopus RCC1 from T. Nishimoto (Kyushu University, Fukuoka, Japan).
In Vitro Binding of Ran to RanBP1, RanBP2, and Karyopherin β Assayed by Gel Transfer Ligand Binding
Filter-binding analysis of interactions with Ran proteins was performed as described by Lounsbury et al. (1994). Briefly, 1-μg samples of recombinant proteins to be tested for interaction with Ran were electrophoresed in duplicate 12% SDS-PAGE gels. One gel was stained with Coomassie blue to confirm that intact proteins were present in the expected amounts. Proteins were transferred from the other gel to a nitrocellulose membrane, immobilized, renatured for 2 h at 4°C in 20 mM 3-N-morpholinopropane-sulfonic acid (MOPS) (pH 7.1)/100 mM sodium acetate/5 mM magnesium acetate/0.25% Tween 20/0.5% bovine serum albumin (BSA)/5 mM DTT, and then incubated for 30 min at RT in 20 mM MOPS (pH 7.1)/100 mM potassium acetate/5 mM magnesium acetate/0.05% Tween 20/0.5% BSA/5 mM DTT/100 μM GTP. The filters were equilibrated briefly in the same buffer without GTP before the addition of Ran protein.
Two to three micrograms of purified Ran protein (wild type or T42A) were incubated with 10 μCi of [α-32P]GTP (3000 Ci/mmol, Dupont/New England Nuclear, Boston, MA) in 20 μl of 10 mM MOPS (pH 7.1)/1 mM EDTA/0.1% BSA for 15 min on ice. (In those cases where the probe contained RanBP1 + Ran, a twofold excess (4–6 μg) of GST-RanBP1 was added to the solution before adding the GTP.) The reaction was then adjusted to 5 mM magnesium acetate in a final volume of 0.5 ml, and the excess GTP was removed with Microcon-10 units (Amicon, Beverly, MA) spun at 10,000 × g for 10 min. Aliquots of the loaded proteins were counted in a scintillation counter and equal counts of loaded proteins (about 5 × 106 cpm) were added to 15 ml of 20 mM MOPS (pH 7.1)/100 mM potassium acetate/5 mM magnesium acetate/0.05% Tween 20/0.5% BSA/5 mM DTT and incubated with replicate blots for 30 min at RT. Filters were then washed five times at RT in the same buffer and autoradiographed. Autoradiographs were digitized using a XRS 12cx flatbed scanner connected to an Apple 8100 Macintosh computer. Images were printed on a Tektronix Phaser 440 dye sublimation printer.
In Vitro Binding of Ran to RanBP1 and Karyopherin β Assayed by Dot Ligand Blotting
Dot ligand blotting was performed exactly as described for gel transfer blotting except 0.4- to 1-μg samples of native (nondenatured) RanBP1 and karyopherin β, each in 300 μl of phosphate-buffered saline, were spotted directly onto nitrocellulose, and the 2-h renaturation step was omitted. For experiments using labeled Ran·GDP as a probe, wild-type or T42A-Ran proteins were charged with 40 μCi [β-35S]GDP (1250 Ci/mmol) (DuPont/New England Nuclear) as described above for [α-32P]GTP, and autoradiography was performed at RT without intensifying screens. Reconstruction experiments in which equal cpm of [α-32P]GTP·Ran and [β-35S]GDP·Ran were spotted and autoradiographed showed the 35S signal to be one-fourth that of the 32P signal; therefore, autoradiography times of 35S samples were increased appropriately.
In Vitro Binding of Ran to p10 Assayed by His*Tag Resin Binding
Binding assays were performed using His*Tag fusion proteins immobilized on Ni2+ resin (Novagen). Either His*Tag-Ran or His*Tag-T42A-Ran beads were washed two or three times with 10 volumes of 1× phosphate-buffered saline and collected by centrifugation at 1000 × g. Ran and T42A-Ran were then charged by addition of GTP or GDP to a final concentration of 1 mM and of EDTA to a final concentration of 5 mM and incubation for 20 min at RT. Protein-nucleotide complexes were then stabilized by the addition of Mg(OAc)2 to a final concentration of 20 mM. Unbound nucleotide was removed by washing the beads with binding buffer (20 mM HEPES, pH 6.8, 150 mM KOAc, 2 mM Mg(OAc)2, 2 mM DTT, 0.1% Tween 20, 0.1% casamino acids) (Rexach and Blobel, 1995). Uncharged Ran was prepared using the same procedure except for the absence of added guanine nucleotide and a preliminary wash in the absence of added Mg(OAc)2. Binding assays were carried out in 50-μl volumes of binding buffer containing 2.5 μM immobilized fusion protein and 3 μM free p10 protein. Assay mixes were incubated for 1.5 to 2 h at 4°C, and beads were collected by centrifugation (1000 × g) for 2 min. The supernatant was removed (“unbound” fraction). Beads were washed two to three times in binding buffer, collected by centrifugation (1000 × g), and the bound fraction was released by boiling in an equal volume of SDS-PAGE loading buffer (100 mM Tris-HCl, pH 6.8, 200 mM DTT, 4% SDS, 0.2% bromophenol blue, 20% glycerol). One-half of each “unbound” and all of bound fraction were analyzed on 12–15% SDS-PAGE gels. Proteins were visualized by immunoblotting using a polyclonal rabbit anti-p10 primary antibody, a goat anti-rabbit secondary antibody, and a chemiluminescent substrate kit (KPL).
GAP Assays
To measure GTP hydrolysis by wild-type Ran, T42A-Ran, or GTPase defective dm-Ran, His*Tag fusion forms of these proteins were immobilized on His*Bind Resin (Novagen). The immobilized proteins were equilibrated in 20 mM Tris-HCl (pH 7.5)/50 mM NaCl/1 mM EDTA/10% glycerol by washing the resin three times in 10 volumes of buffer followed by brief centrifugation at 1000 × g. The resin was resuspended in 50–100 μl of the same buffer containing 20 μCi of [γ-32P]GTP (6000 Ci/mmol, Dupont/New England Nuclear) and incubated for 20 min at RT. GTP-loaded protein was stabilized and excess labeled GTP was removed by washing the resin five times in 20 mM Tris-HCl (pH 7.5)/50 mM NaCl/15 mM MgCl2 (GAP buffer). The resin was resuspended in GAP buffer so that 50-μl aliquots contained loaded Ran at a concentration of 1 μM. Individual aliquots were incubated for up to 30 min at 30°C with 1–5 μl of various GST fusion proteins. Reactions were terminated by the addition of 1 ml of ice-cold GAP buffer and centrifugation. Supernatant fractions, containing hydrolyzed radioactive label, were subjected to scintillation counting. Pelleted resin fractions were washed with an additional 1 ml of GAP buffer before recovery for scintillation counting. In each case, the amount of label remaining complexed (bound) to Ran protein (resin fraction) was expressed as a percentage of the total cpm recovered from the resin and supernatant fractions.
Nucleotide Exchange Assays
The exchange of labeled GTP or GDP for unlabeled GTP or GDP, respectively, on wild type or T42A-Ran was also examined using His*Tag fusion proteins. In this case proteins were charged with 20 μCi of [α-32P]GTP (3000 Ci/mmol) or 40 μCi of [β-35S]GDP (1250 Ci/mmol) (DuPont/New England Nuclear), and individual aliquots were incubated at 30°C with 1 mM unlabeled GTP or GDP, in the presence or absence of 0.2 μM purified Xenopus RCC1. Conditions and measurements of bound label were the same as described for GAP assays. All exchange and GAP assays were performed a minimum of three times, with essentially identical results.
Nuclear Import Assay
Protein import assays, in digitonin-permeabilized buffalo rat liver cells, used rhodamine-labeled human serum albumin coupled to nuclear localization sequence peptides as an import substrate and 1 mM GTP as an energy source. Assays at 21°C were performed as described previously (Moore and Blobel, 1993; Ren et al., 1995), except that recombinant human p10 was used in place of purified Xenopus p10, and in some cases recombinant full-length human karyopherins α and β were used in place of Xenopus fraction A. For assays at 4°C, import mixtures were prepared on ice in the cold room and pipetted onto Parafilm-covered glass plates on ice. Coverslips with permeabilized cells were then placed cell side down on the import mixtures.
RESULTS
T42A Mutant Ran Protein (E1-Ran) Interacts Weakly with RanBP1 and not Detectably with RanBP2/Nup358 or Karyopherin β
Wild-type Ran·GTP binds to RanBP1, RanBP2/Nup358, and karyopherin β, but not to p10, while wild-type Ran·GDP binds to p10 and not to the other three proteins. This selective binding to only one nucleotide-charged form of Ran supports the classification of these four proteins as Ran effectors. RAS proteins with alanine substituted for threonine at residue 35 are not sensitive to GAP stimulation and bind poorly to effector proteins (Bourne et al., 1991; Vojtek et al., 1993). The homologous amino acid in Ran is the threonine at residue 42. Both the RAS and the Ran threonine residues are exposed at the surfaces of the proteins on peptide loops near the bases of the proteins’ GTP-binding sites (Scheffzek et al., 1995). To determine whether the region of Ran homologous to the effector binding loop of true RAS proteins is indeed involved in such interactions, we generated the corresponding Ran mutation, T42A. We designate the mutant protein E1-Ran.
When equal amounts of wild-type Ran and E1-Ran were incubated with radiolabeled GTP or GDP, both proteins were labeled to the same specific activity, and the bound nucleotide was essentially equally well retained by both proteins (Figures 4D and 5, below, and our unpublished observations). However, in contrast to wild-type Ran·GTP, E1-Ran·GTP bound weakly or not detectably to RanBP1 and did not bind detectably to RanBP2/Nup358 or to karyopherin β (Figures 1 and 2). Specifically, in ligand blot assays (Figure 1), E1-Ran·GTP bound reproducibly but extremely weakly to full-length RanBP1 protein and to a RanBP1 deletion fragment that interacted with wild-type Ran. In contrast, E1-Ran failed to interact detectably with RanBP1 in multiple yeast double-hybrid assays (our unpublished observations).
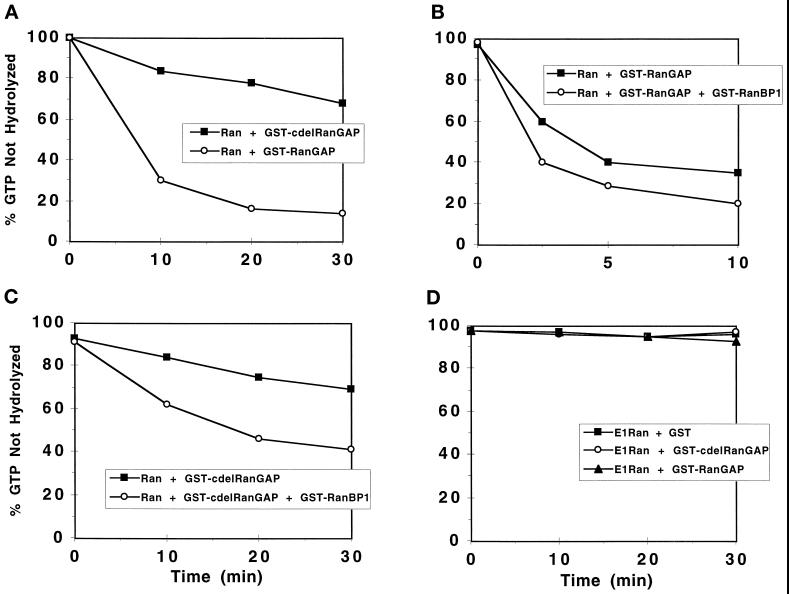
Figure 4.
Stimulation of GTP hydrolysis by wild-type Ran but not E1-Ran, by full-length and carboxyl-terminal-deleted (c-del) RanGAP1 proteins. Recombinant wild-type Ran·[γ-32P]GTP (∼1 μM) was incubated with GST-RanGAP1 (A, ∼0.01 μM, ○ or GST-c-del RanGAP1 (0.15 μM, ▪), GST-RanGAP1 (B, ∼0.01 μM) alone (▪) or GST-RanGAP1 (∼0.01 μM) plus GST-RanBP1 (0.5 μM, ○), and GST-c-del RanGAP1 (C, ∼0.15 μM) alone (▪) or GST-c-del RanGAP1 (∼0.15 μM) plus GST-RanBP1 (0.5 μM, ○). (D) E1-Ran·[γ-32P]GTP (∼1 μM) was incubated with either GST (5.0 μM, ▪), GST-RanGAP1 (∼0.01 μM, ▴), or GST-c-del RanGAP1 (0.15 μM, ○). At the indicated times, the percentage of 32P still bound to Ran was determined (see MATERIALS AND METHODS).
Figure 5.
Stimulation of nucleotide exchange on wild-type and E1-Ran in the presence of RCC1. (A) Recombinant wild-type Ran·[α-32P]GTP (∼1 μM, ○, •) or E1-Ran·[α-32P]GTP (∼1 μM, □, ▪) proteins were incubated in the presence (•, ▪) or absence (○, □) of 0.2 μM RCC1. All reactions contained 1 mM unlabeled GDP. At the indicated times, the percentage of 32P still bound to Ran was determined (see MATERIALS AND METHODS). (B) Recombinant wild-type Ran·[β-35S]GDP (∼1 μM, ○, •) or E1-Ran·[β35S]GDP (∼1 μM, □, ▪) proteins were incubated in the presence (•, ▪) or absence (○, □) of 0.2 μM RCC1. All reactions contained 1 mM unlabeled GTP. At the indicated times, the percentage of 35S still bound to Ran was determined.
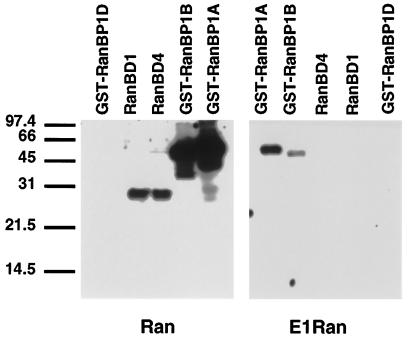
Figure 1.
Interaction of wild-type and E1-Ran with RanBP1 and RanBP2/Nup358, assayed by ligand blotting. One-microgram samples of intact GST-RanBP1 A (full length, aa 1–203), GST-RanBP1 fragments B (aa 1–159) and D (aa 69–203), and 0.3-μg samples of two His*Tag Ran-binding domain fragments from RanBP2/Nup358 (RanBD1 and RanBD4) were resolved by SDS-PAGE, transferred to nitrocellulose, renatured in situ, and probed with wild type and E1-Ran charged with [α-32P]GTP. Filters were subjected to autoradiography for 6 h. Mobilities of size markers, in kDa, are indicated on the left. GST-RanBP1 fragment D, which does not bind to wild-type Ran·GTP, was included as a negative control.
Figure 2.
Interaction of wild type and E1-Ran with RanBP1 and karyopherin β, assayed by ligand blotting. (A) Gel transfer assay. A 0.5-μg sample of GST-RanBP1 and 1-μg samples of GST, karyopherin β, and a truncated karyopherin β lacking its amino terminal 12 residues (Δ karyopherin β) were resolved by SDS-PAGE, transferred to nitrocellulose, renatured in situ, and probed with wild-type Ran·[α32P]GTP alone, wild-type Ran·[α-32P]GTP + RanBP1, or E1-Ran·[α-32P]GTP alone. Filters were subjected to autoradiography for 3 h. Mobilities of size markers in kDa are indicated on the left. The duplicate Coomassie blue-stained gel shows the relative intactness of the karyopherin β preparations. (B) Dot blot assay. One- or 0.4-μg samples of nondenatured GST-RanBP1, GST, karyopherin β, and Δ karyopherin β were spotted directly onto nitrocellulose and probed with wild type and E1-Ran charged with [α-32P]GTP in the presence or absence of added RanBP1 as indicated on the left. Filters were subjected to autoradiography for 4 h. The 1-μg and 0.4-μg labels on the right indicate the protein content of the adjacent eight dots.
No protein of the size of RanBP2/Nup358 could be detected in ligand blots of HeLa cell extracts probed with E1-Ran·GTP, in contrast to replicate blots probed with wild-type Ran·GTP (our unpublished observations). To confirm that E1-Ran failed to interact detectably with RanBP2/Nup358, ligand-binding assays were carried out with recombinant proteins corresponding to two of the four specific Ran-binding domains of RanBP2/Nup358. Under conditions in which wild-type Ran bound strongly to both fragments, E1-Ran failed to bind detectably to either (Figure 1).
Also, in ligand-binding assays, Ran·GTP but not E1-Ran·GTP bound to karyopherin β (Figure 2). These assays were done in two ways. The first (Figure 2A) was a gel transfer, in which denatured proteins were renatured after filter immobilization, and the second (Figure 2B) was a dot assay, in which native proteins were applied directly to the filter. In the case of karyopherin β, we have found the nondenaturing dot-blotting procedure to be much more sensitive than gel transfer, perhaps due to the inefficient renaturation of this protein. As shown in Figure 2, wild-type Ran·GTP bound clearly to karyopherin β, and its binding was further stimulated in the presence of RanBP1, as reported previously (Lounsbury et al., 1996b; Chi et al., 1996, 1997). In contrast, E1-Ran·GTP, either alone or in the presence of RanBP1, failed to bind to karyopherin β. Also as reported previously, the binding of wild-type Ran·GTP to a truncated form of karyopherin β (lacking its 12-amino terminal residues; abbreviated Δ karyopherin β in Figure 2) was weak and, in some circumstances, detectable only in the presence of RanBP1. As expected, when wild type and E1-Ran charged with [β-35S]GDP were used as probes (without added RanBP1) in a series of dot-blotting experiments identical to those shown in Figure 2, neither protein bound detectably to either RanBP1 or karyopherin β (our unpublished observations). The same result was obtained when RanBP1 was added to the probes even though, as noted previously, RanBP1 had been reported to promote an interaction between wild-type Ran·GDP and full-length karyopherin β (Chi et al., 1996, 1997). This discrepancy may reflect a difference in assay conditions, but the fact that RanBP1 did stimulate significant binding between wild-type Ran·GTP and karyopherin β, but not between E1-Ran·GTP and karyopherin β (Figure 2B), supports the conclusion that the E1-Ran–karyopherin β interaction is defective under all conditions.
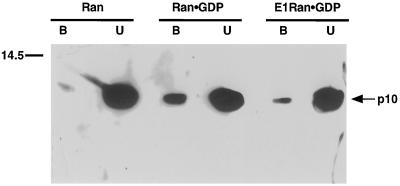
Because of the difficulty in our hands of detecting p10-Ran·GDP binding using filter-immobilized p10, ligand-binding assays were not used to compare the interactions of wild-type and E1-Ran·GDP with p10. Instead, fixed matrix assays were used, with His*Tag-Ran fusion proteins. Under conditions in which essentially nucleotide-free wild-type Ran bound a barely detectable amount of p10, p10 was bound significantly by both E1-Ran·GDP and wild-type Ran·GDP. The amount of binding to E1-Ran was between one-fifth and one-half that observed with wild-type Ran. A representative experiment is shown in Figure 3. In additional fixed matrix assays, neither wild-type nor E1-Ran·GTP interacted with karyopherin α, as expected; and wild-type Ran·GTP but not E1-Ran·GTP, wild-type Ran·GDP, or E1-Ran·GDP interacted directly with karyopherin β (our unpublished observations).
Figure 3.
Interaction of wild-type and E1-Ran with p10, assayed by His*Tag resin binding. His*Tag-wild-type Ran (predominantly nucleotide free, see MATERIALS AND METHODS), His*Tag-wild-type Ran·GDP, and His*Tag-E1-Ran·GDP, bound to Ni2+ resin, were incubated with p10, and the amounts of the latter bound to the resin were determined as described in MATERIALS AND METHODS. Mobility of a size marker is indicated on the left and mobility of p10 on the right. B, bound p10; U, unbound p10.
Taken together, these findings support the classification of E1-Ran as an effector mutant. Under a variety of assay conditions, the ability of E1-Ran to interact with RanBP1 is sharply reduced, and the mutant protein yields no detectable interactions with the possible Ran effectors RanBP2/Nup358 and karyopherin β. The effector mutant retains the ability to interact with p10, albeit to a somewhat reduced extent.
RanGAP1 Does Not Stimulate GTP Hydrolysis by E1-Ran
The low or undetectable interactions of E1-Ran·GTP with RanBP1, RanBP2/Nup358, and karyopherin β suggested that E1-Ran might also not respond to RanGAP1. To validate our GAP assay, we demonstrated that a RanGAP1 fusion protein accelerated hydrolysis of GTP bound to Ran. We then assessed the effects of adding RanBP1 and/or deleting the carboxyl-terminal 231 aa of RanGAP1 (c-del RanGAP1). The deleted amino acids included a highly acidic domain, residues 359–399, common to mammalian and yeast RanGAP proteins, and a carboxyl-terminal region unique to mammalian RanGAP. In multiple trials, fusion Ran reproducibly catalyzed little or no GTP hydrolysis either alone or in the presence of 5 μM GST or 0.5 μM GST-RanBP1 (less than 10% of bound 32P released in the course of a 30-min incubation, our unpublished observations), but was stimulated to catalyze rapid GTP hydrolysis in the presence of wild-type RanGAP1 fusion protein and was stimulated partially in the presence of c-del RanGAP1 fusion protein. A typical result is shown in Figure 4A. Wild-type RanBP1 fusion protein reproducibly interacted with both wild-type RanGAP1 (Figure 4B) and c-del RanGAP1 (Figure 4C) to augment Ran-catalyzed GTP hydrolysis. c-del RanGAP1 also interacted with RanBP1 in a yeast double-hybrid assay (our unpublished observations).
In multiple trials, E1-Ran alone also catalyzed little or no GTP hydrolysis, but, in contrast to wild-type Ran, it showed no additional GTPase activity in the presence of full-length or c-del RanGAP1 (Figure 4D). [The assay could not exclude the possibility of low level (<10%) stimulation.] Additional controls confirmed that GTPase-defective Ran was also insensitive to RanGAP1 and that no guanine nucleotide exchange activity could be detected in any of these assay mixtures (Coutavas et al., 1993, and our unpublished observations). Thus, under conditions where wild-type Ran responded strongly and specifically to RanGAP1 stimulation, E1-Ran showed no detectable response.
RCC1 Stimulates Guanine Nucleotide Exchange by E1-Ran
Both wild-type and E1-Ran fusion proteins interacted weakly with RCC1 in fixed matrix and yeast double-hybrid assays (our unpublished observations), suggesting that RCC1 might stimulate normal guanine nucleotide exchange by E1-Ran. To test this possibility, purified wild-type and E1-Ran proteins charged with either [α-32P]GTP or [β-35S]GDP were incubated with excess unlabeled GDP or GTP in the presence or absence of purified RCC1 protein, and amounts of radioactivity remaining protein bound were measured as a function of time. The results of typical experiments in which labeled GTP was exchanged for unlabeled GDP and in which labeled GDP was exchanged for unlabeled GTP are shown in Figure 5. In the absence of RCC1, neither mutant nor wild-type Ran exchanged extensive amounts of bound for free nucleotide. In contrast, in the presence of RCC1, both proteins underwent extensive exchange within 1 min. Similar results were obtained in multiple trials and in studies of labeled GTP, unlabeled GTP exchange (our unpublished observations). These results indicate that E1-Ran can retain bound guanine nucleotides approximately as well as wild-type Ran (as measured by low intrinsic rates of GTP and GDP release) and that RCC1-stimulated guanine nucleotide exchange rates for E1-Ran and wild-type Ran are similar. [The slightly greater intrinsic [β-35S]GDP release rate, compared with that of [α-32P]GTP (Figure 5), was observed consistently in multiple experiments. It may be a property of the sulfur derivative, as Ran would ordinarily be expected to release GDP more slowly than GTP (Klebe et al., 1995)].
E1-Ran Blocks Nuclear Protein Import in Digitonin-permeabilized Cells
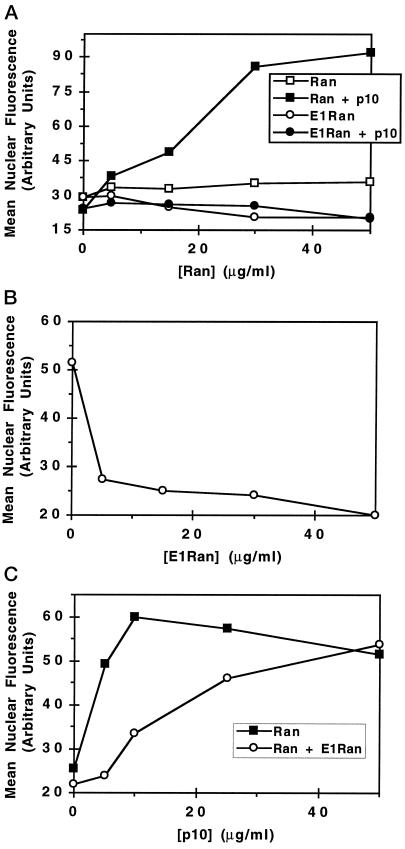
Nuclear protein import in digitonin-permeabilized buffalo rat liver cells requires the addition of karyopherin α, karyopherin β, p10, and Ran. The added Ran protein is thought to bind and hydrolyze GTP and to interact with RanBP2/Nup358, karyopherin β, and p10 in the course of stimulating import. We hypothesized that E1-Ran would not substitute for wild-type Ran in such a reconstitution experiment. To test this hypothesis, we used permeabilized cells supplemented with Xenopus fraction A (which contains karypherins α and β), human recombinant p10, and 1 mM GTP. A titration study with wild-type Ran·GDP showed that it promoted maximal nuclear protein import when added at a concentration of 50 μg/ml (Figure 6A). A parallel titration with E1-Ran·GDP yielded no restoration of import. To test the possibility that E1-Ran might also inhibit protein import reconstituted by the addition of wild-type Ran, we added various amounts of E1-Ran·GDP to import assays reconstituted with a constant amount of wild-type Ran·GDP. E1-Ran inhibited import nearly completely when added at a concentration twice that of the wild-type protein (Figure 6B).
Figure 6.
(A) Effects of wild-type and E1-Ran·GDP on nuclear protein import in digitonin-permeabilized buffalo rat liver cells. Assays were carried out at 21°C as described in MATERIALS AND METHODS on permeabilized cells supplemented with 1.4 mg/ml of Xenopus fraction A (which contains karyopherin α and β), 1.5 μg/ml of purified human nuclear import factor p10, the indicated concentrations of wild-type Ran·GDP (▪) or E1-Ran·GDP (○), 5 μg/ml of NLS-tagged, rhodamine-labeled human serum albumin, and 1 mM GTP. Protein import was measured as mean nuclear fluorescence in arbitrary units. (B) Inhibition of nuclear import by E1-Ran. Assays were performed at 21°C, in the presence of 1.45 mg/ml Xenopus fraction A, 1.5 μg/ml p10, 50 μg/ml wild-type Ran·GDP, 1 mM GTP, and the indicated concentrations of additional wild-type Ran·GDP (▪) or E1-Ran·GDP (○).
Moreover, this inhibition appears to be specific for active transport through the nuclear pore and not for docking (Figure 7). E1-Ran·GDP did not block docking of the import substrate at either 4°C (where import is inhibited by the low temperature) or at 21°C (the standard import assay temperature).
Figure 7.
Effect of E1-Ran on docking of import substrate at the nuclear pore. Permeabilized cells were incubated with complete import mixture (Mix) (karyopherins α and β (Xenopus fraction A), p10, 50 μg/ml wild-type Ran·GDP, and 1 mM GTP) plus an additional 100 μg/ml wild-type Ran·GDP or 100 μg/ml E1-Ran·GDP. Assays were performed at 21°C or at 4°C as described in MATERIALS AND METHODS.
Inhibition of Nuclear Protein Import by E1-Ran Can Be Overcome by the Addition of Excess p10
Since E1-Ran interacts poorly with RanBP1, RanBP2/Nup358, and karyopherin β, but does interact with p10, we next hypothesized that inhibition of import by E1-Ran might be due to inhibitory p10 trapping. To test this hypothesis, we used permeabilized cells supplemented with purified human recombinant karyopherin α, karyopherin β, and p10 proteins, and with 1 mM GTP.
As shown in Figure 8,A and B, permeabilized cells reconstituted with purified components behave essentially the same as ones reconstituted with Xenopus fraction A; E1-Ran·GDP alone does not support import, and E1-Ran inhibits import stimulated by wild-type Ran. However, the E1-Ran inhibition can be overcome by addition of excess p10 (Figure 8C). The E1-Ran inhibition and p10 rescue experiments shown in Figure 8, B and C, respectively, used 15 μg/ml wild-type Ran. Repetition of the inhibition and rescue studies with 30 μg/ml wild-type Ran yielded qualitatively the same result, but with higher levels of E1-Ran and p10 needed to yield inhibition and rescue. (Compare also Figure 6B, where a higher level of E1-Ran is required to yield inhibition of import in the presence of 50 μg/ml wild-type Ran).
Figure 8.
Inhibition of import by E1-Ran can be overcome by the addition of excess p10. Assays were carried out at 21°C on permeabilized cells supplemented with 20 μg/ml human recombinant karyopherin α, 25 μg/ml human recombinant karyopherin β, 1 mM GTP, 20 μg/ml NLS-tagged, rhodamine-labeled human serum albumin, and the indicated concentrations of p10 and wild-type Ran·GDP or E1-Ran·GDP. Import was measured as mean nuclear fluorescence in arbitrary units. (A) Effect of p10 on wild-type Ran·GDP- and E1-Ran·GDP-mediated nuclear protein import. Assays were performed with the indicated amounts of wild-type Ran·GDP (▪, □) or E1-Ran·GDP (•, ○) in the presence (•, ▪) or absence (○, □) of 5 μg/ml p10. (B) Inhibition of nuclear protein import by E1-Ran. Assays were performed with (c 15 μg/ml wild-type Ran·GDP, 5 μg/ml p10, and the indicated concentrations of E1-Ran·GDP. (C) Effect of increasing concentrations of p10 on E1-Ran inhibition of nuclear protein import. Assays were performed with 15 μg/ml wild-type Ran·GDP in the presence (○) or absence (▪) of 30 μg/ml E1-Ran·GDP and the indicated concentrations of p10.
DISCUSSION
Diverse cellular processes are disrupted when the Ran GTPase cycle is perturbed, but it remains unclear which of these processes are normally directly regulated by Ran, and which are only affected secondarily, e.g., as a consequence of a failure in nuclear protein import. As one means of addressing this issue, we and others have constructed missense and deletion mutants of Ran to identify regions involved in modulating one biological process but not another (Dasso et al., 1994; Ren et al., 1995; Carey et al., 1996). These studies have exploited the close structural homology between Ran and RAS proteins (Scheffzek et al., 1995). Here, we have identified a residue, threonine 42, whose structural homologue in RAS can be mutated to perturb the latter protein’s interaction with effectors (Vojtek et al., 1993) and generated a mutant Ran protein with alanine substituted for this threonine (T42A).
The biochemical properties of T42A mutant Ran protein are consistent with its classification as an effector mutant, and we have designated it Ran effector mutant 1, E1-Ran. It appears to bind and exchange GTP and GDP normally, has little or no detectable intrinsic GTPase activity, and also catalyzes little or no GTP hydrolysis in the presence of RanGAP1 (Figures 4 and 5). In vitro, the E1-Ran·GTP complex interacts specifically with RanBP1, but the interaction is extremely weak compared with that between wild-type Ran·GTP and RanBP1 (Figures 1 and 2). E1-Ran does not interact with RanBP1 in a double-hybrid assay. The E1-Ran·GTP complex also fails to interact with karyopherin β or RanBP2/Nup358 (Figures 1 and 2). Except for its ability to exchange GTP and GDP normally, these properties of E1-Ran are essentially the same as those observed for another putative effector mutant, L43E Ran (Lounsbury et al., 1996a).
RanBP1, RanBP2/Nup358, and karyopherin β bind directly to Ran·GTP but not Ran·GDP, and, at least in the cases of RanBP1 and RanBP2, a conserved amino acid sequence motif, the so-called Ran-binding domain, mediates interactions with Ran (Butler and Wolfe 1994; Beddow et al., 1995; Ouspenski et al., 1995; Wu et al., 1995; Yokoyama et al., 1995; Dingwall et al., 1996; Hartmann and Görlich, 1995). Inasmuch as RanBP2/Nup358 and karyopherin β have well-defined roles in nuclear protein import and a budding yeast strain mutant in a RanBP1 homologous gene expresses defects in nuclear RNA and protein trafficking (Schlenstedt et al. 1995b), it seems appropriate to classify RanBP1, RanBP2/Nup358, and karyopherin β as putative effectors and E1-Ran as an effector mutant. The ability of the protein import factor p10 to bind both His*Tag wild-type Ran·GDP and His*Tag E1-Ran·GDP (Figure 3) suggests that p10 may interact with a different region of Ran.
Amino acid residue 42 is located on a polypeptide loop exposed at the surface of the Ran protein, hence readily accessible for interactions with other proteins (Scheffzek et al., 1995). The binding properties of the T42A Ran mutant strongly suggest that this “E1 loop” represents a major interacting domain, especially in regard to several well-characterized proteins that link the Ran GTPase cycle to nuclear protein import.
It was therefore not surprising to find that E1-Ran did not support nuclear protein import in a digitonin permeabilized cell assay (Figures 6, 7, 8). However, the fact that E1-Ran inhibited import stimulated by wild-type Ran was unexpected (Figures 6, 7, 8). Other mutants of Ran trapped in either their GTP- or GDP-bound forms have also been found to inhibit wild-type function in this system (Palacios et al., 1996), and in these cases the inhibition may be attributed to the ability of these mutants to bind, trap, and block the function of known import factors. For example, a nonhydrolyzable Ran·GTP could block RanBP2/Nup358, karyopherin β, or RanBP1 function, a Ran trapped in its GDP-bound form might sequester p10, and a Ran unable to exchange GTP for GDP could block the nucleotide exchange activity of the system (Dasso et al., 1994; Klebe et al., 1995; Rush et al., 1996). The properties of purified E1-Ran in vitro suggest that it might be trapped in a GTP-bound state (Figure 4), but that this E1-Ran·GTP should have little or no ability to compete with wild-type Ran for known effector interactions (Figures 1 and 2). This reasoning raised the possibility that E1-Ran might either bind and trap an additional, nondigitonin-extractable Ran-interacting protein required for nuclear import or that E1-Ran might inhibit Ran-stimulated protein import as a result of its ability to bind p10.
The demonstration that addition of excess p10 can overcome E1-Ran inhibition (Figure 8) certainly supports the latter possibility. However, although the in vitro-permeabilized cell assay has been of inestimable value in identifying and characterizing components of the nuclear protein import process, such as karyopherins α and β and Ran, the interpretation of specific quantitative results from this system is often complicated. For example, p10 is absolutely required in some circumstances (Figure 8; Moore and Blobel, 1994b) but appears to be only stimulatory in others (Paschal and Gerace, 1995; Chi et al., 1996, 1997; ). Moreover, different studies have yielded conflicting results related to the role of Ran in snRNP import (Marshallsay et al., 1996; Palacios et al., 1996), the requirement for a GTPase in addition to Ran (Sweet and Gerace, 1996; Weis et al., 1996), and the identity of the nucleotide- (GTP or GDP) charged form of Ran most effective for import stimulation (Chi et al., 1995, 1996; Melchior et al., 1995; Görlich et al., 1996). Most of these discrepancies probably reflect subtle differences in permeabilized cell preparation and/or assay conditions. With these cautions in mind, it should be noted that while the ability of excess p10 to overcome E1-Ran·GDP inhibition is easily accommodated by current models for nuclear protein import, E1-Ran inhibition of import may not be due only to p10 trapping.
The fact that E1-Ran did not block docking of the import substrate in either an isolated specific docking assay at 4°C or in the process of inhibiting import stimulated by wild-type Ran at 21°C (Figure 7) is consistent with the hypothesis that only karyopherins α and β are required for docking, and the observation that neither of these proteins interacts with E1-Ran (our unpublished observations; Figure 2). All of the nuclear protein import studies presented in this manuscript involved permeabilized cells supplemented with GDP-charged Ran plus excess free GTP. Ran must bind and hydrolyze GTP for nuclear protein import to occur in intact or permeabilized cells. However, the point in the overall import pathway at which Ran-mediated GTP hydrolysis occurs and the function of this hydrolysis,remain controversial (Melchior et al., 1995; Görlich et al., 1996).
Recently, another putative Ran effector mutant, L43E, has been examined for dominant phenotypes following expression in vivo (Carey et al., 1996). The L43E mutant protein inhibited cell proliferation, but appeared not to affect nuclear protein import. Whether the properties of this mutant, at least in terms of import inhibition, differ from those of E1-Ran or reflect different assay conditions, such as excess p10 in vivo, remains to be determined.
In addition to characterizing the interactions of the E1-Ran point mutant, we have also examined the properties of a RanGAP1 deletion mutant. None of the sequence motifs associated with GTPase-activating domains of the GAP proteins of other GTPases has been identified in RanGAP1. Our demonstration that a large carboxyl-terminal region of RanGAP1, well-conserved between mouse and human RanGAP proteins but absent from S. cerevisiae and S. pombe proteins, can be deleted without a drastic loss in GAP activity (Figure 4) suggests that the GTPase-stimulating activity of RanGAP1 will be located wholly or predominantly within the protein’s amino terminal 358 residues. This amino terminal region (c-del RanGAP1) contains a series of leucine-rich repeats that may be responsible for Ran binding, since such repeats in other proteins define regions responsible for protein–protein interactions (Kobe and Deisenhofer, 1995). The isolated carboxyl-terminal fragment, which lacks leucine-rich repeats, does not exhibit GAP activity (our unpublished observations). It might stabilize the Ran–RanGAP interaction or mediate interaction of Ran with downstream targets specific to mammalian systems.
The data presented here indicate that as with other small GTPases, Ran effector mutants are valuable tools for elucidating and confirming the mechanism(s) of Ran function. Specifically, it will be interesting to determine whether E1-Ran is defective in other biological processes attributed to Ran and whether E1-Ran can be used to identify additional Ran effectors.
ACKNOWLEDGMENTS
We thank Drs. J. Oppenheim and E. Coutavas for help with fast protein liquid chromatography. RanBD1 and BD4 His*Tag fusion constructs were a generous gift from Dr. J. Wu. This work was supported by grant CB-100 from the American Cancer Society (to M.G.R.) and grant GM-53678 from the National Institutes of Health (to M.S.M.). G.M. and A.V. were supported by Public Health Service Training grant GM07827. P.P.d.l.O was supported by a fellowship from the Ministerio de Educación y Ciencia, Spain. Computer work was carried out at the Research Computer Resource of New York University Medical Center, supported by National Science Foundation grant DIR-8908095.
Footnotes
Abbreviations used: aa, amino acid; BSA, bovine serum albumin; DTT, dithiothreitol; GST, glutathione S-transferase; His*Tag, histidine-tagged; MOPS, 3-N-morpholinopropane-sulfonic acid; NTF, nuclear transport factor; RanBD, Ran-binding domain; RanGAP1, Ran GTPase-activating protein 1; RCC1, regulator of chromosome condensation 1; RT, room temperature.
REFERENCES
- Adam SA, Sterne-Marr R, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avis JM, Clarke PR. Ran, a GTPase involved in nuclear processes: its regulators and effectors. J Cell Sci. 1996;109:2423–2427. doi: 10.1242/jcs.109.10.2423. [DOI] [PubMed] [Google Scholar]
- Beddow A, Richards SA, Orem NR, Macara IG. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Butler G, Wolfe KH. Yeast homologue of mammalian Ran binding protein 1. Biochim Biophys Acta. 1994;1219:711–712. doi: 10.1016/0167-4781(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Carey KL, Richards SA, Lounsbury KM, Macara IG. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the RAN/TC4 GTPase mediates an essential function independent of nuclear protein import. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Adam SA. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutavas E, Ren M, Oppenheim J, D’Eustachio P, Rush MG. Characterization of proteins that interact with the cell cycle regulatory protein Ran/TC4. Nature. 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- Dasso M. RCC1 in the cell cycle: the regulator of chromosome condensation takes on new roles. Trends Biol Sci. 1993;18:96–101. doi: 10.1016/0968-0004(93)90161-f. [DOI] [PubMed] [Google Scholar]
- Dasso M, Seki T, Azuma Y, Ohba T, Nishimoto T. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 1994;13:5732–5744. doi: 10.1002/j.1460-2075.1994.tb06911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Kandels-Lewis S, Séraphin B. A family of Ran binding proteins that includes nucleoporins. Proc Natl Acad Sci USA. 1995;92:7525–7529. doi: 10.1073/pnas.92.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DJ, Stutz F, Lescure A, Rosbash M. mRNA nuclear export. Curr Opin Genet Dev. 1994;4:305–309. doi: 10.1016/s0959-437x(05)80058-9. [DOI] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Grundmann U, Nerlich C, Rein T, Lottspeich F, Kuepper HA. Isolation of cDNA coding for the placental protein 15 (PP15) Nucleic Acids Res. 1988;16:4721. doi: 10.1093/nar/16.10.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Görlich D. A Ran-binding motif in nuclear pore proteins. Trends Cell Biol. 1995;5:192–193. doi: 10.1016/s0962-8924(00)88992-8. [DOI] [PubMed] [Google Scholar]
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Silver PA. A GTPase controlling nuclear trafficking: running the right way or walking RANdomly? Cell, 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- Lehmeier B, Amann E. Tac promoter vectors incorporating the bacteriophage T7 gene 10 translational enhancer sequence for improved expression of cloned genes in Escherichia coli. J Biotech. 1992;23:153–165. doi: 10.1016/0168-1656(92)90089-r. [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Beddow AL, Macara IG. A family of proteins that stabilize the Ran/TC4 GTPase in its GTP-bound conformation. J Biol Chem. 1994;269:11285–11290. [PubMed] [Google Scholar]
- Lounsbury KM, Richards SA, Carey KL, Macara IG. Mutations within the Ran/TC4 GTPase. Effects on regulatory factor interactions and subcellular localization. J Biol Chem. 1996a;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Richards SA, Perlungher RR, Macara IG. Ran binding domains promote the interaction of Ran with p97/β-karyopherin, linking the docking and translocation steps of nuclear import. J Biol Chem. 1996b;271:2357–2360. doi: 10.1074/jbc.271.5.2357. [DOI] [PubMed] [Google Scholar]
- Marshallsay C, Dickmanns A, Bischoff FR, Ponstingl H, Fanning E, Lührmann R. In vitro and in vivo evidence that protein and U1 snRNP nuclear import in somatic cells differ in their requirement for GTP-hydrolysis, Ran/TC4 and RCC1. Nucleic Acids Res. 1996;24:1829–1836. doi: 10.1093/nar/24.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Gerace L. Mechanisms of nuclear protein import. Curr Opin Cell Biol. 1995;7:310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. A G protein involved in nucleocytoplasmic transport: the role of Ran. Trends Biol Sci. 1994a;19:211–216. doi: 10.1016/0968-0004(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994b;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Forbes DJ. Nuclear import can be separated into distinct steps in vitro, nuclear pore binding and translocation. Cell. 1988;52:641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- Ouspenski II, Mueller UW, Matynia A, Sazer S, Elledge SJ, Brinkley BR. Ran-binding protein-1 is an essential component of the Ran/RCC1 molecular switch system in budding yeast. J Biol Chem. 1995;270:1975–1978. doi: 10.1074/jbc.270.5.1975. [DOI] [PubMed] [Google Scholar]
- Palacios I, Weis K, Klebe C, Mattaj IW, Dingwall C. Ran/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J Cell Biol. 1996;133:485–494. doi: 10.1083/jcb.133.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Villamarin A, Shih A, Coutavas E, Moore MS, LoCurcio M, Clarke V, Oppenheim J, D’Eustachio P, Rush MG. Separate domains of the Ran GTPase interact with different factors to regulate nuclear protein import and RNA processing. Mol Cell Biol. 1995;15:2117–2124. doi: 10.1128/mcb.15.4.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Mills AD, Dilworth SM, Laskey RA, Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988;52:655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Rush MG, Drivas G, D’Eustachio P. The small nuclear GTPase Ran: how much does it run? Bioessays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- Sazer S. The search for the primary function of the Ran GTPase continues. Trends Cell Biol. 1996;6:81–85. doi: 10.1016/0962-8924(96)80992-5. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Klebe C, Fritz-Wolf K, Kabsch W, Wittinghofer A. Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature. 1995;374:378–381. doi: 10.1038/374378a0. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Saavedra C, Loeb JDJ, Cole CN, Silver PA. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and the appearance of poly(A)+ RNA in the cytoplasm. Proc Natl Acad Sci USA. 1995a;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995b;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet DJ, Gerace L. A GTPase distinct from Ran is involved in nuclear protein import. J Cell Biol. 1996;133:971–983. doi: 10.1083/jcb.133.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana T, Imamoto N, Seino H, Nishimoto T, Yoneda Y. Loss of RCC1 leads to suppression of nuclear protein import in living cells. J Biol Chem. 1994;269:24542–24545. [PubMed] [Google Scholar]
- Tartakoff AM, Schneiter R. The nuclear GTPase cycle: promoting peripheralization? Trends Cell Biol. 1995;5:5–8. doi: 10.1016/s0962-8924(00)88925-4. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Weis K, Mattaj I, Lamond A. Identification of hSRP1a as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1051. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- Weis K, Dingwall C, Lamond AI. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran GTP binding sites, zinc fingers, a cyclophilin A homologous domain and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]