Abstract
Quorum sensing (QS) is a communication mechanism exploited by a large variety of bacteria to coordinate gene expression at the population level. In gram-negative bacteria, QS occurs via synthesis and detection of small chemical signals, most of which belong to the acyl-homoserine lactone (AHL) class. In such a system, binding of an AHL signal to its cognate transcriptional regulator (R-protein) often induces stabilization and subsequent dimerization of the R-protein, which results in the regulation of downstream gene expression. Existence of diverse QS systems within and among species of bacteria indicates that each bacterium needs to distinguish among a myriad of structurally similar chemical signals. We show, using a mathematical model, that fast degradation of an R-protein monomer can facilitate discrimination of signals that differentially stabilize it. Furthermore, our results suggest an inverse correlation between the stability of an R-protein and the achievable limits of fidelity in signal discrimination. In particular, an unstable R-protein tends to be more specific to its cognate signal, whereas a stable R-protein tends to be more promiscuous. These predictions are consistent with experimental data on well-studied natural and engineered R-proteins, and thus have implications for understanding the functional design of QS systems.
Keywords: gene regulation, quorum sensing, kinetic proofreading, systems biology
Introduction
Bacteria employ quorum sensing (QS) to regulate diverse cellular functions, such as antibiotic production, biofilm formation, and bioluminescence.1 In gram-negative bacteria, QS often involves the production of acyl-homeserine lactones (AHL), which are a class of small chemical signaling molecules freely diffusible across the cell membrane. Accumulation of the AHL molecules to a threshold concentration, which corresponds to bacteria reaching a critical density via reproduction, results in the full activation of the AHL-binding transcription factor and it is thereby rendered capable of inducing gene expression for functional regulation.1; 2; 3 A classical example of this type of QS is the lux system from the marine bacterium Vibrio fischeri. As the concentration of the small signal, 3OC6HSL in the case of V. fischeri, increases with increasing cell density, it binds to and activates its cognate transcription factor LuxR. Upon binding 3OC6HSL, LuxR dimerizes and subsequently binds the lux promoter leading to downstream gene expression. Employing such a QS motif, many other gram-negative bacteria modulate the expression of a diverse collection of genes with different physiological functions. 4
A salient property of AHL-based QS systems is that in the absence of AHL signal, the transcriptional regulator (R-protein monomer) is often highly unstable and requires its cognate AHL to be stabilized.5; 6; 7; 8; 9; 10 For example, structural studies of several well-characterized LuxR-type proteins indicate that the chemical signals essentially serve as “folding switches”, which, upon binding, enable the transcriptional regulators to fold into stable conformations. Further stabilization may occur as a result of the dimerization of the signal-monomer complex.5; 9 These observations suggest that a cell containing a QS module must continuously expend energy to synthesize and simultaneously degrade the R-protein in the absence of the signaling molecule. This raises the question as to what benefit the cell might receive, in terms of optimizing the function of its QS system, by employing such a rapid turnover rate, as opposed to a slower one, of the QS signal response proteins.
Several explanations have been proposed to address this apparent paradox. Rapid turnover of the monomer can prevent premature activation of the QS system without the cognate signal.3 This property could also enhance effects of nonlinear protein degradation, which could increase the ratio of protein concentrations between the activated and inactivated states, thereby enhancing the “bistable” character of the system.11 This is analogous to the modulation of the dynamic range of signaling proteins by phosphorylation-mediated protein stabilization or destabilization.12 Our bifurcation analysis further validates this hypothesis that the LuxR degradation rate can indeed influence the bistable region of the QS system signal response when LuxI and LuxR are involved in the positive feedback loop determining LuxR production in a coupled fashion (Figure S5C,D in the Supplementary Information (SI)). However, the bistable region of the QS system signal response is independent of the LuxR degradation rate constant when only LuxR expression is subjected to positive auto-regulation (Figure S5A,B, also see ref 13). Finally, rapid turnover of the R-protein may help reduce variability in gene expression controlled by a QS module.14
Here we suggest another plausible explanation based on a kinetic model of the QS signal discrimination process. Specifically, the rapid turnover of the R-protein enables the QS system to distinguish between multiple signaling molecules with increased fidelity. This mechanism is analogous to the well-established “kinetic proofreading” mechanism, which has been adopted to explain the fidelity of tRNA selection, T-cell receptor signal transduction, disentanglement of DNA by topoisomerases, and phosphorylation cascades.15; 16; 17; 18 Since the vast array of small signal molecules present in the environment of naturally occurring bacteria could lead to undesirable crosstalk among species-specific QS systems from different species, increasing the fidelity of the accessible dynamic range of QS signal recognition would likely serve to optimize QS systems that function in a species-specific manner. Our analysis demonstrates that the relationship between signal recognition fidelity and the rate of signal transducing protein degradation may provide a justification for the maintenance of highly unstable R-proteins as generic components of species-specific QS systems.
Results
Fidelity in QS signal recognition
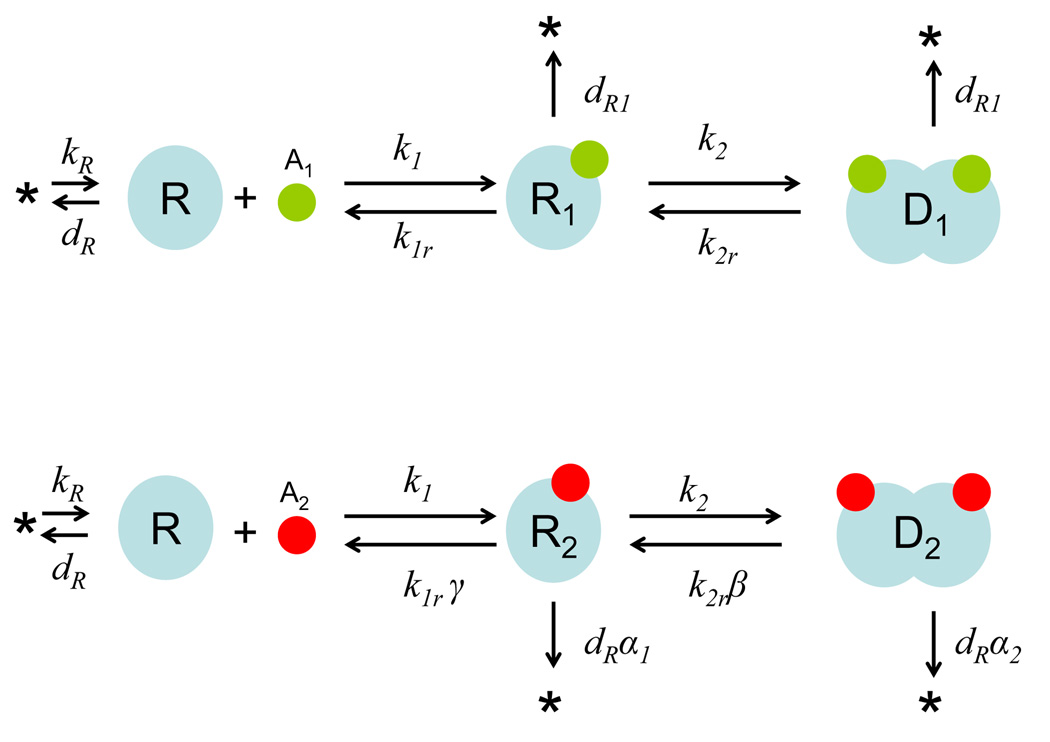
Figure 1 shows the binding of an unstable transcription regulator (R) by either its cognate signal (A1) or a non-cognate signal (A2). The binding of A1 and R forms R1, which dimerizes to form D1. The binding of A2 and R forms R2, which dimerizes to form D2. The two signaling branches are structurally symmetric but differ in the rate constants of constituent reactions, which provide the basis for discrimination. As detailed in Materials and Methods, we have developed a simple model that allows us to predict effects of modulating particular parameters on signaling fidelity (f), defined as:
| (1) |
where D1ss and D2ss are the steady-state concentrations of the dimers resulting from A1 and A2 binding, respectively (Figure 1). f is used to characterize the accuracy of discrimination in the quorum-sensing signaling process. The greater f is, the more preferably the R-protein will respond to the cognate signal (A1) than to the non cognate signal (A2).
Figure 1. Signal discrimination by a quorum sensing regulator (R).
A1 is the cognate signal; A2 is a non-cognate signal. R1 and R2 are the signal-monomer complexes. D1 and D2 are dimerized complexes. See main text and Table S1 for details on parameters.
Based on the kinetic model (Equations 6–8), we derive an analytical solution for f in terms of system parameters (see SI for details) by assuming A1 = A2 = A:
| (2) |
K represents the dissociation constant for both the signal-protein complexes and the dimers (K = k1r/k1 = k2r/k2). Equation (2) represents the limiting case where the binding reactions are much faster than protein synthesis and decay. It is the basis for all numerical calculations unless noted otherwise. If the binding reactions are irreversible (K = 0), then f can be simplified to a concise expression, which is the ratio of the system boundary output for D1 to D2:
| (3) |
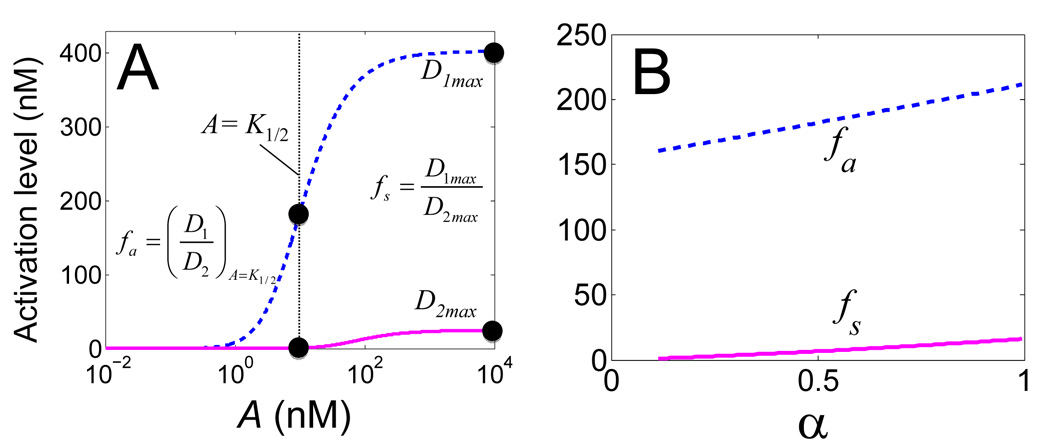
In the following analysis, we consider two fidelity metrics, depending on the signal concentrations at which the fidelity function defined in Equation (1) is evaluated (Figure 2A). The first metric (fa) corresponds to the signal concentration eliciting half-maximal induction of D1 (Equation (4), where A = K1/2). The second metric (fs) corresponds to the saturating induction of both D1 and D2 (Equation (5)).
| (4) |
| (5) |
Figure 2. Quantifying fidelity in signal recognition using two metrics.
(A) Dose responses of D1 and D2 to A1 and A2 respectively. fa is evaluated at the signal concentration (K1/2) that leads to half-maximum induction of D1. fs is evaluated at a saturating signal concentration.
(B) Modulation of fidelity (fa or fs) by α (for β = γ =10), which characterizes the differential protein stabilization by A1 and A2. A similar trend is observed for other values of β and γ (Figure S1).
As shown in Figure 2B, both fa and fs monotonically increase with α. This makes intuitive sense: increasing α causes faster depletion of products resulting from the A2-pathway than that from the A1-pathway, which leads to a decrease in the steady-state concentration of D2 relative to that of D1.
Fidelity increases with asymmetry in signal binding and R-protein stabilization
In this framework, three key parameters determine the potential for signal discrimination (α, β, and γ). Figure S1 shows the dependence of f on α for varying β and γ. As α increases, so do the degradation rates of the non-cognate signal-monomer complex (R2) and dimer (D2), and consequently f. This same trend is observed, albeit with less sensitivity to α, when the binding reactions are asymmetric between the two pathways (β, γ > 1). This intuitive result highlights two aspects of the role of differential stabilization in signal discrimination. First, it acts synergistically with differential binding of A1 (cognate) and A2 (non-cognate) to the R-protein, and subsequent dimerization. Second, its contribution is more pronounced for smaller differences in the binding kinetics between the two pathways.
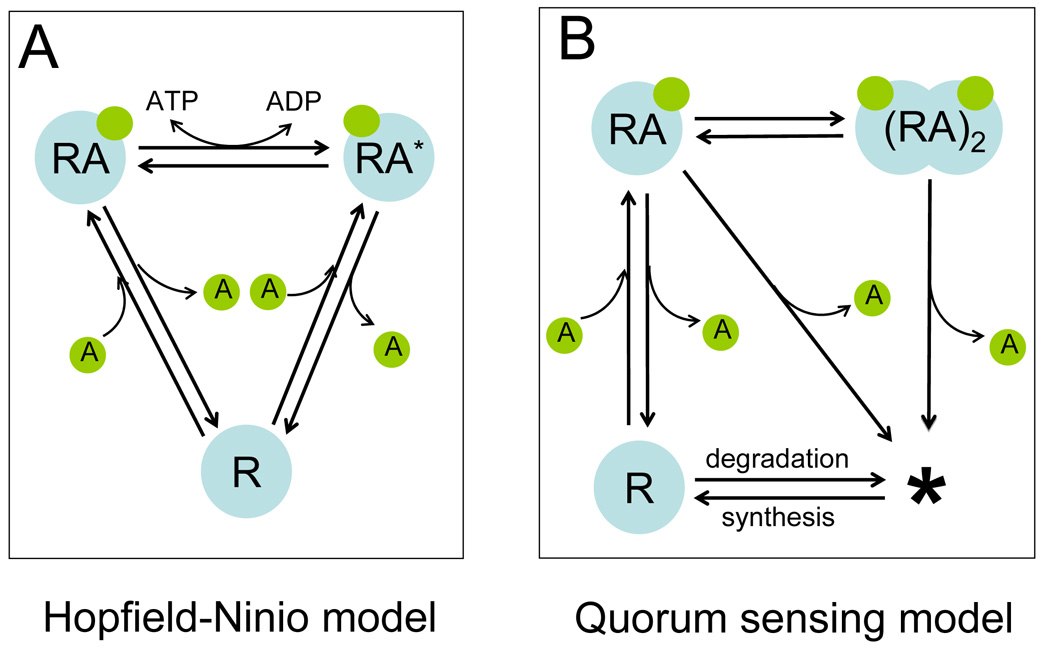
We note that the QS model represents a special case of two-cycle kinetic proofreading that relies on synthesis and degradation as the energy source, rather than direct hydrolysis of ATP, from which fidelity is derived (see Figure 4). Indeed, as indicated in Figure S1B, for increasing β (= γ), the corresponding increase in fs and fa are always less than β3, which would correspond to the thermodynamic limit of fidelity by kinetic proofreading with two cycles, given unlimited available energy to expend.15; 19
Figure 4. QS signal recognition as an example of kinetic proofreading.
(A) The canonical Hopfield-Ninio (HN) model of kinetic proofreading with one cycle.
(B) QS signaling as two-cycle kinetic proofreading.
In QS, the degradation of the R-protein provides a second cycle of proofreading. R is a receptor protein (HN model) or an R-protein (QS model). A is a signaling molecule. RA is the complex of R and A. In the HN model, RA* the activated form RA. The star (*) in the QS model represents unspecified substrates or degradation products.
R-protein degradation sets the limits of fidelity
Our model assumes, by construction, that both signals tend to stabilize the R-protein and that A1 is capable of stabilizing the R-protein to a greater extent than A2. Thus, the asymmetry in the differential protein stabilization is bounded by dR1/dR < α <1. In a real system, dR1 is bounded by cell growth rate. If stabilization by the cognate signal is so strong that R1 and D1 are not degraded inside the cell, their concentrations would only be reduced by dilution due to cell growth. In this scenario, the only way to modulate the dynamic range in differential stabilization is to vary dR, the turnover rate of the unbound R-protein. Thus, we ask how the fidelity in signal recognition can be modulated by varying dR, while fixing the asymmetry in the binding kinetics.
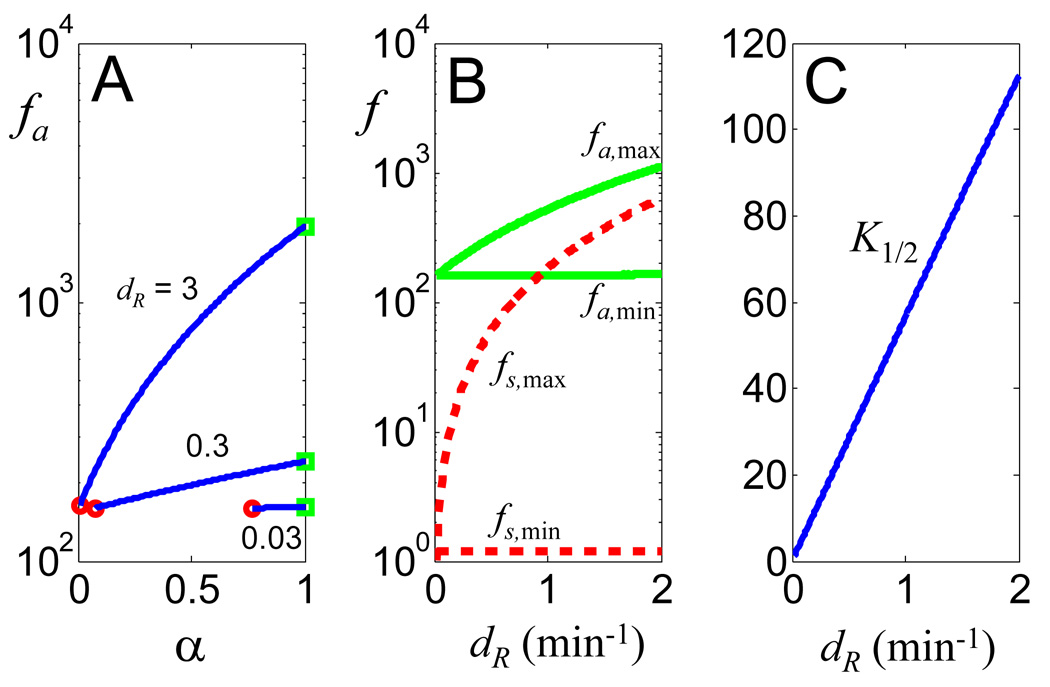
As shown in Figure 3A, for each specific dR > dR1, f always increases with increasing α (constrained between dR1/dR and 1). Furthermore, we note that f on average increases drastically with increasing dR, and that it is bounded for a specific dR value. The fidelity reaches minimum (fmin) when α = dR1/dR (circles, no differential R-protein stabilization between signals) and it reaches maximum (fmax) when α = 1 (squares, A2 does not stabilize the R-protein). As shown in Figure 3B, for fixed β and γ, fmin is essentially independent of dR, while increasing dR leads to increasing fmax for both fidelity metrics. These results indicate that dR uniquely defines the dynamic range of fidelity in signal recognition by exhibiting control over the maximal limit of signal recognition fidelity.
Figure 3. Increasing the R degradation rate constant enhances signaling fidelity.
(A) Dependence of fa on α for varying dR (β = γ = 10). Note that dR determines the range of α for a constant dR1 (= 0.023 min−1).
(B) Modulation of the maximal (α = 1) and minimal (α = dR1/dR) limits of fidelity by dR. For both metrics, the maximal fidelity increases with dR, but the minimal fidelty does not change with dR.
(C) The half-activation threshold (K1/2, nM) by the cognate signal increases with dR.
For fa, this increase in fidelity that results from increasing dR comes with an energetic cost comprised of two components. The first component, shared by the fs metric, is the rapid turnover of the R-protein per se. The second component, unique to fa, is that increasing dR necessitates a greater activation threshold for the cognate signal to induce R-protein function. Figure 3C shows that for the metric fa, the signal concentration that corresponds to the half-maximal activation of the dimer increases approximately linearly with increasing dR. Also, the dependence of fmax on dR is more sensitive as β and γ increase as shown in Figures S3 and S4.
Discussion
Quorum sensing has been identified as a potential evolutionary basis for both inter- and intracellular signaling pathways.20 Mechanisms that account for accuracy in quorum sensing may provide insights into the understanding of these systems and facilitate their use in constructing synthetic gene circuits.
QS signal discrimination as an example of kinetic proofreading
Many receptor proteins display highly specific binding affinity to their ligands with nanomolar accuracy. The tuning of binding affinity can lead to high fidelity signal recognition. However, the efficacy of tuning binding affinity towards a particular signal is limited, if multiple ligands have similar chemical structures. QS seems to employ a complementary approach to achieve high fidelity for signal discrimination. A highly unstable R-protein has a certain binding affinity for its cognate signal. To improve fidelity beyond this level, the R-protein employs a mechanism analogous to kinetic proofreading15; 21 that takes advantage of its high instability in the absence of its cognate signal.
Since its initial discovery, kinetic proofreading has been found to be implicated in diverse cellular processes. In each case, fidelity in recognizing the correct substrate relies largely on asymmetry between binding reactions to the correct substrate and those to incorrect substrates. In QS, binding of the R-protein to the signal and subsequent R-protein dimerization provide a sequential mechanism similar to that of the Hopfield-Ninio model of kinetic proofreading (Figure 4A). This is reflected by the fact that increasing the asymmetry in binding and dimerization processes directly leads to increased fidelity in recognizing the cognate signal (Figure 3 & Figure S1–4). In QS, the degradation of the R-protein essentially provides another cycle of proofreading to enhance signal recognition (Figure 4B).
Consequently, QS signal recognition critically depends on differential stabilization of the R-protein upon signal binding. Given fixed asymmetry in binding reactions, the fidelity in recognizing the cognate signal can be increased due to its ability to stabilize the R-protein to a greater extent than the non-cognate signal. This aspect is illustrated in Figure 3B, Figure S3 and S4: the degradation rate constant of the monomer directly determines the theoretical bounds of the signaling fidelity. In particular, increasing the monomer degradation rate constant serves to amplify the fidelity of the dynamic range within which the fidelity of signal discrimination can be modulated by differential stabilization.
The range of achievable fidelity is sensitive to changes in the monomer degradation rate constant near its base value (an R-protein with a half-life of ~3.5 minutes, see Table S1). Differences in the binding kinetics between the cognate and non-cognate signal pathways serve to amplify the effect of modulating the monomer degradation rate with respect to its ability to alter fidelity (Figure S3). The fidelity is necessarily enhanced by differential binding kinetics; however, simultaneous increase in the R-protein degradation rate results in further enhancement of QS signaling fidelity beyond that achievable as a result of differential binding kinetics alone. As mentioned, our results indicate that this conclusion is particularly relevant near physiologically achievable R-protein degradation rates based on the experimental characterization of QS signal proteins.
We have highlighted two properties of QS where signal discrimination can be modulated: the binding kinetics of the signal to the R-protein and subsequent degradation of the resulting complexes. Beyond these means of altering signaling fidelity, it is intuitive that there may exist a differential ability between cognate and non-cognate signal bound dimers of LuxR-type proteins to bind their target promoters and thus affect transcription.22 Signal discrimination at this level will likely serve to enhance the effects of the mechanisms that we have identified.
The stability of an R-protein as a major determinant of its specificity
In our model, we have assumed that the major determinant of QS-mediated gene expression is the accumulation of the active dimer. Under this assumption, our model predicts an inverse correlation between the R-protein stability and signal recognition fidelity. In particular, increasing dR would lead to an increase in the achievable fidelity in recognizing the cognate signal. Based on this, we expect that a highly unstable R-protein tends to be specific to its cognate signal, whereas a stable R-protein tends to be more promiscuous in its response to signaling molecules.
To evaluate this notion, we examined published data on several well-studied LuxR-type QS activators. As shown in Table 2, we indeed observe a significant inverse correlation between the R-protein stability and its specificity (SI). For example, several R-proteins with high specificity to their cognate signals, such as LuxR, LasR, and TraR, are all highly unstable in the absence of their respective cognate signals (see Table 2). These quorum sensing systems may be suitable for intraspecies communication that requires high accuracy and specificity in signal recognition. In contrast, the LuxR homolog SdiA is particularly promiscuous, as demonstrated by its response to multiple signaling molecules.7; 23; 24; 25; 26 Thereby, it is actually impractical to define any specific signal as SdiA’s cognate signal. SdiA inclusion bodies obtained via expression in the absence of a signaling molecule have been shown to be able to refold in the presence of C8-HSL.7 This is in stark contrast to signal-specific R-proteins (e.g. LuxR, LasR, and TraR), which are highly unstable without signaling molecules. Similarly, CarR, responsible for activating carbapenem antibiotic production in E. carotovora, can dimerize by itself and can respond to multiple signals.27 In fact, in the absence of AHL signals, mere overexpression of CarR can lead to the activation of its cognate promoter resulting in carbapenem antibiotic production.28 These quorum sensing systems may serve as an interspecies, extraspecies, or at least less signal-specific detection system, where promiscuous activation of the downstream genes is either beneficial or fails to bear a cost significant enough to result in negative selection.
Remarkably, this correlation between stability and signal specificity also appears to exist among mutant R-proteins engineered by directed evolution or site-directed mutagenesis. Collins et al previously generated, by directed evolution, a mutant LuxR able to respond promiscuously to multiple AHLs and another mutant tuned to respond specifically to a non-cognate AHL (for the wild type LuxR). Both mutants display significant alteration in their stability upon binding to AHL(s).29; 30 It has also been shown, by site-directed mutagenesis of TraR, that mutations decreasing the ability of TraR to bind its cognate signal result in enhanced susceptibility to degradation.31 Previous searches for constitutively active forms of LuxR and its analogs have shown that deletion of the N-terminal half of these proteins results in AHL-independent activation of the lux promoter. This observation led to the speculation that the N-terminal AHL binding domain of LuxR-type proteins is responsible for “masking” the DNA-binding activity of the C-terminal domain.32 However, a search for a constitutively active form of TraR via random mutagenesis resulted in N-terminal fusion of TraR to an aminoglycoside N-acetyltransferase that enhanced the overall protein stability and allowed TraR to activate downstream gene expression with its cognate signal.33 This indicates a potential clarification to the previously mentioned theory on the function of the N-terminus of LuxR analogs: the ability of the N-terminus to “mask” the DNA-binding activity of the C-terminus is potentially due to the destabilization or otherwise enhanced affinity for proteolysis it confers upon the protein rather than the capacity to physically cover the DNA-binding domain alone. Together, these results indicate that, for the case of LuxR-type proteins, protein stability and signal specificity are likely to be interrelated. The identification of such a relationship in LuxR-type proteins might be progressed towards through examining methods of classifying such proteins according to these characteristics.
Phylogenetic analysis is helpful for elucidating evolutionary relationships within classes of proteins.34 It has been used to analyze the evolutionary trajectory of Lux-type quorum sensing proteins (including both R-proteins, responsible for the signal response, and I-proteins, the signal synthases). In contrast, both our modeling analysis and validation by experimental data suggest a dynamic and functional dichotomy between two types of R-proteins. Our model predicts that one class, functionally optimized by high specificity, would also be more likely to be inherently less stable than the other class, functionally optimized by exhibiting a non-specific signal response. Such a functional classification is useful for understanding the role of QS in controlling cellular behavior, and for using them to program population dynamics.35; 36; 37; 38; 39 Within this classification, it is intuitive that generic detection of chemical signals is likely to be energetically less costly than specific detection. According to this principle and based on our analysis, it appears that by expending energy in rapid protein turnover—that is by synthesizing and subsequently degrading regulatory proteins at relatively high rates—a QS module can significantly increase its potential for accuracy in distinguishing among structurally similar signals.
Materials and methods
As shown in Figure 1, we assume that the R-protein is synthesized at a constant rate (kR) and degrades following first-order kinetics (dR). Without loss of generality, we further assume that the association constants of A1 or A2 binding to R are identical (k1) but the dissociation constants differ by a factor of γ. The parameter γ modulates the binding kinetics of the non-cognate signal to R relative to that of the cognate signal. We only consider the case where γ > 1, which indicates that the non-cognate signal A2 binds less favorably to the R-protein than does the cognate signal A1, but not vice versa. Similarly, we assume identical association constants (k2) for dimerization reactions but that the dissociation constants differ by a factor of β (>1). β modulates the dimerization kinetics of the non-cognate signaling pathway relative to that of the cognate pathway. Based on these assumptions, we model QS signal recognition with the following equations:
| (6) |
| (7) |
| (8) |
With this model, recognition of the cognate signal (Figure 1) corresponds to: Recognition of a non-cognate signal corresponds to: The base parameter set for our analysis is detailed in Table S1 (SI). Note that the signal concentrations, A1 and A2, are represented as free parameters and are not treated dynamically.
Compared with R, R1 degrades with a reduced degradation rate constant (dR1 < dR), which accounts for stabilization by A1. The degradation rate constant of R2 (dR α1) is greater than dR1 (α1 > dR1/dR) but smaller than dR (α1 < 1). The parameters α1 and α2 modulate the protein degradation rates within the non-cognate signal pathway of the signal-monomer complex and dimer relative to the degradation rate of the protein monomer itself. Thus, the non-cognate signal is able to stabilize the R-protein but does so less effectively than does the cognate signal. We allow, in general, for differential stabilization between the signal-binding and dimerization steps by distinguishing between the parameter that modulates the degradation of the signal-monomer complex (α1) and the analogous parameter that modulates the degradation rate of the dimer (α2) in order to account for this possibility. Analysis including independent modulation of α1 and α2 showed that its effect on the ability of dimerization to perform an additional layer of proofreading is a mere question of degree and not one of mechanistic significance (results not shown). Therefore, the analysis presented herein assumes α1= α2 = α.
Supplementary Material
Table 1.
Selected quorum sensing transcriptional activators
| Protein Name | Native hosts | Cognate signal | Other inducing signals | Conformation without signals | Source |
|---|---|---|---|---|---|
| LuxR | V. fischeri | 3OC6HSL | - | Insoluble inclusion bodies | 6 |
| LuxR-G 2A-H |
Mutants of LuxR | - | 3OC6HSL C8HSL |
Unknown | 29; 30 |
| TraR | A. tumefaciens | 3OC8HSL | - | Insoluble inclusion bodies | 8; 9 |
| LasR | P. aeruginosa | 3OC12HSL | - | Insoluble inclusion bodies | 22; 23 |
| SdiA |
S. enterica E. coli |
- | 3OC6HSL 3OC8HSL C6-HSL C8-HSL |
Insoluble inclusion bodies | 7; 25 |
| CepR | B. cenocepacia | C8-HSL | - | Insoluble inclusion bodies | 40 |
| RhlR | P. aeruginosa | C4HSL | - | Dimer | 41; 42 |
| ExpR | E. carotovora | 3OC6HSL | - | Dimer | 43; 44 |
| CarR | E. carotovora | 3OC8HSL | 3OC6HSL 3OC4HSL 3OC10HSL 3OC12HSL |
Dimer | 27; 28 |
Acknowledgments
We thank Yu Tanouchi for helpful discussions. We also thank two anonymous reviewers for their comments and suggestions, which have improved presentation of our results and their interpretations. In particular, Figure 4 was drawn based on a draft figure suggested by one of the reviewers. This work was partially supported by the National Science Foundation (BES- 0625213), the National Institutes of Health (5R01CA118486), the David and Lucile Packard Foundation, and a Duke University Pratt School of Engineering Fellowship for undergraduate research (to CS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller MB, Bassler BL. Quorum sensing in bacteria. Annual Review of Microbiology. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 2.Fuqua C, Parsek MR, Greenberg EP. Regulation of Gene Expression By Cell-To-Cell Communication: Acyl-Homoserine Lactone Quorum Sensing. Annual Review of Genetics. 2003:6. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 3.Waters CM, Bassler BL. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annual Review of Cell and Developmental Biology. 2005;216:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 4.Fuqua C, Winans SC, Greenberg EP. Census and Consensus in Bacterial Ecosystems: The LuxR-LuxI Family of Quorum-Sensing Transcriptional Regulators. Annual Review of Microbiology. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 5.Qin Y, Luo ZQ, Smyth AJ, Gao P, Beck von Bodman S, Farrand SK. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO. 2000;19:5212–5221. doi: 10.1093/emboj/19.19.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbanowski ML, Lostroh CP, Greenberg EP. Reversible Acyl-Homoserine Lactone Binding to Purified Vibrio fischeri LuxR Protein. J. Bacteriol. 2004;186:631–637. doi: 10.1128/JB.186.3.631-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol. 2006;355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, Winans SC. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weingart CL, White CE, Liu S, Chai Y, Cho H, Tsai CS, Wei Y, Delay NR, Gronquist MR, Eberhard A, Winans SC. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol Microbiol. 2005;57:452–467. doi: 10.1111/j.1365-2958.2005.04656.x. [DOI] [PubMed] [Google Scholar]
- 11.Buchler NE, Gerland U, Hwa T. Nonlinear protein degradation and the function of genetic circuits. Proceedings of the National Academy of Sciences of the United States of America. 2005;1026:9559–9564. doi: 10.1073/pnas.0409553102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee T, Yao G, Nevins J, You L. Sensing and integration of Erk and PI3K signals by Myc. PLoS Comput Biol. 2008;4:e1000013. doi: 10.1371/journal.pcbi.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song H, You L. Evolving sensitivity. ACS Chem Biol. 2006;1:681–682. doi: 10.1021/cb6004596. [DOI] [PubMed] [Google Scholar]
- 14.Tanouchi Y, Tu D, Kim J, You L. Noise reduction by diffusional dissipation in a minimal quorum sensing motif. PLoS Computational Biology. 2008 doi: 10.1371/journal.pcbi.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopfield JJ. Kinetic Proofreading: A New Mechanism for Reducing Errors in Biosynthetic Processes Requiring High Specificity. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swain PS, Siggia ED. The Role of Proofreading in Signal Transduction Specificity. Biophysical Journal. 2002;82:2928–2933. doi: 10.1016/S0006-3495(02)75633-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- 19.Qian H. Reducing Intrinsic Biochemical Noise in Cells and Its Thermodynamic Limit. Journal of Molecular Biology. 2006;362:387–392. doi: 10.1016/j.jmb.2006.07.068. [DOI] [PubMed] [Google Scholar]
- 20.March JC, Bentley WE. Quorum sensing and bacterial cross-talk in biotechnology. Current Opinion in Biotechnology. 2004;15:495–502. doi: 10.1016/j.copbio.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 22.Schuster M, Urbanowski ML, Greenberg EP. Promoter Specificity in Pseudomonas aeruginosa Quorum Sensing Revealed by DNA Binding of Purified LasR. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottomley MJ, Muraglia E, Bazzo R, Carfì A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. Journal of Biological Chemistry. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 24.Janssens JCA, Metzger K, Daniels R, Ptacek D, Verhoeven T, Habel LW, Vanderleyden J, De Vos DE, De Keersmaecker SCJ. Synthesis of N-acyl homoserine lactone analogues reveals strong activators of SdiA, the Salmonella typhimurium LuxR homologue. Applied and Environmental Microbiology. 2006 doi: 10.1128/AEM.01451-06. AEM.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. Journal of Bacteriology. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith JN, Ahmer BMM. Detection of Other Microbial Species by Salmonella: Expression of the SdiA Regulon. Journal of Bacteriology. 2003;182:1357–1366. doi: 10.1128/JB.185.4.1357-1366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch M, Todd DE, Whitehead NA, McGowan SJ, Bycroft BW, Salmond GP. N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO. 2000;19:631–641. doi: 10.1093/emboj/19.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGowan S, Sebaihia M, Jones S, Yu B, Bainton N, Chan PF, Bycroft B, Stewart GS, Williams P, Salmond GP. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator [published erratum appears in Microbiology 1995 May;141(Pt 5):1268] Microbiology. 1995;141:541–550. doi: 10.1099/13500872-141-3-541. [DOI] [PubMed] [Google Scholar]
- 29.Collins CH, Leadbetter JR, Arnold FH. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotech. 2006;24:708–712. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- 30.Collins CH, Arnold FH, Leadbetter JR. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Molecular Microbiology. 2005;55:712–723. doi: 10.1111/j.1365-2958.2004.04437.x. [DOI] [PubMed] [Google Scholar]
- 31.Chai Y, Winans SC. Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: alteration of autoinducer specificity. Molecular Microbiology. 2004;51:765–776. doi: 10.1046/j.1365-2958.2003.03857.x. [DOI] [PubMed] [Google Scholar]
- 32.Choi SH, Greenberg EP. The C-Terminal Region of the Vibrio fischeri LuxR Protein Contains an Inducer-Interdependent lux Gene Activating Domain. Proc Natl Acad Sci USA. 1991;88:11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai Y, Winans SC. Amino-Terminal Protein Fusions to the TraR Quorum-Sensing Transcription Factor Enhance Protein Stability and Autoinducer-Independent Activity. Journal of Bacteriology. 2005;187:1219–1226. doi: 10.1128/JB.187.4.1219-1226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerat E, Moran NA. The Evolutionary History of Quorum-Sensing Systems in Bacteria. Mol Biol Evol. 2004;21:903–913. doi: 10.1093/molbev/msh097. [DOI] [PubMed] [Google Scholar]
- 35.Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balagadde FK, You L, Hansen CL, Arnold FH, Quake SR. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 37.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 38.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci U S A. 2007;104:17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weingart CL, White CE, Liu S, Chai Y, Cho H, Tsai C-S, Wei Y, Delay NR, Gronquist MR, Eberhard A, Winans SC. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Molecular Microbiology. 2005;57:452–467. doi: 10.1111/j.1365-2958.2005.04656.x. [DOI] [PubMed] [Google Scholar]
- 41.Medina G, Juarez K, Valderrama B, Soberon-Chavez G. Mechanism of Pseudomonas aeruginosa RhlR Transcriptional Regulation of the rhlAB Promoter. Journal of Bacteriology. 2003;185:5976–5983. doi: 10.1128/JB.185.20.5976-5983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventre I, Ledgham F, Prima Vr, Lazdunski Ae, Foglino M, Sturgis JN. Dimerization of the quorum sensing regulator RhlR: development of a method using EGFP fluorescence anisotropy. Molecular Microbiology. 2003;48:187–198. doi: 10.1046/j.1365-2958.2003.03422.x. [DOI] [PubMed] [Google Scholar]
- 43.Castang S, Reverchon S, Gouet P, Nasser W. Direct Evidence for the Modulation of the Activity of the Erwinia chrysanthemi Quorum-sensing Regulator ExpR by Acylhomoserine Lactone Pheromone. Journal of Biological Chemistry. 2003;281:29972–29987. doi: 10.1074/jbc.M601666200. [DOI] [PubMed] [Google Scholar]
- 44.Reverchon S, Bouillant ML, Salmond G, Nasser W. Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Molecular Microbiology. 1998;29:1407–1418. doi: 10.1046/j.1365-2958.1998.01023.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.