Abstract
Phosphatidylinositol 4,5-bisphosphate (PIP2) is a prevalent phosphoinositide in cell membranes, with important functions in cell signaling and activation. A large fraction of PIP2 associates with the detergent-resistant membrane “raft” fraction, but the functional significance of this association remains controversial. To measure the properties of raft and nonraft PIP2 in cell signaling, we targeted the PIP2-specific phosphatase Inp54p to either the raft or nonraft membrane fraction using minimal membrane anchors. Interestingly, we observed that targeting Inp54p to the nonraft fraction resulted in an enrichment of raft-associated PIP2 and striking changes in cell morphology, including a wortmannin-sensitive increase in cell filopodia and cell spreading. In contrast, raft-targeted Inp54p depleted the raft pool of PIP2 and produced smooth T cells void of membrane ruffling and filopodia. Furthermore, raft-targeted Inp54p inhibited capping in T cells stimulated by cross-linking the T cell receptor, but without affecting the T cell receptor-dependent Ca2+ flux. Altogether, these results provide evidence of compartmentalization of PIP2-dependent signaling in cell membranes such as predicted by the membrane raft model.
The lipid phosphatidylinositol 4,5-bisphosphate (PIP2)2 is important for cell growth and viability (1), exemplified by its role as the precursor for the second messengers inositol 1,4,5-triphosphate, diacylglycerol, and phosphatidylinositol 3,4,5-bisphosphate (PIP3) (2, 3). PIP2 is also a cofactor for activation of select membrane proteins (4); this includes proteins that function in tethering actin filaments to the plasma membrane, such as the ERM (ezrin-radixin-moesin) proteins (5) and the filamins (6). Other PIP2-regulated membrane proteins include WASP and WAVE, which serve as effectors for the Rho GTPases in actin polymerization (7). Altogether, the properties of PIP2 in regulating actin-associated membrane proteins underlie its important role in establishing interactions between the plasma membrane and actin cytoskeleton (8).
The diverse and important functions of PIP2 underscore the importance of maintaining proper regulation of this lipid. As cell membranes are structurally heterogeneous (9), spatially concentrating PIP2 in discrete domains could be one mechanism for regulating PIP2 functions. Notably, a significant pool of the PIP2 associates with the detergent-resistant membrane (DRM) fraction postulated to represent cholesterol-dependent membrane domains, or membrane “rafts” (10–12). As membrane rafts serve as “reaction vessels” in the plasma membrane (13), enrichment of PIP2 in rafts may augment PIP2-dependent pathways and production of second messengers. Alternatively, rafts may function in regulating PIP2 signaling by sequestering it from activators in steady-state conditions, such as evidenced with certain membrane-associated kinases (14, 15).
Membrane domains such as rafts are often below the resolution of light microscopy, making direct observation of these structures difficult. Accordingly, many of the properties of the rafts have been deciphered from measurements of DRMs. One shortcoming with this approach is that the detergents used to prepare the DRMs also perturb the physical properties of the bilayer. Accordingly, it has been suggested the detergent-insoluble membranes are an artifact of sample preparation and not representative of domains in intact membranes (16, 17). Similarly, studies of rafts often utilize cholesterol-depleting agents such as methyl-β-cyclodextrin, and these compounds can cause nonspecific changes in membrane structure and function (16–19). In one example, it was shown that treating cells with methyl-β-cyclodextrin alters the membrane distribution of PIP2 and its availability to PIP2-binding proteins (19). However, it is not known whether this property represents a cholesterol-dependent property of PIP2 functions or a nonspecific effect from the drug treatment.
Notably, other data show that DRMs are in fact representative of discrete membrane domains (20). For example, imaging studies measuring the plasma membrane distribution of labeled proteins and lipids show a specific clustering of DRM-associated molecules. In one recent study, using fluorescence resonance energy transfer, we showed a specific cholesterol-dependent co-clustering of reporter molecules in the plasma membrane by membrane-anchoring signals that target fluorescent proteins to DRMs (21). One example was the N-terminal membrane-anchoring signal of the Src family kinase Lck. However, an alternative membrane-anchoring signal from Src, which does not target molecules to DRMs, also did not cause a cholesterol-dependent clustering of fluorescent proteins. In total, results from imaging studies show that membrane rafts are heterogeneous in nature, ranging in size from nanoclusters that are on the order of a few nanometers in size to nanodomains that have a diameter of ∼50 nm and to micron-size macrodomains (22, 23). Imaging experiments have also shown colocalization of PIP2 with raft markers in both the plasma membrane and intracellular trafficking vesicles (24–27), thus consistent with the notion that PIP2 is enriched in rafts.
Although both membrane fractionation experiments and imaging studies show that PIP2 is enriched in rafts, the biological significance of its raft association is poorly understood. A useful approach to measuring PIP2 functions in cells while avoiding chemical treatments is to alter membrane PIP2 levels using membrane-targeted PIP2-specific phosphatases (1). One example has been targeting the catalytic domain of the yeast PIP2-specific phosphatase Inp54p to the plasma membrane using the membrane-anchoring signal of the Src family kinase Lyn (Lyn-Inp54p) (8, 28). Expression of membrane-anchored Inp54p has demonstrated a role for PIP2 in maintaining interactions between the plasma membrane and underlying actin cytoskeleton (8). Interestingly, the membrane-anchoring signal of Lyn, which consists of myristoylation of an N-terminal glycine and palmitoylation of an adjacent cysteine, is also an effective raft-targeting sequence (29, 30). Accordingly, the discrete phenotypes evidenced by expression of Lyn-Inp54p may reflect changes specific to the raft-associated pool of PIP2.
Using membrane-targeting sequences from separate Src family kinases to target Inp54p to cell membranes, we observed that raft- and nonraft-targeted Inp54p caused distinct changes in the membrane pools of PIP2 and cell phenotype. For example, enrichment of the raft pool of PIP2 by nonraft Inp54p increased membrane ruffling and cell spreading, and depletion of this pool by raft-targeted Inp54p inhibited the ruffling as well as capping in stimulated T cells. Altogether, our findings provide evidence of sequestering of PIP2-dependent functions between separate membrane fractions in a manner consistent with the membrane raft model but while avoiding artifacts associated with detergent treatments of cell membranes.
MATERIALS AND METHODS
Gene Construction and Expression—The Inp54p constructs each contained a minimal membrane-anchoring signal, followed by green fluorescent protein (GFP) and finally the soluble domain of Inp54p (residues 1–331) that included its active site (see Fig. 1A). The membrane anchors were either the first 10 amino acids of Lck (L10) or the first 15 amino acids of c-Src (S15) (31). The following oligonucleotides were used for amplifying the DNA sequence encoding Inp54p: 5′-ACATCAGAATTCAACAACAACAACAACAAAACGAATTGGAAGGT (sense) and 5′-TCTAGTCTCGAGTTACGGCACTGGCGTCCCTGTAG (antisense). The L10-GFP and S15-GFP coding sequences were amplified from previously described genes (31) using the following primers: 5′-ACATCAGCATCCAGAATGGGGAGCAAGAGCAA (S15, sense), 5′-ACATCACCGCGGAGAATGGGCTGTGTCTGCAGC (L10, sense), and 5′-GCTAGTAAGCTTCTTGTAACAGCTCGTCCAT (GFP, antisense). The underlined nucleotides identify the restriction sites for EcoRI, XhoI, BamHI, SacII, and HindIII, respectively, which were used for directional cloning in pLenti (Invitrogen). The genes were subcloned into pLenti and oriented for control of expression by a 5′-long terminal repeat.
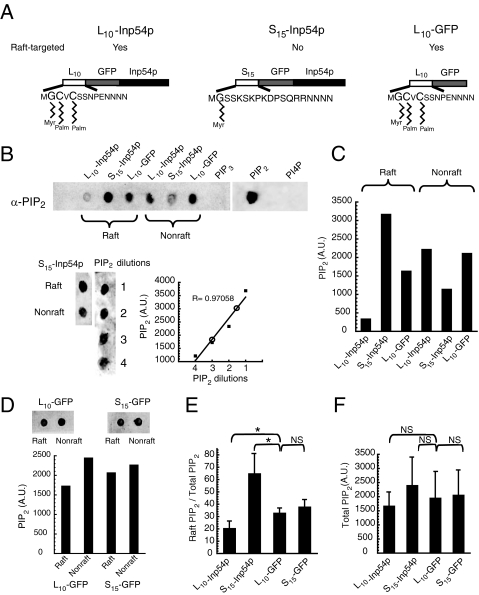
FIGURE 1.
Membrane-targeted Inp54p molecules cause distinct changes in raft and nonraft PIP2. A, targeting of the PIP2-specific phosphatase Inp54p to raft and nonraft membrane fractions. The constructs contained either the first 10 amino acids of Lck (L10) or the first 15 amino acids of c-Src (S15) for membrane association, followed by GFP and finally the soluble domain of Inp54p (L10-Inp54p and S15-Inp54p, respectively). L10 targets proteins to the detergent-resistant raft fraction, and S15 restricts proteins to the detergent-soluble nonraft fraction (21). Myr and Palm indicate sites of myristoylation and palmitoylation, respectively, of the indicated residues in the membrane-anchoring signals. A third construct consisting of membrane-anchored GFP (L10-GFP) was used as a control. B, measurement of changes in membrane PIP2 pools by L10-Inp54p and S15-Inp54p (upper panel). 293T cells expressing either L10-Inp54p or S15-Inp54p were lysed with Triton X-100, and the raft and nonraft membrane fractions were separated by sucrose gradient equilibrium centrifugation. Following separation, the gradient fractions containing the respective membrane fractions were pooled and extracted, and PIP2 was measured in each by immunoblotting. PIP3, PIP2, and phosphatidylinositol 4-phosphate (PI4P) represent purified lipids that were measured in parallel as controls for antibody specificity. Immunoblotting a range of the PIP2 standards together with a set of samples from cells that expressed S15-Inp54p showed that PIP2 extracted from cells was within the dynamic range of the measure (lower panel). In the accompanying plot, values for the S15-Inp54p raft and nonraft fractions are represented by open circles. A.U. denotes arbitrary units. C, quantitation of the immunoblot in B. D, measurement of membrane PIP2 pools for cells expressing L10-GFP and S15-GFP as a control for any sequestering of PIP2 by the S15 sequence. Quantitation of the dots is shown in the graph below. E, average of measurements from three separate trials. These results are represented as the fraction of total PIP2 that was raft-associated. F, total lipids measured for PIP2.In E and F, the error bars represent S.E. *, p ≤ 0.05 by Student's t test; NS (not significant), p > 0.38.
Cell Cultures and Transfections—Jurkat T cells (clone E6-1) were cultured in RPMI 1640 medium with 10% fetal bovine serum and supplemented with l-glutamine and antibiotics. 293T cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and supplemented with antibiotics. All cells were maintained at 37 °C in the presence of 5% CO2.
For gene expression, 293T cells were transfected by CaPO4 (Invitrogen) using 10 μg of plasmid DNA for a 50% confluent 6-cm plate. Jurkat cells were transfected by electroporation as described (32). Following transfection, Jurkat cells were cultured in AIM V medium (Invitrogen) for 48 h and then harvested for experimentation.
Membrane Fractionation—Approximately 107 cells were lysed by incubation at 4 °C for 20–30 min with 750 μl of 1% Triton X-100. Next, the samples were homogenized in a Dounce homogenizer (eight strokes) and centrifuged for 5 min at 4000 rpm (Eppendorf Model 5417C, Brinkmann Instruments). The supernatant was collected, and the raft and nonraft membrane fractions were separated by equilibrium centrifugation using a discontinuous sucrose gradient (14). Following centrifugation, the gradients were fractionated from the top.
PIP2 Measurement—Phosphoinositide extraction was performed as described (33) with minor adjustments. In brief, 3 ml of ice-cold chloroform/methanol/HCl (2:4:0.1) was added to the pooled gradient fractions corresponding to the raft (gradient fractions 2–4) (see Fig. 1 in Ref. 21) and nonraft (gradient fractions 7–10) (see Fig. 1 in Ref. 21) membrane fractions. The samples were mixed vigorously for 30 s and then incubated on ice for 15 min. To induce a phase separation, 1 ml of ice-cold chloroform was added with 1 ml of 1.76% KCl, 100 mm citric acid, 100 mm Na2HPO4, 5 mm EDTA, and 5 mm tetrabutylammonium hydrogen sulfate. Samples were mixed for 30 s and incubated on ice for 5 min. Samples were then centrifuged for 10 min at 2000 × g.
After centrifugation, the organic phase (bottom) was collected, dried, and resuspended in a minimal volume (∼10 μl) of solvent (4:3:1 chloroform/methanol/water). Samples were spotted onto nitrocellulose membrane using a Bio-Rad dot blot vacuum manifold. PIP2 was detected by immunoblotting using a monoclonal antibody (Echelon Biosciences Inc., Salt Lake City, UT) at a final concentration of 1 μg/ml, followed by a biotinylated secondary antibody. The final step was horseradish peroxidase conjugated to streptavidin (Vector Laboratories, Burlingame, CA). The membrane was developed using enhanced chemiluminescence (GE Healthcare, Buckinghamshire, UK) and detected with a Lumi-Imager F1 workstation (Roche Applied Science, Mannheim, Germany).
Cell Labeling and Imaging—Imaging was performed using either a Zeiss LSM510 microscope or a Zeiss Axioplan 2i upright fluorescence microscope (Oklahoma Medical Research Foundation Cell Imaging Core Facility). The images were collected using a 100× oil objective (1.4 numerical aperture). Three-dimensional projection images were generated from confocal stacks that were collected by optically sectioning along the z axis at an interval of 0.24 μm. Before three-dimensional reconstruction, the confocal stacks were deconvolved using AutoDeblur (AutoQuant Imaging, Inc., Watervliet, NY). The projection images were produced using iVision (BioVision Technologies, Exton, PA). All other image processing and quantitation were performed using iVision.
For imaging of T cell capping, cells were stimulated using OKT3-coated polystyrene beads as described (34). Following stimulation, the samples were fixed using 2% paraformaldehyde and then permeabilized with 0.2% Triton X-100 in phosphate-buffered saline. F-actin was labeled by incubation with 0.1 μg/ml Texas Red-labeled phalloidin (Invitrogen) for 30–45 min at 37 °C. Capping was quantitated by dividing the average fluorescence intensity of the plasma membrane in contact with the OKT3-coated bead by the average fluorescence intensity of the remaining plasma membrane. Cells were scored positive for capping if the ratio was 1.5 or greater. Histograms of relative fluorescence enrichment in the caps were prepared using IGOR Pro software (WaveMetrics, Inc., Lake Oswego, OR).
Ca2+ Flux Measurements—For detection of Ca2+ during stimulation, Jurkat T cells were labeled by incubation for 30 min in 2 μm Indo-1 (Invitrogen) at 37 °C. Following labeling, the cells were suspended in 15 mm HEPES (pH 7.4) supplemented with 140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2, and 10 mm glucose. The Ca2+ flux was measured by flow cytometry (MOFLO, DakoCytomation, Fort Collins, CO) based on the change in the fluorescence emission at 475 and 400 nm. Excitation for Indo-1 was at 390 nm. To limit the measurements to transfected cells, the samples were gated on those expressing GFP. GFP was detected using 488 nm excitation with a green band-pass filter centered at 530 nm for emission. The temperature of the instrument was maintained at 37 °C, and the flow rate was maintained by a base sheath pressure of 60 ± 1 p.s.i. A base line was achieved by passing the cells for ∼1 min prior to addition of OKT3. After the OKT3-dependent flux was complete, ionomycin was added to determine the maximal flux capacity of the cells. Store-operated channel Ca2+ flux was measured in the same manner as the total flux, except the buffer contained 0.05 μm EGTA rather than CaCl2. Following the initial flux from release of intracellular stores, Ca2+ (1 mm stock) was added back to the sample to a final concentration of 10 mm.
RESULTS
Membrane Raft- and Nonraft-targeted Inp54p Molecules Cause Distinct Changes in PIP2 Membrane Pools—The membrane-targeted Inp54p molecules used in this study are illustrated in Fig. 1A. Specifically, we targeted Inp54p to cell membranes using either the N-terminal 10 residues of Lck (L10) or the N-terminal 15 residues of Src (S15). We have described the L10 and S15 membrane-anchoring signals previously (31) and showed that the L10 sequence efficiently targets peptides to the DRM raft fraction and that the S15 sequence limits peptides to the detergent-soluble nonraft fraction. As a control to identify changes in the PIP2 pools and cell phenotype that were specific to the phosphatase activity, we expressed a third construct containing the L10 anchor and GFP but no Inp54p (L10-GFP).
To determine the effect of the targeted Inp54p molecules on the raft and nonraft pools of PIP2, we measured the PIP2 present in each membrane fraction after expressing L10-Inp54p, S15-Inp54p, or L10-GFP. For this experiment, we extracted the lipids of the raft and nonraft fractions following separation of the membrane fractions by sucrose gradient equilibrium centrifugation (see “Materials and Methods”). In Fig. 1B is an immunoblot measuring the PIP2 present in each membrane fraction of transfected cells, and in Fig. 1C are the results from quantitating the blot. These data show that expression of L10-Inp54p decreased the raft-associated pool of PIP2, whereas S15-Inp54p both increased the amount of raft PIP2 and decreased the PIP2 in the nonraft fraction. Furthermore, the changes in raft and nonraft PIP2 by S15-Inp54p were specific to the Inp54p domain because expression of a GFP molecule containing the S15 anchor alone (S15-GFP) demonstrated similar levels of PIP2 in each fraction as L10-GFP (Fig. 1, D and E). By averaging the results from several trials, we determined that L10-Inp54p decreased by 10% the fraction of total PIP2 that was raft-associated, whereas S15-Inp54p increased this ratio by 2-fold (Fig. 1E). Notably, neither Inp54p construct caused a statistically significant change in the total PIP2 levels of the cells (Fig. 1F).
T Cells Demonstrate Distinct Morphologies and Cell Spreading Based on Targeted Inp54p Expression—In T cells, PIP2 and its effectors are necessary for cell activation (35–37). To determine whether raft and nonraft pools of PIP2 in T cells are functionally distinct, we measured Jurkat T cells expressing L10-Inp54p, S15-Inp54p, or control L10-GFP. Interestingly, we observed distinct morphologies from expression of the separate Inp54p molecules (Fig. 2A). For example, T cells that expressed raft-targeted L10-Inp54p (Fig. 2A, left panel) often exhibited a smooth morphology that was void of the membrane ruffles and filopodia that occurred in control cells that expressed L10-GFP (right panel). We also observed membrane blebs in the L10-Inp54p samples (Fig. 2C), similar to those described for cells expressing Lyn-Inp54p (8). The blebs represent regions of the plasma membrane that bubble outward and can be due to poor association of the membrane with the underlying actin cytoskeleton (38).
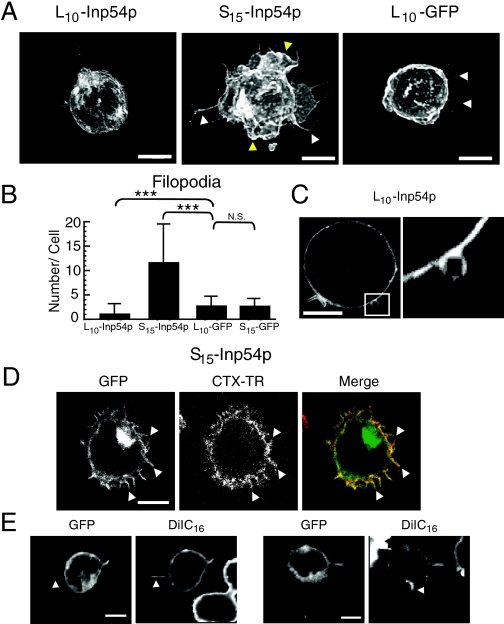
FIGURE 2.
Cell morphologies from expression of the membrane-targeted Inp54p molecules. A, projection images of Jurkat T cells that expressed L10-Inp54p, S15-Inp54p, or L10-GFP. The images were acquired using confocal microscopy and detected using GFP fluorescence. The white and yellow arrowheads indicate filopodia and membrane ruffling, respectively. B, quantitation of filopodia in transfected T cells. Filopodia in cells that expressed L10-Inp54p, S15-Inp54p, L10-GFP, or S15-GFP were counted using wide-field microscopy. The graph represents the average of 90 cells measured in three independent trials. The error bars represent S.E. ***, p ≤ 0.005 by Student's t test; N.S., not significant. C, representative example of the membrane blebs observed in cells that expressed L10-Inp54p. The right panel is a higher magnification of the region indicated by the square in the left panel. D, confocal images of a T cell double-labeled with S15-Inp54p and cholera toxin conjugated to Texas Red (CTX-TR). The arrowheads indicate examples of double-labeled filopodia. E, confocal images of Jurkat cells double-labeled with S15-Inp54p and DiIC16. The arrowheads indicate filopodia labeled with DiIC16 but not S15-Inp54p. Scale bars = 5 μm.
In contrast to L10-Inp54p, T cells that expressed S15-Inp54p exhibited a striking morphology that contained numerous filopodia and extensive membrane folding compared with that of the control samples (Fig. 2A, middle panel). We quantitated the effects of each Inp54p construct on cell morphology by counting the number of filopodia in T cells expressing either L10-Inp54p or S15-Inp54p. As controls, we expressed either L10-GFP or S15-GFP. These measurements showed an increase in filopodial number upon S15-Inp54p expression, whereas L10-Inp54p caused a decrease in filopodial number relative to the control samples (Fig. 2B).
The increase in cell filopodia upon S15-Inp54p expression coincided with an increase in raft-associated PIP2 (Fig. 1), suggesting that the filopodia may contain raft-enriched membrane. To test this hypothesis, we stained T cells expressing S15-Inp54p with biotinylated cholera toxin B subunit and secondary streptavidin conjugated to Texas Red. In Fig. 2D is an example showing labeling of filopodia by Texas Red-conjugated cholera toxin. Interestingly, we often noted that Texas Red-conjugated cholera toxin appeared as puncta localized along the processes, suggestive of rafts localized on the filopodia. We also detected staining of the filopodia with the raft marker DiIC16 (Fig. 2E), although in this case, the labeling was not enriched in puncta. Furthermore, in some cases, we observed efficient labeling of filopodia by DiIC16 that were barely detectable with the S15-Inp54p label (Fig. 2E, white arrowheads). This finding is again suggestive of enrichment of raft membranes in the cell processes. We conclude from these data that the filopodia in cells that expressed S15-Inp54p contained membrane rafts, and this may reflect enrichment of PIP2 in membrane rafts upon S15-Inp54p expression.
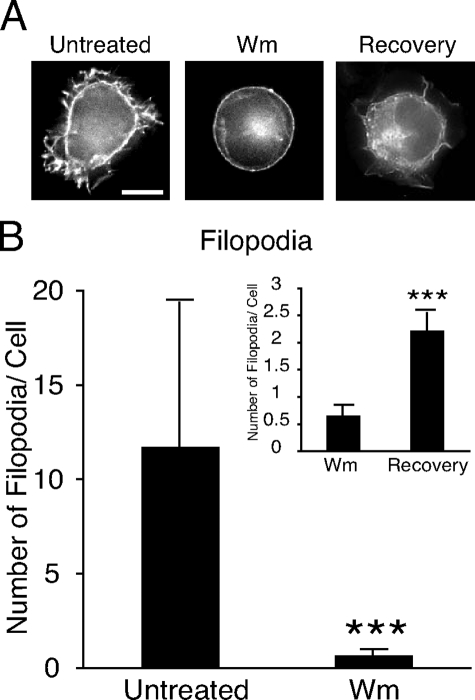
S15-Inp54p-dependent Changes in Cell Morphology Are Phosphatidylinositol 3-Kinase (PI3K)-dependent—An important effector of PIP2 signaling in membrane-actin interactions is the PI3K product PIP3 (3). To determine whether the T cell morphology upon S15-Inp54p expression was PIP3-dependent, we measured cells expressing S15-Inp54p and treated with wortmannin. Cell imaging showed that wortmannin inhibited the membrane ruffling and filopodia that occurred in the untreated cells (Fig. 3A), and quantitation showed that wortmannin reduced the average number of filopodia by 10-fold (Fig. 3B). We also observed that washing away the wortmannin following drug treatment resulted in a return of the membrane ruffling and filopodia 2 h later (Fig. 3A, right panel), albeit not to the number measured in untreated cells (Fig. 3B). We interpret these data as evidence that the cell morphologies that occurred upon S15-Inp54p expression were PI3K-dependent.
FIGURE 3.
S15-Inp54p-dependent filopodial growth is PI3K-dependent. A, representative images of T cells expressing S15-Inp54p that were treated with wortmannin (Wm) or treated with wortmannin, washed, and allowed to recover in growth media for 2 h (Recovery). Scale bar = 5 μm. B, quantitation of the number of filopodia in each set of conditions. The data represent the average of 90 cells measured in three separate trials. ***, p ≤ 0.005 by Student's t test.
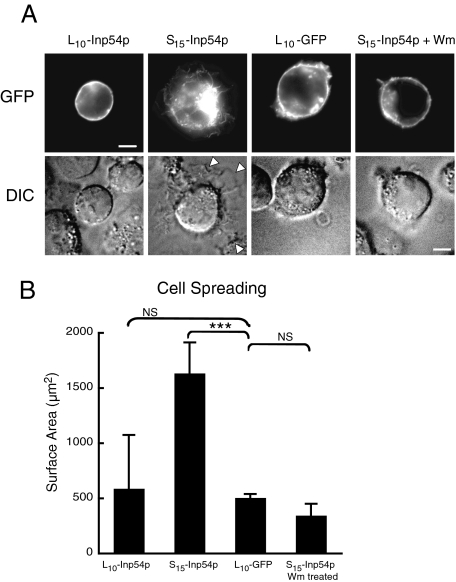
Altered Cell Spreading by Targeted Inp54p Expression—Cell spreading occurs by activation of surface adhesion molecules, many of which are PIP2-dependent (39). Similarly, we observed in T cells expressing S15-Inp54p an increase in cell spreading on poly-l-lysine, often represented by an extension of a thin sheet of membrane from the cell body (Fig. 4A, arrowheads). We quantitated the cell spreading by measuring the area of the cell where it contacted the substratum, and this showed that S15-Inp54p caused a 2–3-fold increase in the cell area over that of cells expressing either L10-GFP or L10-Inp54p. Furthermore, wortmannin inhibited the S15-Inp54p-dependent spreading (Fig. 4, A and B). Thus, the increased spreading from S15-Inp54p expression was also a PI3K-dependent property.
FIGURE 4.
T cells expressing S15-Inp54p exhibit increased cell spreading in a PI3K-dependent manner. A, confocal (upper panels) and differential interference contrast (DIC; lower panels) images of T cells expressing L10-Inp54p, S15-Inp54p, or L10-GFP. One set of cells expressing S15-Inp54p was treated with wortmannin (Wm) before fixation. The arrowheads in the S15-Inp54p sample indicate the edges of cell spreading. Scale bars = 5 μm. B, averaged results measuring the surface area of transfected Jurkat T cells seeded onto poly-l-lysine-coated coverslips. The data are from 60 cells measured in three independent trials using differential interference contrast imaging. ***, p ≤ 0.005 by Student's t test; NS (not significant), p > 0.05.
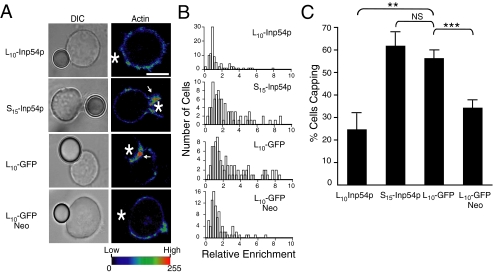
Raft-targeted Inp54p Inhibits T Cell Capping following Stimulation—T cells stimulated by cross-linking the T cell receptor (TCR) generate actin-rich membrane caps at the site of receptor cross-linking (40). To determine the effect of the membrane-targeted Inp54p molecules on membrane capping, transfected T cells were stimulated using 6-μm polystyrene beads coated with an antibody specific to human CD3 (OKT3). To detect the capping, samples were fixed and stained with Texas Red-conjugated phalloidin and measured by confocal microscopy.
In Fig. 5A are representative images of bead-cell conjugates that show an inhibition of capping by L10-Inp54p expression. For example, the cells that expressed either S15-Inp54p or control L10-GFP exhibited a 2–3-fold enrichment of F-actin at the bead-cell interface, but the cells that expressed L10-Inp54p demonstrated no such capping. Furthermore, pretreating cells expressing L10-GFP with neomycin to sequester PIP2 (41) also inhibited the T cell capping, and this is consistent with the inhibition by L10-Inp54p occurring upon phosphatase-dependent changes in the PIP2 pools. The inhibition of T cell capping by L10-Inp54p and neomycin is also represented by the histograms in Fig. 5B, which represents data from measuring the relative fluorescence enrichment at the bead-cell interface of ∼100 conjugates. Finally, in a separate analysis, cells were scored as capped if the average intensity at the bead-cell interface was 50% or greater than the remaining membrane. The fraction of cell-bead conjugates in each sample that exhibited capping under this criterion is plotted in Fig. 5C, and these data show that the inhibition by L10-Inp54p was both specific and statistically significant.
FIGURE 5.
L10-Inp54p inhibits actin capping in stimulated T cells. A, differential interference contrast (DIC) and confocal images of stimulated Jurkat T cells expressing L10-Inp54p, S15-Inp54p, L10-GFP, or L10-GFP treated with neomycin (Neo). Following stimulation, the samples were stained with Texas Red-conjugated phalloidin to detect F-actin. The fluorescence intensities are represented by pseudocolor, and the color assignment for intensity values is shown at the bottom. The asterisks indicate the positions of the antibody-coated beads. The arrows indicate actin-enriched membrane caps at the cell-bead interface. B, histograms of the relative enrichment of F-actin at the bead-cell interface measured in ∼100 bead-cell conjugates. C, quantitation of the frequency of F-actin capping of each sample measured using confocal microscopy. A cell was scored as positive for capping when the mean fluorescence intensity of the bead-cell interface divided by the mean fluorescence intensity of the rest of the cell was 1.5 or greater. Percent cells positive for capping for each sample is graphed. The results were averaged from 90 separate bead-cell conjugates measured in three separate trials. **, p ≤ 0.01 by Student's t test; ***, p ≤ 0.005; NS (not significant), p > 0.05.
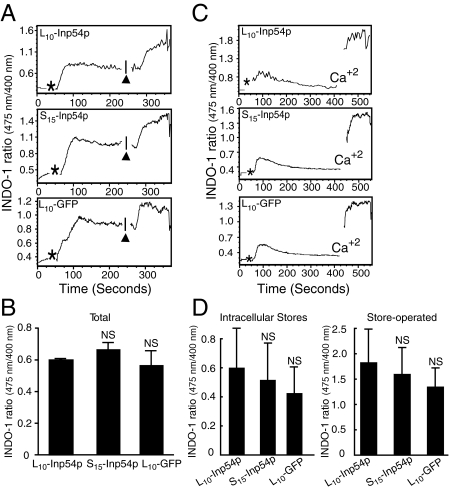
PIP2 is important for a TCR-dependent Ca2+ flux that drives remodeling of the cell cytoskeleton and associated membrane (35). Accordingly, one interpretation of the results in Fig. 5 is that the reduced actin capping by L10-Inp54p and neomycin is due to an inhibition of the TCR-dependent Ca2+ flux. However, in contrast to the capping, we observed that neither L10-Inp54p nor S15-Inp54p significantly affected the total Ca2+ flux relative to control cells (Fig. 6, A and B). Furthermore, the add-back assays in Fig. 6 (C and D) show that the Inp54p constructs also did not affect the release of either intracellular Ca2+ stores or store-operated channels. We therefore conclude that the inhibition of capping by L10-Inp54p did not occur by blocking Ca2+ signals downstream of the TCR.
FIGURE 6.
Ca2+ flux in stimulated T cells is not altered by membrane-targeted Inp54p expression. A, intracellular Ca2+ flux following TCR stimulation of Jurkat T cells expressing L10-Inp54p, S15-Inp54p, or L10-GFP. T cells labeled with Indo-1 were measured by flow cytometry and stimulated with OKT3 antibody (asterisks). Ionomycin addition is indicated by the black arrowheads. B, The total intracellular Ca2+ flux averaged from three independent trials. C, The store-operated Ca2+ flux was measured the same as (A), but cells were measured in buffer containing 0.05 μm EGTA to discriminate between flux from intracellular stores and flux from store-operated channels. Ca2+ indicates the time point when Ca2+ was added. D, The Ca2+ flux from store-operated channels averaged from three separate trials. In B and (D), NS, p > 0.05 by Student's t test.
DISCUSSION
Membrane fractionation experiments show that a significant pool of cellular PIP2 associates with the cholesterol-dependent membrane rafts (10, 11). However, the functional significance of PIP2 compartmentalization has been unclear and even controversial. In this study, we used separate targeted forms of the PIP2-specific phosphatase Inp54p to measure the properties of PIP2 in raft and nonraft fractions. Specifically, Inp54p was either targeted to the raft fraction using the membrane anchor of the Src family kinase Lck or restricted to the nonraft fraction using the membrane anchor of Src. Consistent with the membrane raft model, our data show phenotypes that correspond to specific changes in the raft-associated pool of PIP2. For example, expression of S15-Inp54p increased the fraction of PIP2 in the raft fraction, and this coincided with increased membrane ruffling, cell spreading, and growth of surface filopodia. Conversely, L10-Inp54p caused a decrease in raft-associated PIP2, and this coincided with cells that were smooth in appearance and void of membrane ruffles and filopodia as well as inhibited in their TCR-dependent capping.
Neither Inp54p molecule significantly changed the amount of total PIP2, further indicating that the phenotypes reported here were due to changes in discrete pools of PIP2 rather than its global levels in the membrane. In the case of S15-Inp54p, the enrichment of raft PIP2 was offset by a decrease in nonraft PIP2. However, no such offset that accommodated a decrease in raft PIP2 by L10-Inp54p expression was observed. One explanation for this discrepancy is that L10-Inp54p decreased the total PIP2, but below a level that was detectable by our method.
One possible mechanism for the enrichment of PIP2 in rafts by S15-Inp54p is an increase in the expression of protein factors such as MARCKS (myristoylated alanine-rich C kinase substrate) and GAP-43. These proteins bind PIP2 and cause its partitioning into raft-like liquid-ordered phase lipids (49, 50). Alternatively, depletion of nonraft PIP2 may activate synthesis of PIP2 specifically in the membrane rafts. Overall, the similar level of total PIP2 in cells expressing the targeted phosphatases shows a coupling of PIP2 synthesis with its consumption by Inp54p. PIP2 levels in cell membranes in general undergo extensive regulation, underscored by the quick re-establishment of steady-state levels following its hydrolysis by phospholipase C (PLC) during cell activation (51). Substantial reductions of PIP2 by drug treatment have global effects on plasma membrane structure (19), and this reflects the multiplicity of functions of PIP2 in establishing and maintaining membrane architecture. Our data show that at least some of the functions of PIP2 are regulated by its compartmentalization between raft and nonraft membrane fractions.
Interestingly, the membrane ruffling and cell spreading brought about by S15-Inp54p were sensitive to wortmannin, showing that these changes occur through a PIP3-dependent mechanism. This could occur through basal PI3K activity acting on the elevated raft-associated PIP2, thereby elevating the PIP3 content of the membrane and activating PIP3-dependent enzymes. Jurkat cells lack expression of the PIP3-specific phosphatase PTEN, and this will allow for an accumulation of PIP3 (42). However, the S15-Inp54p-dependent morphology was not restricted to Jurkat cells because we also observed extensive membrane ruffling and filopodia in HeLa cells transfected with this construct (data not shown). Interestingly, expression of dominant-negative and constitutively active forms of the GTPase Rac in Jurkat cells generates cell morphologies similar to those we report here for L10-Inp54p and S15-Inp54p, respectively (43). Rac is a downstream effector of the guanine nucleotide exchange factor Vav (44), which itself is a PIP3-dependent enzyme. Accordingly, the changes in cell phenotype that we evidenced with the membrane-targeted Inp54p molecules may reflect signaling in the Vav-Rac pathway affected by the altered levels of raft PIP2.
In stimulated T cells, PLCγ1 is recruited to the plasma membrane through binding to LAT (linker of activated T cells) (45, 46). Membrane recruitment of PLCγ1 is followed by production of inositol 1,4,5-trisphosphate through hydrolysis of PIP2, which then serves as a second messenger to initiate the TCR-dependent Ca2+ flux. LAT is constitutively associated with membrane rafts (47), suggesting that much of the PLCγ1 is proximal to raft pools of PIP2. However, our data show that reducing these pools using L10-Inp54p did not significantly affect the Ca2+ flux. One interpretation of this finding is that PLCγ1 utilizes nonraft PIP2 more than predicted based on the proximity of LAT to raft PIP2. Utilization of nonraft PIP2 by LAT-associated PLCγ1 may also account for the ability of a nonraft form of LAT to fully restore the TCR-dependent Ca2+ flux when expressed in LAT-deficient thymocytes (48). Altogether, these findings illustrate the caution that should be exercised in interpreting membrane fractionation data as well as the complexities relating to regulation and activation of PIP2-dependent signaling.
In contrast to the notion of membrane domains enriched with PIP2, one recent study using the pleckstrin homology (PH) domain of PLCδ1 to detect PIP2 did not observe clustering of the lipid in the plasma membrane (51). PH domains are a useful tool in qualitative measurements of phosphoinositides, such as detecting their turnover during cell signaling (52, 53). However, their utility in quantitative measurements is less certain. For example, the level of detection of the lipids by the probes is likely proportional to their level of expression, which itself will vary from cell to cell in an individual trial and between separate trials in each experiment. Furthermore, PH domains are often nonspecific in nature because many bind efficiently to more than one phosphoinositide species or their metabolites (54). For example, the PH domain from PLCδ1 binds both PIP2 and the PLC product inositol 1,4,5-trisphosphate (55). Accordingly, additional experiments using more quantitative probes may demonstrate properties for the PIP2 membrane distribution that are more consistent with its enrichment in membrane rafts.
Altogether, the factors that affect protein and lipid distributions in cell membranes remain an ongoing topic of studies of membrane structure. The membrane localization of PIP2 is a particularly important question because of its pivotal role in many membrane functions and cell signaling events. Membrane fractionation experiments show that a significant fraction of PIP2 occurs in membrane rafts. We provide evidence here showing that this represents a compartmentalization of PIP2 functions in T cells, where raft-associated PIP2 corresponds to a discrete pool of molecules that confer distinct phenotypes on T cells when modified using targeted PIP2-specific phosphatases. Further experiments are, however, necessary to better define the character of raft-associated PIP2 in biological membranes and the mechanism for PIP2 enrichment in these structures.
Acknowledgments
We thank Jacob Bass (Oklahoma Medical Research Foundation Flow Cytometry Core Facility) for flow cytometry assistance, L. Tsiokas for helpful discussions, and K. Moore and M. Coggeshall for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM070001. This work was also supported by American Heart Association Grant 0625648Z. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PIP2, phosphatidylinositol 4,5-bisphosphate; PIP3, phosphatidylinositol 3,4,5-trisphosphate; DiIC16, 1,1′-dihexadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; DRM, detergent-resistant membrane; GFP, green fluorescent protein; PI3K, phosphatidylinositol 3-kinase; TCR, T cell receptor; PLC, phospholipase C; PH, pleckstrin homology.
References
- 1.Di Paolo, G., and De Camilli, P. (2006) Nature 443 651-657 [DOI] [PubMed] [Google Scholar]
- 2.Majerus, P. W., Connolly, T. M., Deckmyn, H., Ross, T. S., Bross, T. E., Ishii, H., Bansal, V. S., and Wilson, D. B. (1986) Science 234 1519-1526 [DOI] [PubMed] [Google Scholar]
- 3.Rameh, L. E., and Cantley, L. C. (1999) J. Biol. Chem. 274 8347-8350 [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin, S., Wang, J., Gambhir, A., and Murray, D. (2002) Annu. Rev. Biophys. Biomol. Struct. 31 151-175 [DOI] [PubMed] [Google Scholar]
- 5.Ilani, T., Khanna, C., Zhou, M., Veenstra, T. D., and Bretscher, A. (2007) J. Cell Biol. 179 733-746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stossel, T. P., Condeelis, J., Cooley, L., Hartwig, J. H., Noegel, A., Schleicher, M., and Shapiro, S. S. (2001) Nat. Rev. Mol. Cell Biol. 2 138-145 [DOI] [PubMed] [Google Scholar]
- 7.Takenawa, T., and Suetsugu, S. (2007) Nat. Rev. Mol. Cell Biol. 8 37-48 [DOI] [PubMed] [Google Scholar]
- 8.Raucher, D., Stauffer, T., Chen, W., Shen, K., Guo, S., York, J. D., Sheetz, M. P., and Meyer, T. (2000) Cell 100 221-228 [DOI] [PubMed] [Google Scholar]
- 9.Engelman, D. M. (2005) Nature 438 578-580 [DOI] [PubMed] [Google Scholar]
- 10.Pike, L. J., and Casey, L. (1996) J. Biol. Chem. 271 26453-26456 [DOI] [PubMed] [Google Scholar]
- 11.Pike, L. J., and Miller, J. M. (1998) J. Biol. Chem. 273 22298-22304 [DOI] [PubMed] [Google Scholar]
- 12.Laux, T., Fukami, K., Thelen, M., Golub, T., Frey, D., and Caroni, P. (2000) J. Cell Biol. 149 1455-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian, T., Harding, A., Inder, K., Plowman, S., Parton, R. G., and Hancock, J. F. (2007) Nat. Cell Biol. 9 905-914 [DOI] [PubMed] [Google Scholar]
- 14.Rodgers, W., and Rose, J. K. (1996) J. Cell Biol. 135 1515-1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young, R. M., Zheng, X., Holowka, D., and Baird, B. (2005) J. Biol. Chem. 280 1230-1235 [DOI] [PubMed] [Google Scholar]
- 16.Heerklotz, H. (2002) Biophys. J. 83 2693-2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heerklotz, H., Szadkowska, H., Anderson, T., and Seelig, J. (2003) J. Mol. Biol. 329 793-799 [DOI] [PubMed] [Google Scholar]
- 18.Pizzo, P., Giurisato, E., Tassi, M., Benedetti, A., Pozzan, T., and Viola, A. (2002) Eur. J. Immunol. 32 3082-3091 [DOI] [PubMed] [Google Scholar]
- 19.Kwik, J., Boyle, S., Fooksman, D., Margolis, L., Sheetz, M. P., and Edidin, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13964-13969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock, J. F. (2006) Nat. Rev. Mol. Cell Biol. 7 456-462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chichili, G. R., and Rodgers, W. (2007) J. Biol. Chem. 282 36682-36691 [DOI] [PubMed] [Google Scholar]
- 22.Rodgers, W., Farris, D., and Mishra, S. (2005) Trends Immunol. 26 97-103 [DOI] [PubMed] [Google Scholar]
- 23.Jacobson, K., Mouritsen, O. G., and Anderson, R. G. (2007) Nat. Cell Biol. 9 7-14 [DOI] [PubMed] [Google Scholar]
- 24.Parmryd, I., Adler, J., Patel, R., and Magee, A. I. (2003) Exp. Cell Res. 285 27-38 [DOI] [PubMed] [Google Scholar]
- 25.Huang, S., Lifshitz, L., Patki-Kamath, V., Tuft, R., Fogarty, K., and Czech, M. P. (2004) Mol. Cell. Biol. 24 9102-9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golub, T., and Caroni, P. (2005) J. Cell Biol. 169 151-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozelle, A. L., Machesky, L. M., Yamamoto, M., Driessens, M. H., Insall, R. H., Roth, M. G., Luby-Phelps, K., Marriott, G., Hall, A., and Yin, H. L. (2000) Curr. Biol. 10 311-320 [DOI] [PubMed] [Google Scholar]
- 28.Wiradjaja, F., Ooms, L. M., Whisstock, J. C., McColl, B., Helfenbaum, L., Sambrook, J. F., Gething, M. J., and Mitchell, C. A. (2001) J. Biol. Chem. 276 7643-7653 [DOI] [PubMed] [Google Scholar]
- 29.Resh, M. D. (1994) Cell 76 411-413 [DOI] [PubMed] [Google Scholar]
- 30.Field, K. A., Holowka, D., and Baird, B. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9201-9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodgers, W. (2002) BioTechniques 32 1044-1051 [DOI] [PubMed] [Google Scholar]
- 32.Jordan, S., and Rodgers, W. (2003) J. Immunol. 171 78-87 [DOI] [PubMed] [Google Scholar]
- 33.Pettitt, T. R., Dove, S. K., Lubben, A., Calaminus, S. D., and Wakelam, M. J. (2006) J. Lipid Res. 47 1588-1596 [DOI] [PubMed] [Google Scholar]
- 34.Rodgers, W., and Zavzavadjian, J. (2001) Exp. Cell Res. 267 173-183 [DOI] [PubMed] [Google Scholar]
- 35.Penninger, J. M., and Crabtree, G. R. (1999) Cell 96 9-12 [DOI] [PubMed] [Google Scholar]
- 36.Acuto, O., and Cantrell, D. (2000) Annu. Rev. Immunol. 18 165-184 [DOI] [PubMed] [Google Scholar]
- 37.Villalba, M., Bi, K., Rodriguez, F., Tanaka, Y., Schoenberger, S., and Altman, A. (2001) J. Cell Biol. 155 331-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charras, G. T., Hu, C. K., Coughlin, M., and Mitchison, T. J. (2006) J. Cell Biol. 175 477-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caroni, P. (2001) EMBO J. 20 4332-4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roelants, G., Forni, L., and Pernis, B. (1973) J. Exp. Med. 137 1060-1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabev, E., Kasianowicz, J., Abbott, T., and McLaughlin, S. (1989) Biochim. Biophys. Acta 979 105-112 [DOI] [PubMed] [Google Scholar]
- 42.Shan, X., Czar, M. J., Bunnell, S. C., Liu, P., Liu, Y., Schwartzberg, P. L., and Wange, R. L. (2000) Mol. Cell. Biol. 20 6945-6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arrieumerlou, C., Randriamampita, C., Bismuth, G., and Trautmann, A. (2000) J. Immunol. 165 3182-3189 [DOI] [PubMed] [Google Scholar]
- 44.Jaffe, A. B., and Hall, A. (2005) Annu. Rev. Cell Dev. Biol. 21 247-269 [DOI] [PubMed] [Google Scholar]
- 45.Zhang, W., Sloan-Lancaster, J., Kitchen, J., Trible, R. P., and Samelson, L. E. (1998) Cell 92 83-92 [DOI] [PubMed] [Google Scholar]
- 46.Zhang, W., Irvin, B. J., Trible, R. P., Abraham, R. T., and Samelson, L. E. (1999) Int. Immunol. 11 943-950 [DOI] [PubMed] [Google Scholar]
- 47.Zhang, W., Trible, R. P., and Samelson, L. E. (1998) Immunity 9 239-246 [DOI] [PubMed] [Google Scholar]
- 48.Zhu, M., Shen, S., Liu, Y., Granillo, O., and Zhang, W. (2005) J. Immunol. 174 31-35 [DOI] [PubMed] [Google Scholar]
- 49.Wang, J., Arbuzova, A., Hangyas-Mihalyne, G., and McLaughlin, S. (2001) J. Biol. Chem. 276 5012-5019 [DOI] [PubMed] [Google Scholar]
- 50.Tong, J., Nguyen, L., Vidal, A., Simon, S. A., Skene, J. H., and McIntosh, T. J. (2008) Biophys. J. 94 125-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rheenen, J., Achame, E. M., Janssen, H., Calafat, J., and Jalink, K. (2005) EMBO J. 24 1664-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stauffer, T. P., Ahn, S., and Meyer, T. (1998) Curr. Biol. 8 343-346 [DOI] [PubMed] [Google Scholar]
- 53.Varnai, P., and Balla, T. (1998) J. Cell Biol. 143 501-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemmon, M. A., and Ferguson, K. M. (2001) Biochem. Soc. Trans. 29 377-384 [DOI] [PubMed] [Google Scholar]
- 55.Varnai, P., Lin, X., Lee, S. B., Tuymetova, G., Bondeva, T., Spat, A., Rhee, S. G., Hajnoczky, G., and Balla, T. (2002) J. Biol. Chem. 277 27412-27422 [DOI] [PubMed] [Google Scholar]