Abstract
KLF6 (Kruppel-like factor 6) is a zinc finger transcription factor and a tumor suppressor that is frequently mutated in prostate cancer. KLF6 suppresses tumor growth and induces apoptosis in cancer cells through mechanisms still not defined. Here we show that KLF6 induces apoptosis in prostate cancer cells by ATF3 (activating transcription factor 3) expression. KLF6 binds directly to and activates the ATF3 promoter. ATF3 induced apoptosis when ectopically expressed in cells, whereas knockdown of ATF3 by small interference RNA blocked KLF6-induced apoptosis. KLF6 mutants derived from clinical prostate cancers failed to activate the ATF3 promoter and were unable to induce apoptosis. Furthermore, stress conditions (exposure to staurosporine and hypoxia induced by sodium azide) caused significant increase in ATF3 expression and induced apoptosis, whereas knockdown of KLF6 by small interference RNA blocked the increase of ATF3 as well as the induction of apoptosis in these conditions. Thus, ATF3 is a key mediator of KLF6-induced apoptosis in prostate cancer cells.
KLF6 (Kruppel-like factor 6) is a tumor suppressor protein that is down-regulated or mutated in several types of cancers, including prostate cancer (1–3). KLF6 is a zinc finger transcription factor that binds to a GC box and regulates the expression of target genes. It has been shown that KLF6 suppresses tumor growth through activating p21WAF1/Cip1, an inhibitor of the cyclin-dependent kinases, in both cultured cells and a transgenic mouse model (2, 4). KLF6 also directly interacts with cyclin D1 to suppress cyclin-dependent kinase 4 and causes cell cycle arrest (5). Besides growth inhibition, KLF6 has also been shown to induce apoptosis in non-small cell lung cancer cells (1). However, the mechanism of KLF6-induced apoptosis is still not known.
ATF3 (activating transcription factor 3) is a member of the ATF/cAMP-response element-binding protein family of transcription factors (6). ATF3 is rapidly up-regulated under various stress conditions, including hepatotoxicity, UV/ionizing radiation, and exposure to DNA-damaging agents (7). ATF3 can also be induced by ischemia and hypoxia (8–10). ATF3 is a pro-apoptotic protein. It induces apoptosis in ovarian cancer cells (11) and enhances etoposide- or camptothecin-induced apoptosis in HeLa cells (12). Although transgenic mice expressing ATF3 in beta cells develop abnormal islets and defects secondary to beta cell apoptosis, primary islets derived from ATF3 knock-out mice were partially protected from cytokine- and nitric oxide-induced apoptosis (13). Additionally, fibroblasts from ATF3 knock-out mice were partially protected from UV-induced apoptosis (10). On the other hand, it was shown that ATF3 overexpression promoted invasiveness of prostate tumor cells in vitro and significantly enhanced spontaneous lung metastasis without affecting primary tumorigenicity in a severe combined immunodeficient mouse model (14). Interestingly, it was also shown that ATF3 has a dichotomous role in cancer development, while enhancing apoptosis in the untransformed MCF10A cells, ATF3 inhibited apoptosis in the more aggressive MCF10CA1a cells and enhanced cell mobility (15).
We found that KLF6 induces apoptosis when ectopically expressed in prostate cancer cells. To understand the mechanism of KLF6-induced apoptosis, we employed microarray gene expression analysis and identified ATF3 as one of the target genes regulated by KLF6. We further demonstrated that ATF3 is a key mediator of KLF6-induced apoptosis in prostate cancer cells. KLF6 and ATF3 are required for cancer cell apoptosis in stress conditions.
EXPERIMENTAL PROCEDURES
Cells and Transfection—Human prostate cancer cell lines PC-3 and LNCaP were purchased from American Type Culture Collection. The cells were cultured in RPMI 1640 media containing 10% fetal bovine serum. For transient transfection, plasmids were transfected into cells using Lipofectamine™Plus reagent (Invitrogen) following the manufacturer's protocol.
Drugs and Chemicals—Staurosporine and sodium azide were purchased from Sigma. Staurosporine was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 1 mm. Sodium azide was dissolved in PBS3 at concentration of 4 m. In all studies, an equivalent amount of diluent (DMSO or PBS) was added to the culture media as a negative control.
Plasmid Construction—Human cDNAs encoding full-length KLF6 gene and SV2 variant were obtained by PCR amplification using an EST clone (I.M.A.G.E. clone ID 3623401) as template. KLF6 and KLF6-SV2 cDNAs were subcloned into pCMV-Tag2 (Stratagene) vector to express a FLAG-tagged protein. For doxycycline-inducible expression using the Tet-On advanced system, KLF6 was subcloned into pTRE-Tight vector (Clontech). The pCG and pCG-ATF3 expression vectors were kindly provided by Dr. Tsonwin Hai at Ohio State University. The ATF3 promoter reporter plasmids Luc-1850, Luc-632, Luc-111, and Luc-84 were kindly provided by Dr. Shigetaka Kitajima of Tokyo Medical and Dental University. Luc-258 was generated by PCR. Luc-632 and Luc-258 carrying KLF6-binding site mutants (CC to AA) and KLF6 mutants were created by PCR using the QuickChange II site-directed mutagenesis kit (Stratagene), following the supplied protocol.
Cell Viability and Apoptosis Assays—Trypan blue dye exclusion assay was used to measure cell viability. Briefly, cells were suspended 1:1 with 0.4% trypan blue solution. Dead cells (stained blue) were counted under a microscope using a hemocytometer. Apoptosis was measured using the Cell Death Detection ELISAPLUS kit (Roche Applied Science) following the manufacturer's protocol. This assay determines apoptosis by measuring mono- and oligonucleosomes in the lysates of apoptotic cells. The cell lysates were placed into a streptavidin-coated microplate and incubated with a mixture of anti-histone/biotin and anti-DNA/peroxidase. The amount of peroxidase retained in the immunocomplex was photometrically determined with 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid as the substrate. Absorbance was measured at 405 nm.
Western Blot Analysis—Cells were lysed in RIPA buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS in PBS). Complete protease inhibitor mixture (Roche Applied Science) was added to lysis buffer before use. Protein concentration was determined by the DC protein assay (Bio-Rad). Protein samples were subjected to SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked in 5% nonfat milk in PBS overnight and incubated with primary antibody and subsequently with appropriate horseradish peroxidase-conjugated secondary antibody. Signals were developed with ECL reagents (Amersham Biosciences) and exposure to x-ray films. Anti-ATF3 polyclonal antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-β-tubulin monoclonal antibody was purchased from Sigma. Anti-KLF6 monoclonal antibody was purchased from Invitrogen. Anti-KLF6 polyclonal antibody was purchased from Santa Cruz Biotechnology. Anti-cleaved caspase-9 and anti-cleaved PARP polyclonal antibodies were purchased from Cell Signaling Technologies (Danvers, MA).
siRNAs and Transfection—Silencer™ pre-designed siRNAs and negative control siRNA (catalog number 4611) were purchased from Ambion (Austin, TX). The sequence for KLF6 siRNA is: sense (5′-GCCCGAGCUUUUGUUACAAtt-3′) and antisense (5′-UUGUAACAAAAGCUCGGGCtg-3′). The sequence for ATF3 siRNA is sense (5′-UCACAAAAGCCGAGGUAGCtt-3′) and antisense (5′-GCUACCUCGGCUUUUGUGAtg-3′). The sequence for KLF6-SV1 siRNA is sense (5′GCUUUUCUCCUUCCCUGGCtt-3′) and antisense (5′-GCCAGGGAAGGAGAAAAGCct-3′). The siRNAs were transfected into PC-3 cells using X-tremeGENE siRNA transfection reagent (Roche Applied Science) following the manufacturer's protocol. Cells were cultured and transfected in 6-well plates (1 × 105 cells per well), and the final siRNA concentrations were 100 nm. Protein samples were collected 48 h after transfection for Western blot analysis.
Real-time PCR—The mRNA level of ATF3 was measured by real-time PCR using TaqMan® gene expression assay (catalog number Hs00231069) from Applied Biosystems (Foster City, CA). Total RNA was isolated from PC-3 cells using RNeasy® kit (Qiagen). 5 μg of total RNA was used in reverse transcription reaction. The cDNAs were used as templates to perform PCR on an Applied Biosystems 7500 real-time PCR system following the manufacturer's protocol.
Luciferase Assay—PC-3 or LNCaP cells in 6-well plates were cotransfected with reporter plasmids and pCMV-Tag2-KLF6 or empty expression vectors. To reduce serum-induced activation of the ATF3 promoter (16), cells were incubated in media containing 0.1% serum after transfection. Caspase inhibitor benzyloxycarbonyl-VAD was added to the culture media (20 μm) to prevent cell death caused by KLF6 expression. Supernatants of cell extracts were assayed for luciferase activity using a luciferase assay system (Promega).
Chromatin Immunoprecipitation Assay—ChIP assay was performed using the ChIP assay kit from Millipore (Billerica, MA), following the supplied protocol. Immunoprecipitations were performed using anti-KLF6 or control IgG antibodies. PCR was performed with the following primers for two regions on the ATF3 promoter: an upstream randomly selected region (RND) from –1418 to –1219 as the negative control, 5′-CTGCGGCCGCGCAGGTCTCC-3′ and 5′-GATTCGAGCTGAGACCTCAG-3′; and KLF6 binding region (KB) from –370 to –120, 5′-GCCGGTAACCGTGTGGATTC-3′ and 5′-GACTAGGTGAGGCTGGGAAG-3′.
Electrophoretic Mobility Shift Assay (EMSA)—Recombinant KLF6 protein (Abnova, Taiwan) was incubated with the following biotin-labeled DNA probes for 30 min: –167TGCCCCCTCTCTCCACCCCTTCGGC–191; –202CTGGGCTGGCTCCTCCCCGAACTT–225. EMSA was performed using LightShift Chemiluminescent EMSA Kit (Pierce) following the manufacturer's protocol.
Statistical Analysis—Differences between the mean values were analyzed for significance using the unpaired two-tailed Student's test for independent samples; p ≤ 0.05 was considered to be statistically significant.
RESULTS
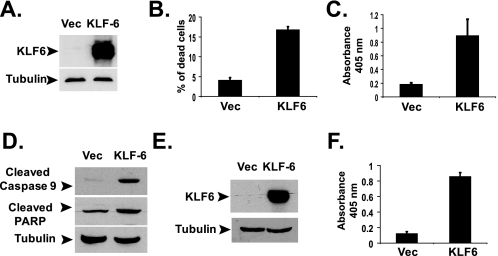
KLF6 Induces Apoptosis in PC-3 and LNCaP Cells—When ectopically expressed, KLF6 induced apoptosis in non-small cell lung cancer cells (1). Because KLF6 is frequently mutated in prostate cancer (2, 17), we determined whether KLF6 could also induce apoptosis in prostate cancer cells. KLF6 was overexpressed in PC-3 prostate cancer cells by transient transfection using pCMV-Tag2-KLF6 expression vector (Fig. 1A). KLF6 expression significantly reduced the viability of PC-3 cells (Fig. 1B). PC-3 cells died through apoptosis as evidenced by the enzyme-linked immunosorbent assay measuring mono- and oligonucleosomes in the lysates of apoptotic cells (Fig. 1C) and Western blotting of cleaved PARP and cleaved (activated) caspase-9 (Fig. 1D). Overexpression of KLF6 also induced apoptosis in another prostate cancer cell line LNCaP cells (Fig. 1, E and F), suggesting a common pro-apoptotic activity of KLF6 in prostate cancer cells.
FIGURE 1.
KLF6 induces apoptosis in prostate cancer cells. A, overexpressing KLF6 by transient transfection. PC-3 cells were transfected with 1.0 μg of empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids in 6-well plates. Western blotting was done using anti-FLAG antibody with cell lysates collected at 24 h after transfection. Vec, vector. B, KLF6 expression reduced the viability of PC-3 cells. Cells were transfected with 1.0 μg of empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids in 6-well plates. Cell viability was determined by trypan blue assay 24 h after transfection. C, KLF6 induced apoptosis in PC-3 cells. Cells were transfected with 1.0 μg of empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids in 6-well plates. Apoptosis was analyzed using the Cell Death Detection ELISAPLUS kit at 24 h after transfection. The average results from three independent experiments are shown. D, PC-3 cells were transfected with 1.0 μg of empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids. Cell lysates were analyzed by Western blotting with anti-cleaved caspase-9 and anti-cleaved PARP antibodies. E, LNCaP cells were transfected with 1.0 μg of empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids in 6-well plates. Western blotting was done using monoclonal anti-KLF6 antibody with cell lysates collected at 24 h after transfection. F, LNCaP cells were transfected with 1.0 μg of empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids in 6-well plates. Apoptosis was analyzed as described in C.
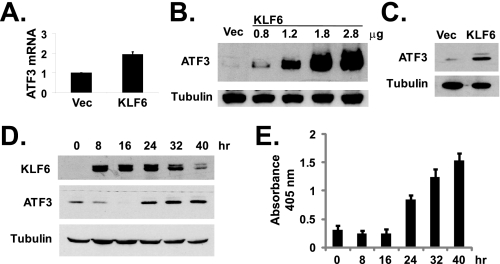
KLF6 Up-regulates the Expression of ATF3—To investigate the molecular mechanism of KLF6-induced apoptosis in prostate cancer cells, we compared the gene expression profiles between empty pCMV-Tag2 vector and pCMV-Tag2-KLF6-transfected PC-3 cells using microarray analysis. Among the genes that were up-regulated by KLF6 overexpression, we selected ATF3 for further study because of its known involvement in apoptosis (10, 13). We verified the up-regulation of ATF3 expression by KLF6 using real-time PCR. As shown in Fig. 2A, overexpression of KLF6 by transient transfection caused about 94% increase of ATF3 mRNA expression, compared with cells transfected with empty vector. Furthermore, KLF6 transfection also caused significant increases in ATF3 protein expression in a dose-dependent manner, as determined by Western blotting (Fig. 2B). Overexpression of KLF6 also induced ATF3 expression in LNCaP cells (Fig. 2C). Using the Tet-On inducible gene expression system (Clontech), we were able to control the expression of KLF6 in PC-3 cells with doxycycline. As shown in Fig. 2D, doxycycline-induced KLF6 expression (started at 8 h after adding doxycycline) preceded the induction of ATF3 protein expression (which occurred at 24 h after doxycycline addition). Consistently, measurable apoptosis was only detected at the same time when the ATF3 protein level was increased at 24 h after adding doxycycline but not when KLF6 was initially induced at 8- or 16-h time points (Fig. 2E).
FIGURE 2.
KLF6 up-regulates ATF3 expression. A, KLF6-activated transcription of ATF3 mRNA. PC-3 cells were transfected with empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids. Total RNA was isolated from cells at 24 h after transfection and analyzed by real-time PCR as described under “Experimental Procedures.” The average results from three independent experiments are shown. B, KLF6 up-regulated ATF3 protein expression. PC-3 cells were transfected with empty pCMV-Tag2 or various amounts of pCMV-Tag2-KLF6 vectors (Vec). ATF3 and β-tubulin protein levels were detected by Western blotting. C, LNCaP cells were transfected with empty pCMV-Tag2 or pCMV-Tag2-KLF6 plasmids. ATF3 and β-tubulin protein levels were detected by Western blotting. D, PC-3 cells were transfected with pTet-On Advanced and pTRE-Tight-KLF6 for doxycycline-inducible expression of KLF-6. At various time points after adding 1 μg/ml doxycycline, cell lysates were collected for Western blot analysis. E, PC-3 cells were transfected with plasmids as described in D, and KLF6 expression was induced by adding 1 μg/ml doxycycline. At various time points after doxycycline addition, apoptosis was measured using the Cell Death Detection ELISAPLUS kit. The average results from three independent experiments are shown.
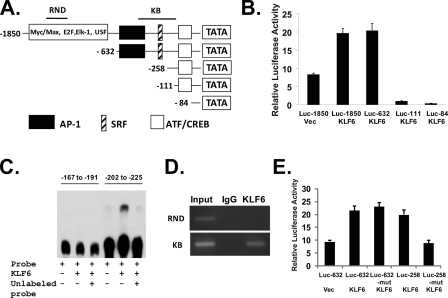
KLF6 Binds to and Activates the ATF3 Promoter—To determine whether KLF6 directly regulates the expression of the ATF3 gene, we performed an ATF3 gene reporter assay. Fig. 3A illustrates the promoter elements of the ATF3 gene promoter and the structures of the ATF3 promoter reporters containing various deletions. As shown in Fig. 3B, when KLF6 was expressed in LNCaP cells by co-transfection with the reporter plasmids, the ATF3 promoter was activated. The deletion mutant down to –632 was still activated by KLF6. In contrast, further deletions down to –111 and –84 completely abolished the effects of KLF6. The region between –632 and –111 contains the putative AP-1 and SRF motifs. Similarly in PC-3 cells, KLF6 was able to activate the ATF3 promoter, and deletions down to –111 and –84 abolished the effects of KLF6 (supplemental Fig. 1A). By carefully examine the DNA sequence in the region of –632 to –111, we identified a potential KLF6-binding site (–202 to –225) that contains the sequence of CCTCCCC (18). Electrophoretic mobility shift assay confirmed that KLF6 can directly bind to this sequence, whereas a nearby sequence (–167 to –191) failed to interact with KLF6 (Fig. 3C). To further determine whether KLF6 binds directly to this promoter region in vivo, we performed a chromatin immunoprecipitation (ChIP) assay, using PCR primers designed to target the endogenous ATF3 promoter region. In LNCaP cells overexpressing KLF6 (by transient transfection), anti-KLF6 antibody was able to immunoprecipitate the –370 to –120 region of the ATF3 promoter (KB), but not the randomly selected upstream region (RND) between –1418 and –1219 (Fig. 3D). A similar result was obtained in PC-3 cells (supplemental Fig. 1B). The control IgG did not immunoprecipitate with any fragments of the DNA. We mutated the KLF6-binding motif CCTCCCC to CCTCCAA and tested the effects of this mutation on KLF6-induced ATF3 promoter activity. To determine the role of AP-1 site, we also created a reporter plasmid Luc-258, in which the AP-1 site is deleted. As shown in Fig. 3E, in the absence of AP-1 site, KLF6 was able to activate the Luc-258 reporter, but Luc-258 KLF6-binding motif mutant reporter was not activated by KLF6. In the presence of AP-1 site, mutation of the KLF6-binding motif failed to block KLF6 activation of the reporter, indicating other transcription factors could also activate this promoter (potentially through binding to the AP-1 site).
FIGURE 3.
KLF6 binds to and activates ATF3 promoter. A, structures of ATF3 promoter reporter plasmid and those containing various deletions. The positions of regions RND and KB in the ChIP assay are shown on the map. B, KLF6 activated the ATF3 promoter. LNCaP cells were cotransfected with reporter plasmids and pCMV-Tag2-KLF6 or empty pCMV-Tag2. Cells were incubated in media containing 0.1% serum after transfection. Cell extracts were assayed for luciferase activity. The experiment has been repeated three times. C, recombinant KLF6 protein was incubated with biotin-labeled DNA probes (–167 to –191 and –202 to –225 on the ATF3 promoter) for gel shift assay. In the 3rd and 6th lanes, 200-fold molar excess unlabeled probes were used to compete with biotin-labeled probes. D, KLF6 binds directly to the ATF3 promoter. ChIP assay was performed with KLF-6-transfected LNCaP cells using anti-KLF6 antibody. Promoter regions RND (–1418 to –1219) and KB (–370 to –120) were amplified by PCR. E, LNCaP cells were cotransfected with wild-type or mutant reporter plasmids and pCMV-Tag2-KLF6 or empty pCMV-Tag2. Cells were incubated in media containing 0.1% serum after transfection. Cell extracts were assayed for luciferase activity. The experiment has been repeated three times. Mutation of CCTCCCC to CCTCCAA was created in the KLF6-binding sites of the Luc-632 and Luc-258 reporter plasmids.
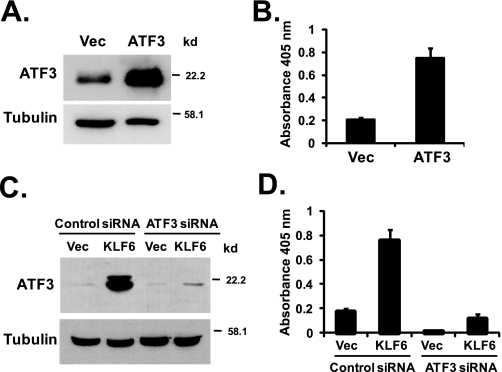
ATF3 Induces Apoptosis in PC-3 Cells—To determine whether ATF3 plays a role in apoptosis in PC-3 cells, we transiently overexpressed ATF3 in PC-3 cells. Transient transfection of pCG-ATF3 plasmids into PC-3 cells resulted in a significant increase in the ATF3 protein level (Fig. 4A), which was accompanied by a significant induction of apoptosis in these cells (Fig. 4B). Thus, ATF3 has strong pro-apoptotic activity in PC-3 cells.
FIGURE 4.
ATF3 mediates KLF6-induced apoptosis. A, overexpression of ATF3 in PC-3 cells. PC-3 cells were transfected with empty vector (Vec) or pCG-ATF3 vectors. ATF3 and β-tubulin protein levels were detected by Western blotting. B, ATF3 induces apoptosis in PC-3 cells. PC-3 cells were transfected with empty vector or pCG-ATF3 vectors. Apoptosis was analyzed using the Cell Death Detection ELISAPLUS kit 24 h after transfection. The average result from three independentexperiments is shown. C, knocking down of ATF3 by siRNA blocked KLF6-induced ATF3 expression. ATF3 targeting siRNA or negative control siRNA were transfected into PC-3 cells as described under “Experimental Procedures.” At 24 h post-siRNA transfection, empty pCMV-Tag2 or pCMV-Tag2-KLF6 vectors were transfected into cells. Down-regulation of ATF3 was confirmed by Western blotting of protein samples collected at 48 h after siRNA transfection. D, knocking down of ATF3 by siRNA blocked KLF6-induced apoptosis. PC-3 cells were transfected with ATF3 siRNA or negative control siRNA, followed by empty pCMV-Tag2 or pCMV-Tag2-KLF6 vectors as described in C, and apoptosis was analyzed 48 h after siRNA transfection using the Cell Death Detection ELISAPLUS kit.
ATF3 Mediates KLF6-induced Apoptosis—To determine whether ATF3 is a critical factor that mediates KLF6-induced apoptosis in PC-3 cells, we employed an ATF3-targeting siRNA to specifically knock down the expression of ATF3 in PC-3 cells. As shown in Fig. 4C, ATF3 siRNA was able to significantly down-regulate the level of ATF3 induction by KLF6 (in which cells were transfected with pCMV-Tag2-KLF6 vectors and control siRNA or ATF3 siRNA, compare 2nd and 4th lanes). Although KLF6 induced apoptosis in cells transfected with the negative control siRNA, it failed to induce apoptosis in cells where ATF3 was knocked down by - siRNA (Fig. 4D).
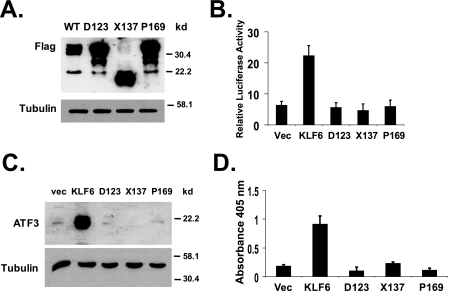
KLF6 Mutants Are Unable to Up-regulate ATF3 or Induce Apoptosis—KLF6 mutations have been identified in as high as 71% of human prostate cancer samples (2). Functional studies have shown that these KLF6 mutants have lost the ability to inhibit cell proliferation, a key function of the wild-type KLF6 as a tumor suppressor (2). To determine the effects of tumor-derived mutations on KLF6 function in ATF3 regulation and apoptosis, we generated cDNAs encoding three KLF6 mutants that have been detected in clinical prostate cancer samples, A123D, S137X, and L169P (2). When transiently transfected into PC-3 cells, all three KLF6 mutant constructs expressed their respective proteins at levels higher than that of the wild-type KLF6 protein (Fig. 5A). Although the wild-type KLF6 was able to activate the ATF3 promoter, none of the three mutants was active in the promoter reporter assay (Fig. 5B). In contrast to the wild-type KLF6, none of these mutant proteins was able to up-regulate ATF3 in PC-3 cells (Fig. 5C). Consistently, all three KLF6 mutants failed to induce apoptosis when transiently transfected into PC-3 cells (Fig. 5D).
FIGURE 5.
KLF6 mutants fail to up-regulate ATF3 and are unable to induce apoptosis. A, expression of KLF6 mutant proteins. FLAG-tagged KLF6 mutant expression plasmids (in pCMV-Tag2) were transfected into PC-3 cells, KLF6 and β-tubulin protein levels were detected by Western blotting. B, KLF6 mutants failed to activate the ATF3 promoter. LNCaP cells were cotransfected with reporter plasmid Luc-1850 and pCMV-Tag2-KLF6 or KLF6 mutant constructs (in pCMV-Tag2). Luciferase assay was performed as described in Fig. 3. C, KLF6 mutants failed to up-regulate ATF3 protein expression. PC-3 cells were transfected with empty pCMV-Tag2, pCMV-Tag2-KLF6, or KLF6 mutant constructs in pCMV-Tag2 vectors. ATF3 and β-tubulin protein levels were detected by Western blotting. D, KLF6 mutants failed to induce apoptosis in PC-3 cells. Cells were transfected with empty pCMV-Tag2, pCMV-Tag2-KLF6, or KLF6 mutant constructs in pCMV-Tag2 vectors. Apoptosis was analyzed using the Cell Death Detection ELISAPLUS kit. The average result from three independent experiments is shown.
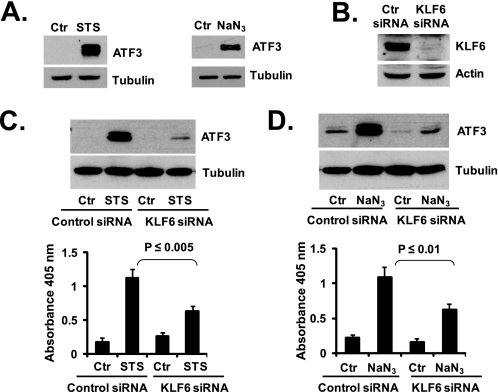
KLF6 Mediates Stress-induced Apoptosis through Up-regulation of ATF3—ATF3 plays an important role in physiological stress conditions (6, 7). We hypothesized that by controlling ATF3 expression, KLF6 may have a critical function in stress-induced apoptosis in prostate cancer cells. To test this hypothesis, we examined the effects of KLF6 knockdown by siRNA on PC-3 cell apoptosis induced by two agents as follows: staurosporine, a relatively nonselective protein kinase inhibitor; and sodium azide, an inducer of hypoxia condition. Exposure to staurosporine or sodium azide induced significant increases in the level of ATF3 protein in PC-3 cells (Fig. 6A), which were accompanied by the induction of apoptosis (Fig. 6, C and D). To determine the role of KLF6 in PC-3 cell apoptosis in these conditions, we selected a KLF6 siRNA that was able to down-regulate KLF6 protein almost completely (Fig. 6B). The KLF6 siRNA was able to block ATF3 induction caused by exposure to staurosporine or sodium azide, as well as to abolish apoptosis caused by these agents (Fig. 6, C and D). Thus, KLF6 and its downstream target ATF3 play important roles in PC-3 cell apoptosis in stress conditions.
FIGURE 6.
KLF6 mediates stress-induced apoptosis through ATF3. A, staurosporine (STS) and NaN3 up-regulated ATF3 in PC-3 cells. Cells were treated with 1 μm staurosporine for 8 h or 50 mm NaN3 for 24 h. ATF3 protein levels were determined by Western blotting. Ctr, control. B, knocking down of KLF6 by siRNA. KLF6 targeting siRNA and negative control siRNA were transfected into PC-3 cells. Protein samples were collected at 48 h after siRNA transfection. Western blot was performed with anti-KLF6 antibody. C, KLF6 siRNA blocked STS-induced ATF3 expression and apoptosis. PC-3 cells were transfected with KLF6 targeting siRNA or negative control siRNA. At 24 h after siRNA transfection, cells were treated with 1 μm STS for an additional 8 h. ATF3 protein level was determined by Western blotting. Apoptosis was analyzed using the Cell Death Detection ELISAPLUS kit. D, KLF6 siRNA blocked NaN3-induced ATF3 expression and apoptosis. PC-3 cells were transfected with KLF6 targeting siRNA or negative control siRNA. At 24 h after siRNA transfection, cells were treated with 50 mm NaN3 for additional 24 h. ATF3 protein level was determined by Western blotting. Apoptosis was determined as described above. The average result from three independent experiments is shown.
KLF6 Splicing Variants Also Up-regulate ATF3—KLF6 variant proteins, KLF6-SV1 and KLF6-SV2, are expressed in prostate cancer cells through alternative splicing (19). The KLF6-targeting siRNA that we employed in the above studies will knock down the expression of both wild-type KLF6 and the variant proteins. To determine whether the KLF6 splicing variants also regulate ATF3 expression, we transiently transfected PC-3 cells with pCMV-Tag2-KLF6-SV2. As shown in supplemental Fig. 2A, overexpression of KLF6-SV2 also induced ATF3 protein expression. KLF6-SV2 was also able to induce apoptosis in PC-3 cells, although to a lower degree compared with wild-type KLF6 (supplemental Fig. 2B). Furthermore, using a previously designed siRNA targeting KLF6-SV1 (20), we were able to down-regulate KLF6-SV1 in PC-3 cells (supplemental Fig. 2C). Down-regulation of KLF6-SV1 by siRNA also blocked staurosporine-induced ATF3 expression, as well as apoptosis in PC-3 cells (supplemental Fig. 2D).
DISCUSSION
The KLF6 gene is located on human chromosome 10p, a region deleted in about 55% of sporadic prostate adenocarcinoma (21, 22). Mutations of KLF6 gene have been identified in clinical prostate cancer samples (2, 17), suggesting a tumor suppressor role for KLF6 in prostate cancer. Indeed, decreased KLF6 expression has been shown to correlate with clinical outcome in prostate cancer patients (23). One mechanism by which KLF6 inhibits tumor proliferation is through activation of p21WAF1/cip1 (4), a function that is independent of p53 (2). KLF6 also has pro-apoptotic activity in non-small cell lung cancer cells (1), indicating that apoptosis induction can be another mechanism through which KLF6 suppresses tumor formation. However, the molecular mechanism of KLF6-mediated apoptosis has not been defined.
In this study, we found that KLF6 also induced apoptosis when ectopically expressed in prostate cancer cells. Thus, apoptosis induction could play a key role in its tumor suppressing activity in prostate cancers. Our data indicate that the direct activation of ATF3 mRNA expression and resulted increase of ATF3 protein level are the key mechanisms through which KLF6 induces apoptosis in prostate cancer cells. We showed that not only increased level of ATF3 could induce apoptosis in PC-3 cells, but also knocking-down of ATF3 by siRNA blocked KLF6-induced apoptosis. Furthermore, KLF6 mutants derived from clinical prostate cancers failed to activate ATF3 expression or to induce apoptosis, suggesting a critical role for the loss of the pro-apoptotic activity of KLF6 in clinical prostate cancer development. Although KLF6 could regulate the expression of other pro-apoptotic proteins, our data (especially the siRNA knockdown experiments) strongly support a critical role of ATF3 in KLF6-induced apoptosis in prostate cancer cells. However, the exact mechanism of how ATF3 regulates apoptosis in PC-3 cells remains to be determined. Although LNCaP cells express negligible levels of KLF6 and are p53 wild type, PC-3 cells express relatively high levels of KLF6 and are p53 null. However, similar results were obtained in the promoter reporter assay and ChIP assay in the two cell lines, indicating the effect of KLF6 on ATF3 promoter is independent of the status of p53 expression. Because p53 induces apoptosis by activating the expression of other pro-apoptotic proteins, such as Bax and Puma (24, 25), mutations in KLF6 and p53 may complement each other in the loss of tumor suppression by these two transcription factors.
ATF3 is a stress sensor, which is maintained at low levels in quiescent cells but rapidly up-regulated by a variety of stresses (26). ATF3 induces apoptosis in stress conditions, and its proapoptotic activity has been demonstrated in cultured cells as well as transgenic animal models (11–13). We found that KLF6 and ATF3 played a key role in PC-3 cell apoptosis caused by stress conditions, including staurosporine exposure and sodium azide-induced hypoxia. Knocking down of KLF6 by siRNA abolished ATF3 induction and apoptosis in these stress conditions. In large solid tumors, tumor cells are often exposed to hypoxic conditions (27, 28). Having loss of function mutations of KLF6 gene (as the ones that were used in our study) will render tumor cells a survival advantage, thus contributing to tumor growth and progression. Interestingly, we did not observe an up-regulation of KLF6 by the stress inducers (staurosporine and sodium azide). However, the siRNA knockdown experiments clearly indicate the absolute requirement of KLF6 for stress-induced ATF3 expression. Thus, we propose that other mechanisms must contribute to stress-induced KLF6 activation, such as post-translational modifications. A recent study suggests that acetylation of lysine 209 is critical to its function as a tumor suppressor (29). In addition, various potential phosphorylation sites (cAMP-dependent protein kinase, protein kinase C, casein kinase II, and p38 MAPK) exist on the KLF6 protein, providing other possible mechanisms of activation of this transcription factor.
Recent reports have demonstrated a role of ATF3 in promoting tumor invasion and metastasis (14), as well as in promoting cell proliferation (30). The apparent discrepancy from our data could be explained by the recently reported dichotomous feature of ATF3 action (15). Although ATF3 may induce apoptosis in certain cells under certain conditions, it may also stimulate growth and metastasis in other cells or under other conditions. Other proteins that have similar dual roles in regulating proliferation and apoptosis include c-Myc, which stimulates cancer cell proliferation but at the same time enhances apoptosis (31).
The KLF6 targeting siRNA that we employed for the knockdown experiments also target the splicing variants of KLF6, which were shown to antagonize the ability of wild-type KLF6 in suppressing cell proliferation (19). Interestingly, both variants SV1 and SV2 were able to up-regulate ATF3 and induce apoptosis in PC-3 cells (supplemental Fig. 2). Because KLF6 suppresses tumor growth by up-regulating p21 (4), these variant proteins could still contain the amino acids sequences that are required for activating ATF3 gene expression, whereas the domains/sequences necessary for p21 promoter binding and activation could be lost through alternative splicing.
In conclusion, we have identified ATF3 as a key target of KLF6 in tumor suppression. ATF3 mediates KLF6-induced apoptosis in stress conditions. Mutations in the KLF6 gene can contribute to resistance to apoptosis in prostate cancer cells.
Supplementary Material
Acknowledgments
We thank Dr. Tsonwin Hai (Ohio State University) for kindly providing us the pCG and pCG-ATF3 expression vectors. We also thank Dr. Shigetaka Kitajima (Tokyo Medical and Dental University) for providing the ATF3 promoter reporter plasmids Luc-1850, Luc-632, Luc-111, and Luc-84.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R21CA111765 (from NCI) and 2P20RR015566 (from the NCRR). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: PBS, phosphate-buffered saline; siRNA, small interference RNA; ChIP assay, chromatin immunoprecipitation assay; EMSA, electrophoretic mobility shift assay; KB, KLF6 binding region; PARP, poly(ADP-ribose) polymerase.
References
- 1.Ito, G., Uchiyama, M., Kondo, M., Mori, S., Usami, N., Maeda, O., Kawabe, T., Hasegawa, Y., Shimokata, K., and Sekido, Y. (2004) Cancer Res. 64 3838–3843 [DOI] [PubMed] [Google Scholar]
- 2.Narla, G., Heath, K. E., Reeves, H. L., Li, D., Giono, L. E., Kimmelman, A. C., Glucksman, M. J., Narla, J., Eng, F. J., Chan, A. M., Ferrari, A. C., Martignetti, J. A., and Friedman, S. L. (2001) Science 294 2563–2566 [DOI] [PubMed] [Google Scholar]
- 3.Kremer-Tal, S., Reeves, H. L., Narla, G., Thung, S. N., Schwartz, M., Difeo, A., Katz, A., Bruix, J., Bioulac-Sage, P., Martignetti, J. A., and Friedman, S. L. (2004) Hepatology 40 1047–1052 [DOI] [PubMed] [Google Scholar]
- 4.Narla, G., Kremer-Tal, S., Matsumoto, N., Zhao, X., Yao, S., Kelley, K., Tarocchi, M., and Friedman, S. L. (2007) Oncogene 26 4428–4434 [DOI] [PubMed] [Google Scholar]
- 5.Benzeno, S., Narla, G., Allina, J., Cheng, G. Z., Reeves, H. L., Banck, M. S., Odin, J. A., Diehl, J. A., Germain, D., and Friedman, S. L. (2004) Cancer Res. 64 3885–3891 [DOI] [PubMed] [Google Scholar]
- 6.Liang, G., Wolfgang, C. D., Chen, B. P., Chen, T. H., and Hai, T. (1996) J. Biol. Chem. 271 1695–1701 [DOI] [PubMed] [Google Scholar]
- 7.Hai, T., and Hartman, M. G. (2001) Gene (Amst.) 273 1–11 [DOI] [PubMed] [Google Scholar]
- 8.Allen-Jennings, A. E., Hartman, M. G., Kociba, G. J., and Hai, T. (2001) J. Biol. Chem. 276 29507–29514 [DOI] [PubMed] [Google Scholar]
- 9.Chen, B. P., Wolfgang, C. D., and Hai, T. (1996) Mol. Cell. Biol. 16 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu, D., Wolfgang, C. D., and Hai, T. (2006) J. Biol. Chem. 281 10473–10481 [DOI] [PubMed] [Google Scholar]
- 11.Syed, V., Mukherjee, K., Lyons-Weiler, J., Lau, K. M., Mashima, T., Tsuruo, T., and Ho, S. M. (2005) Oncogene 24 1774–1787 [DOI] [PubMed] [Google Scholar]
- 12.Mashima, T., Udagawa, S., and Tsuruo, T. (2001) J. Cell. Physiol. 188 352–358 [DOI] [PubMed] [Google Scholar]
- 13.Hartman, M. G., Lu, D., Kim, M. L., Kociba, G. J., Shukri, T., Buteau, J., Wang, X., Frankel, W. L., Guttridge, D., Prentki, M., Grey, S. T., Ron, D., and Hai, T. (2004) Mol. Cell. Biol. 24 5721–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandyopadhyay, S., Wang, Y., Zhan, R., Pai, S. K., Watabe, M., Iiizumi, M., Furuta, E., Mohinta, S., Liu, W., Hirota, S., Hosobe, S., Tsukada, T., Miura, K., Takano, Y., Saito, K., Commes, T., Piquemal, D., Hai, T., and Watabe, K. (2006) Cancer Res. 66 11983–11990 [DOI] [PubMed] [Google Scholar]
- 15.Yin, X., Dewille, J. W., and Hai, T. (2008) Oncogene 27 2118–2127 [DOI] [PubMed] [Google Scholar]
- 16.Tamura, K., Hua, B., Adachi, S., Guney, I., Kawauchi, J., Morioka, M., Tamamori-Adachi, M., Tanaka, Y., Nakabeppu, Y., Sunamori, M., Sedivy, J. M., and Kitajima, S. (2005) EMBO J. 24 2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, C., Hyytinen, E. R., Sun, X., Helin, H. J., Koivisto, P. A., Frierson, H. F., Jr., Vessella, R. L., and Dong, J. T. (2003) Am. J. Pathol. 162 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiambaretta, F., Nakamura, H., De Graeve, F., Sakai, H., Marceau, G., Maruyama, Y., Rigal, D., Dastugue, B., Sugar, J., Yue, B. Y., and Sapin, V. (2006) Investig. Ophthalmol. Vis. Sci. 47 582–590 [DOI] [PubMed] [Google Scholar]
- 19.Narla, G., Difeo, A., Reeves, H. L., Schaid, D. J., Hirshfeld, J., Hod, E., Katz, A., Isaacs, W. B., Hebbring, S., Komiya, A., McDonnell, S. K., Wiley, K. E., Jacobsen, S. J., Isaacs, S. D., Walsh, P. C., Zheng, S. L., Chang, B. L., Friedrichsen, D. M., Stanford, J. L., Ostrander, E. A., Chinnaiyan, A. M., Rubin, M. A., Xu, J., Thibodeau, S. N., Friedman, S. L., and Martignetti, J. A. (2005) Cancer Res. 65 1213–1222 [DOI] [PubMed] [Google Scholar]
- 20.Narla, G., DiFeo, A., Yao, S., Banno, A., Hod, E., Reeves, H. L., Qiao, R. F., Camacho-Vanegas, O., Levine, A., Kirschenbaum, A., Chan, A. M., Friedman, S. L., and Martignetti, J. A. (2005) Cancer Res. 65 5761–5768 [DOI] [PubMed] [Google Scholar]
- 21.Ittmann, M. (1996) Cancer Res. 56 2143–2147 [PubMed] [Google Scholar]
- 22.Trybus, T. M., Burgess, A. C., Wojno, K. J., Glover, T. W., and Macoska, J. A. (1996) Cancer Res. 56 2263–2267 [PubMed] [Google Scholar]
- 23.Glinsky, G. V., Glinskii, A. B., Stephenson, A. J., Hoffman, R. M., and Gerald, W. L. (2004) J. Clin. Investig. 113 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyashita, T., and Reed, J. C. (1995) Cell 80 293–299 [DOI] [PubMed] [Google Scholar]
- 25.Yu, J., Zhang, L., Hwang, P. M., Kinzler, K. W., and Vogelstein, B. (2001) Mol. Cell 7 673–682 [DOI] [PubMed] [Google Scholar]
- 26.Hai, T., Wolfgang, C. D., Marsee, D. K., Allen, A. E., and Sivaprasad, U. (1999) Gene Expr. 7 321–335 [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle, R. G., and Travers, S. (2006) Anticancer Agents Med. Chem. 6 281–286 [DOI] [PubMed] [Google Scholar]
- 28.Vaupel, P., and Harrison, L. (2004) Oncologist 9 Suppl. 5, 4–9 [DOI] [PubMed] [Google Scholar]
- 29.Li, D., Yea, S., Dolios, G., Martignetti, J. A., Narla, G., Wang, R., Walsh, M. J., and Friedman, S. L. (2005) Cancer Res. 65 9216–9225 [DOI] [PubMed] [Google Scholar]
- 30.Pelzer, A. E., Bektic, J., Haag, P., Berger, A. P., Pycha, A., Schafer, G., Rogatsch, H., Horninger, W., Bartsch, G., and Klocker, H. (2006) J. Urol. 175 1517–1522 [DOI] [PubMed] [Google Scholar]
- 31.Jamerson, M. H., Johnson, M. D., and Dickson, R. B. (2000) Oncogene 19 1065–1071 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.