Abstract
Indolopyridones are potent inhibitors of reverse transcriptase (RT) of the human immunodeficiency virus type 1 (HIV-1). Although the structure of these compounds differs from established nucleoside analogue RT inhibitors (NRTIs), previous studies suggest that the prototype compound INDOPY-1 may bind in close proximity to the polymerase active site. NRTI-associated mutations that are clustered around the active site confer decreased, e.g. M184V and Y115F, or increased, e.g. K65R, susceptibility to INDOPY-1. Here we have studied the underlying biochemical mechanism. RT enzymes containing the isolated mutations M184V and Y115F cause 2–3-fold increases in IC50 values, while the combination of the two mutations causes a >15-fold increase. K65R can partially counteract these effects. Binding studies revealed that the M184V change reduces the affinity to INDOPY-1, while Y115F facilitates binding of the natural nucleotide substrate and the combined effects enhance the ability of the enzyme to discriminate against the inhibitor. Studies with other strategic mutations at residues Phe-61 and Ala-62, as well as the use of chemically modified templates shed further light on the putative binding site of the inhibitor and ternary complex formation. An abasic site residue at position n, i.e. opposite the 3′-end of the primer, prevents binding of INDOPY-1, while an abasic site at the adjacent position n+1 has no effect. Collectively, our findings provide strong evidence to suggest that INDOPY-1 can compete with natural deoxynucleoside triphosphates (dNTPs). We therefore propose to refer to members of this class of compounds as “nucleotide-competing RT inhibitors” (NcRTIs).
The polymerase active site of the reverse transcriptase (RT)3 enzyme of the human immunodeficiency virus type 1 (HIV-1) is a target for two classes of approved antiretroviral drugs referred to as nucleoside analogue RT inhibitors (NRTIs) and non-nucleoside analogue RT inhibitors (NNRTIs). Once phosphorylated, NRTIs act as chain-terminators that compete with natural nucleotide substrates while NNRTIs comprise a structurally diverse family of compounds that bind to a hydrophobic pocket near the active site of RT and appear to affect the chemical step of the reaction and not nucleotide binding (reviewed in Refs. 1–4).
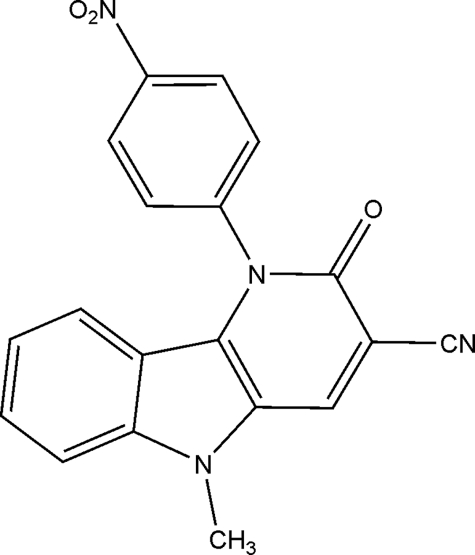
Indolopyridones represent a newly discovered class of inhibitors that interfere with RT function through a mechanism of action that is distinct from that described for NRTIs and NNRTIs (5). The prototype compound INDOPY-1 (Fig. 1) has been shown to be active against NNRTI-resistant HIV strains (6). INDOPY-1, unlike NNRTIs, but like natural deoxyribonucleoside triphosphates (dNTPs), can bind to and stabilize RT-DNA/DNA complexes (5). Footprinting experiments and binding studies revealed that the complex with INDOPY-1 is trapped in the post-translocational state that likewise allows dNTP binding. However, in contrast to NRTI or dNTP substrates, binding of INDOPY-1 depends on the chemical nature of the ultimate base pair at the 3′-end of the primer and not on the chemical nature of the templated base that is engaged in classic base pairing. INDOPY-1 binds preferentially following pyrimidines (thymidines > cytidines).
FIGURE 1.
Chemical structure of INDOPY-1. 5-Methyl-1-(4-nitrophenyl)-2-oxo-2,5-dihydro-1H-pyrido[3,2-b]indole-3-carbonitrile.
Steady-state kinetic analysis with homopolymeric substrates revealed competitive (5), or mixed type (competitive/non-competitive) inhibition (6) with respect to the nucleotide. The non-nucleosidic compound aphidicolin shows a similar effect on calf thymus DNA polymerase α and ε (7, 8). Together, these findings suggest that the binding site for indolopyridones and nucleotide substrates can at least partially overlap. The resistance profile of INDOPY-1 provides further independent evidence for this notion (5, 6). In vitro selection experiments and phenotypic susceptibility measurements with clinical isolates and constructs generated by site-directed mutagenesis suggest that most mutations associated with decreased susceptibility to INDOPY-1 are clustered around the dNTP binding site. These mutations include the NRTI-associated change M184V that confers high level resistance to lamivudine (3TC) and emtricitabine (FTC) (3). The combination of M184V and Y115F is associated with decreased susceptibility to guanosine analogue abacavir (ABC) (9). Of note, K65R, which is associated with decreased susceptibility to tenofovir (TFV) (10), confers increased susceptibility to INDOPY-1 (5, 6). The inhibitor is generally sensitive against a background of thymidine analogue-associated mutations (TAMs) or NNRTI-associated mutations, respectively, with the exception of the novel mutation L234F that is located in close proximity to the NNRTI-binding pocket (11).
M184V and Y115F show relatively moderate 5–8-fold increases in half-maximal effective concentrations (EC50). However, the combination of mutations M184V and Y115F appears to amplify the effects of the individual mutations, and cause >100 fold increases in the EC50 values when compared with wild-type HIV-1 (5). Here, we studied the underlying mechanism. We show that mutant RT enzymes containing M184V can diminish binding of INDOPY-1, while binding of the natural dNTP substrate remains largely unchanged. In contrast, Y115F increases binding of the natural nucleotide substrate. Thus, the combined properties appear to amplify the ability of the enzyme to discriminate against the inhibitor. Our biochemical studies provide strong support for the notion that the binding sites for INDOPY-1 and the natural dNTP substrate can at least partially overlap, and the mechanism of inhibition is predominantly competitive in nature.
EXPERIMENTAL PROCEDURES
Materials—Heterodimeric (p66/p51) HIV-1 RT enzymes were expressed in Escherichia coli and purified as previously described (12). Site-directed mutagenesis was applied to generate RT mutants of the HXB2 strain using the Stratagene QuikChange procedure according to the manufacturer's protocol. “WT RT” refers to wild-type enzyme. M184V, K65R, Y115F, and F61A RT enzymes each contain a single mutation at the indicated residues and the presence of multiple mutations is indicated likewise. The RT inhibitor indolopyridone-1 (INDOPY-1) was synthesized as described (4), and was obtained from Tibotec BVBA, Mechelen, Belgium. DNA oligonucleotides used in this study were obtained from Invitrogen. The long RNA template PBS-250 was synthesized through in vitro transcription with T7 RNA polymerase (13). Nucleic acid substrates were 32P-radiolabeled at their 5′-end with [γ-32P]ATP and T4 polynucleotide kinase (Fermentas) (14). Reactions were allowed to proceed for 1 h at 37 °C. The radiolabeled material was purified on 12% polyacrylamide gels containing 50 mm Tris-borate pH 8.5, 1 mm EDTA, and 7 m urea. Oligonucleotide samples were eluted overnight in a buffer containing 500 mm ammonium acetate and 0.1% SDS.
Primer Extension Assay—A 2-fold molar excess of PBS-250 RNA template was hybridized to 50 nm 5′-radiolabeled PBS-28 DNA primer (5′-CTTTCAGGTCCCTGTTCGGGCGCCACTG-3′). DNA/RNA hybrids were formed in a buffer containing 50 mm Tris-HCl, pH 7.8 and 50 mm NaCl. Samples were heated to 95 °C for 3 min followed by a gradual decrease to room temperature (45 min of incubation). The hybrid was then incubated with 125 nm of RT in a buffer containing 50 mm Tris-HCl, pH 7.8, 50 mm NaCl, and 10 μm of each of dATP, dTTP, dGTP, and dCTP. The samples were then incubated with 6 μm INDOPY-1. DNA synthesis was initiated by the addition of 6 mm MgCl2 at 37 °C. The reaction was stopped at defined time points (0, 15, and 30 s, and 1, 2, 5, and 10 min) with formamide-loading dye containing xylene cyanol and bromphenol blue. Samples were resolved on an 8% denaturing polyacrylamide gel and analyzed with a phosphor-imager (Amersham Biosciences) using Quantity One and ImageQuant software. DNA synthesis assays with the F61A RT mutant used a similar protocol with DNA template PBS-57 (5′-GTTCGAACAAATCTCTAGCAGTGGCGCCCGAACAGGGACCTGAAAGCGAAAGCTAC-3′). The RT concentration was increased to 250 nm.
Band Shift Experiments—50 nm of radiolabeled DNA primer (5′-TTAAAAGAAAAGGGGGGA-3′) was annealed to 3-fold molar excess of the DNA template (5′-CCTTCCCATCCCCCCTTTTCTTTTAAAAAGT-3′) as described above. The 3′-end of the primer was blocked for 30 min with 10 μm ddTTP and 500 nm RT in a buffer containing 6 mm MgCl2, 50 mm Tris-HCl, pH 7.8, and 50 mm NaCl. Increasing concentrations of INDOPY-1 or dGTP, i.e. the next complementary nucleotide, ranging from 0 to 6 μm, were added, and the complex was then challenged by adding 4 μg/μl heparin and 1 h incubation at room temperature. The samples were fractionated through 6% non-denaturing polyacrylamide gels and visualized as described above. GraphPad Prism software was used to obtain apparent Kdvalues for INDOPY-1, and the nucleotide using the one-site binding (hyperbola) equation. Similar experiments were performed with oligonucleotides containing abasic sites, whereby the DNA primer (5′-TTAAAAGAAAAGGGGGGACT-3′) was annealed to each of the following DNA templates where the abasic residue is indicated by a line: control template (5′-CCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′), the abasic template at position n (5′-CCCTTCC_GTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′), and the abasic template at position n+1 (5′-CCCTTC_ AGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′).
Filter-based DNA Synthesis Inhibition Assay—61.54 nm annealed poly(rA)/(dT)20 hybrid duplex was incubated for 30 min at room temperature with 7 nm HIV-1 RT in a buffer containing 50 mm Tris-HCl pH 7.8 and 50 mm NaCl. 500 nm [3H]dTTP and INDOPY-1 (ranging from 8 nm to 16 μm) were added to the samples, and the reaction was initiated by adding 6 mm MgCl2. The samples were then incubated for 2 h at 37 °C, and the reaction was quenched with the addition of 10% trichloroacetic acid. Samples were then filtered and scintillation analysis was used to measure the amount of remaining radioactivity. IC50 values were obtained through analysis with GraphPad Prism 4.1 software.
Single Nucleotide Incorporation Assay—100 nm of a radiolabeled DNA primer (5′-TTCTGACTAAAAGGGTCTGAGGGAT-3′) was annealed to a 3-fold molar excess of DNA template (5′-GTAACTAGAGATCCCTCAGACCCTTTTAGTCAGAAT-3′) as described above. The hybrid was then incubated with 500 nm HIV-1 RT and increasing concentrations of INDOPY-1 (0–30 μm) in a buffer of 50 mm Tris-HCl pH 7.8 and 50 mm NaCl. The reaction was started by adding 6 mm MgCl2, 4 μg/μl of heparin and the next complementary nucleotide, i.e. dCTP. The two different dCTP concentrations used were 0.15 μm and 0.50 μm, respectively. Nucleotide incorporation was allowed to proceed for 1 min at 37 °C before being stopped by the addition of gel-loading buffer. The samples were fractionated through a 12% polyacrylamide gel and analyzed as described above. Normalized inhibition by INDOPY-1 was plotted using the sigmoidal dose response (variable slope) equation in GraphPad Prism 4.1 software.
RESULTS
Effects of Mutations M184V, Y115F, and K65R on Inhibition of DNA Synthesis with INDOPY-1—Initially, we studied whether the changes in phenotypic susceptibility to INDOPY-1 translate directly in altered biochemical properties of HIV-1 RT. For this purpose, we generated RT enzymes containing mutations M184V, Y115F, and K65R, as well as the double mutant M184V/Y115F. We found that the changes in EC50 values measured in cell-based, phenotypic susceptibility assays followed consistent trends as changes in 50% inhibitory concentrations (IC50) in a cell-free RT assay (Table 1). We measured inhibition on homopolymeric poly(rA)-(dT)20 primer/template substrates and observed that enzymes containing mutations M184V and Y115F showed ∼2-fold increases in IC50 values. These effects are markedly increased with the M184V/Y115F double mutant that shows >15-fold increases in IC50 values, which points to a synergistic effect. In contrast, K65R appears to enhance the inhibitory effects of INDOPY-1, which is consistent with the hypersusceptible phenotype in this mutational context.
TABLE 1.
Changes in EC50 and IC50 values for INDOPY-1 against HIV-1 RT containing M184V, Y115F, and K65R
| Mutational context | Cell-based assay fold-changeain EC50b | Cell-free assay fold-changeain IC50c |
|---|---|---|
| K65R | 0.5 | 0.74 ± 0.13 |
| M184V | 5.0 | 2.0 ± 0.58 |
| Y115F | 7.9 | 2.5 ± 2.6 |
| M184V/Y115F | >100 | >15 |
Values were obtained by comparing mutant enzymes with WT RT
EC50 values as previously reported by Jochmans et al. (5)
Each reported value is the outcome of at least three separate experiments
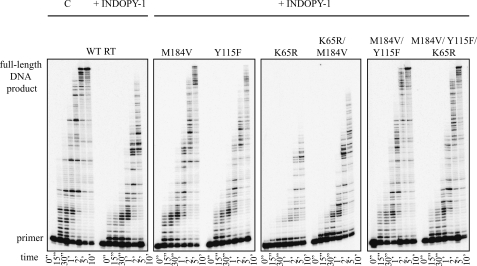
Inhibition was also studied in time course experiments with a heteropolymeric DNA/RNA substrate that better mimics physiologically relevant conditions. We have previously reported that INDOPY-1 increases enzyme pausing following incorporation of pyrimidines (5). Here we employed the same assay to assess the effects of mutations M184V, Y115F, and K65R on RT pausing and DNA synthesis in the presence of inhibitor (Fig. 2). The amount of enzyme was adjusted such that WT and mutant enzymes produced the same amount of full-length product in the absence of inhibitor. These data show preferential inhibition following pyrimidines with wild-type HIV-1 RT and all mutant enzymes used in this study. We therefore conclude that the mutant enzymes do not appear to affect sequence specificity of INDOPY-1. In the presence of the inhibitor, enzymes containing either M184V or Y115F mutations showed diminished enzyme pausing, which results in higher yields of the full-length product. The amount of the full-length product obtained with the M184V/Y115F double mutant in the presence of inhibitor was almost identical with that obtained by WT RT in the absence of the inhibitor. The K65R mutation, when present against a background of M184V/Y115F, is unable to counteract the resistance conferring effects of the double mutant. However, K65R appears to neutralize M184V alone. INDOPY-1 is as active against the M184V/K65R double mutant as against WT RT. Together, the biochemical findings are in good agreement with cell-based susceptibility measurements.
FIGURE 2.
Effect of RT mutations M184V, Y115F, and K65R on INDOPY-1 inhibition of DNA synthesis. DNA synthesis was observed on a DNA/RNA substrate in the presence of 6 μm INDOPY-1 over a period of time (0–10 min). The lowest band shows the non-elongated radiolabeled primer and the highest band shows the full-length DNA product. The mutant RT enzymes studied are indicated on each corresponding panel. Lane C shows control DNA synthesis with WT RT in the absence of the inhibitor.
Effects of M184V, Y115F, and K65R Mutations on Complex Formation with INDOPY-1—We next asked whether differences in the inhibitory effects of INDOPY-1 correlate with differences in inhibitor binding. To address this question, we employed band shift experiments that allow us to monitor the stability of RT-primer/template complexes in the presence of inhibitor and nucleotide substrate, respectively. Like natural dNTPs (15), INDOPY-1 is capable of stabilizing the RT-DNA/DNA complex (5). Ternary complexes composed of HIV-1 RT, a DNA/DNA primer/template, and the nucleotide substrate or INDOPY-1 are resistant to dissociation in the presence of an enzyme trap such as heparin. While binary complexes are not stable under these conditions, ternary complexes with the nucleotide or the inhibitor can be visualized by non-denaturing polyacrylamide gel electrophoresis. Ternary, or “Dead-end complex” formation is therefore a measure for the ability of INDOPY-1 or the nucleotide to bind to pre-formed RT-primer/template complexes. Using this assay, we measured apparent Kd values (Kd(app)) for ligand binding in the context of the various RT mutants.
WT RT shows similar apparent affinities for INDOPY-1 and the natural nucleotide substrate dGTP with Kd(app) values of ∼100 and 160 nm, respectively (Table 2). Of note, nucleotide incorporation was prevented through use of a chain-terminated primer (“Experimental Procedures”). The M184V mutant shows higher values for both the inhibitor (Kd(app) = 620 nm), and the nucleotide substrate (Kd(app) = 270 nm). The Y115F mutant shows a different effect. The apparent affinity to the inhibitor is only slightly decreased, while binding of the natural nucleotide substrate is enhanced (Kd(app) = 50 nm as compared with 160 nm with WT RT). These findings suggest that both mutations together may efficiently discriminate against the inhibitor through (i) diminished binding of INDOPY-1 with M184V and (ii) increased binding of the natural nucleotide substrate with Y115F.
TABLE 2.
The apparent Kd value for INDOPY-1 with WT and mutant RT enzymes
|
Enzyme
|
Kd(app)a
|
Selectivityb(fold-changec)
|
||
|---|---|---|---|---|
| INDOPY-1 | dGTP | |||
| nm | ||||
| WT | 97 ± 11 | 156 ± 30 | 0.6 | |
| K65R | 61 ± 8 | 365 ± 39 | 0.2 (0.3) | |
| M184V | 618 ± 77 | 270 ± 27 | 2.3 (3.8) | |
| Y115F | 156 ± 44 | 48 ± 10 | 3.3 (5.5) | |
| M184V/Y115F | 1201 ± 355 | 124 ± 15 | 9.7 (16) | |
| M184V/K65R | 452 ± 57 | 355 ± 51 | 1.3 (2.2) | |
| M184V/Y115F/K65R | 1298 ± 432 | 278 ± 21 | 4.7 (7.8) | |
Each reported value is the average of at least three separate experiments
Selectivity is defined as [Kd(app)INDOPY-1]/[Kd(app)dGTP]
Fold-change of selectivity is defined as Mutant Kd ratio/WT Kd ratio
The double mutant M184V/Y115F is severely compromised with regards to inhibitor binding, while the ability to bind the natural dGTP substrate is largely maintained (Table 2). The ratio in selectivity of mutant enzymes versus WT RT provides a measure for the ability of a given mutant to discriminate against the nucleotide substrate solely on the basis of differences in ligand binding. Our data suggest that discrimination against the inhibitor is 16-fold increased when the M184V/Y115F double mutant is compared with WT RT. This effect is much stronger than as seen with M184V (3.8-fold increase) and Y115F (5.5-fold) alone. In contrast, the K65R mutant shows enhanced binding of the inhibitor and diminished binding of dGTP as compared with WT RT (Table 2), which overall results in discrimination against the natural nucleotide. Introducing the K65R mutation against a background of M184V neutralizes their individual opposing effects. The M184V/K65R double mutant shows a small 2.2-fold increase in discrimination against the inhibitor. However, K65R cannot efficiently counteract the effects of the double M184V/Y115F mutant. The ability to discriminate against INDOPY-1 remains relatively high with a 7.8-fold increase as compared with the WT. Collectively, these binding studies are in good agreement with the inhibition data in both cell-free and cell-based assays.
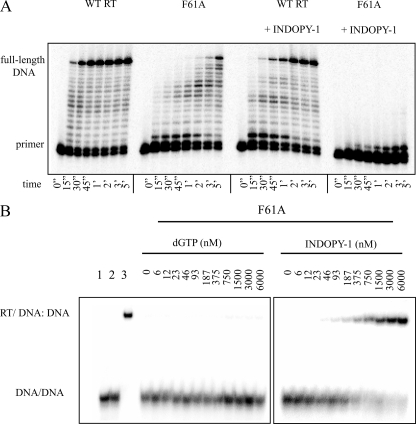
Effects of F61A on Both Nucleotide and INDOPY-1 Binding—The aforementioned findings suggest that differences among the various mutant enzymes in their ability to bind the natural nucleotide substrate can affect susceptibility to INDOPY-1. Enhanced binding for the incoming dNTP in the context of Y115F correlates with decreased inhibitory effects of INDOPY-1, while diminished binding for the nucleotide in the context of K65R correlates with increased inhibitory effects. However, both mutations also affect inhibitor binding, which makes it difficult to assess the contribution of the nucleotide substrate on the observed phenotype. To further address this problem, we included the F61A mutation in this analysis. The F61A mutant enzyme is severely compromised with regards to nucleotide binding while primer/template binding is largely unaffected. HIV variants containing this mutation do not replicate, and the enzyme shows marked reductions in DNA synthesis (16–19). Here we monitored DNA synthesis with the RT mutant in excess over primer/template substrate to generate reasonable yields of the product (Fig. 3A, left panels). However, in the presence of INDOPY-1, DNA synthesis is almost completely blocked with the F61A mutant as the inhibitor causes strong pausing during the first nucleotide incorporation events (Fig. 3A, right panels). Equivalent results were obtained in filter-based RT assays (Table 3).
FIGURE 3.
Effect of mutation F61A on INDOPY-1 inhibition of DNA synthesis complex formation. A, DNA synthesis on a DNA/DNA substrate by WT or F61A mutant RT is monitored in the absence or presence of INDOPY-1 over a time course (0–5 min). The lowest band shows the non-elongated radiolabeled primer, and the highest band shows the full-length DNA product. B, band shift assays were performed on a DNA/DNA substrate with the F61A mutant RT. A ternary complex was formed with the addition of a concentration gradient (0–6000 nm) of either dGTP (left panel) or INDOPY-1 (right panel). Lane 1 represents the negative RT control and lanes 2 and 3 are pre-RT heparin addition and negative heparin controls, respectively. For obtained Kd(app) values refer to Table 2.
TABLE 3.
IC50 values for INDOPY-1
| Enzyme | Filter-based IC50a |
|---|---|
| nm | |
| WT | 843.0 ± 131 |
| F61A | 19.7 ± 12 |
| A62V | 1123.0 ± 192 |
| F61A/A62V | 45.5 ± 31 |
Each reported value is the average of at least three separate experiments
In agreement with published data (16–18), the natural nucleotide substrate fails to form a stable ternary complex with the F61A mutant (Fig. 3B, left panel). In contrast, INDOPY-1 is capable of forming stable ternary complex (Fig. 3B, right panel). The Kd(app) value is comparable with WT RT (Kd(app) = 200 nm versus 100 nm with WT RT) (Table 2). Thus, the diminished binding of the nucleotide alone is sufficient to increase the inhibitory effects of INDOPY-1. Of note, a change at the neighboring residue, i.e. A62V, has been identified among other mutations in HIV isolates that show decreased susceptibility to INDOPY-1 (5, 6). However, increases in IC50 values for INDOPY-1 are generally subtle when comparing the A62V mutant enzyme with WT RT (Table 3).
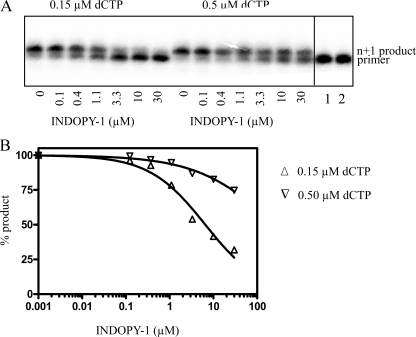
Effect of the Nucleotide Concentration on Inhibition with INDOPY-1—The combined data of our mutational analysis provide strong evidence to suggest that both differences in nucleotide binding and differences in inhibitor binding can affect the susceptibility to INDOPY-1. These findings support the notion of competitive inhibition. If correct, increasing concentration of the nucleotide substrate should diminish binding of INDOPY-1, and, in turn, its inhibitory effects. To address this issue, we studied single nucleotide incorporation events at different concentrations of the dNTP substrate in the presence of increasing concentrations of INDOPY-1. DNA synthesis was initiated in the presence of trap to ensure single turnover conditions (Fig. 4A). Thus, any effects of the nucleotide and/or the inhibitor on complex dissociation that can complicate the interpretation of the data, are excluded under these conditions. The NNRTI nevirapine is not competitive with respect to the dNTP substrate, and, as expected, the IC50 value for nevirapine was independent of the nucleotide concentration (data not shown). In contrast, the IC50 values for INDOPY-1 increased in accordance with increases in concentrations of the dNTP substrate (Fig. 4B), which is indicative of a competitive mode of inhibition.
FIGURE 4.
Effect of various dCTP concentrations on inhibition with INDOPY-1. A, single nucleotide incorporation was monitored in the presence of increasing concentrations of INDOPY-1 (ranging from 0 to 30 μm) where INDOPY-1-mediated inhibition is indicated by the disappearance of the n+1 product band. The experiment was performed in the presence of either 0.15 μm dCTP or 0.5 μm dCTP, respectively. Lane 1 shows the pre-RT heparin addition control. B, logarithmic graph represents normalized percentage of product formed plotted as a function of INDOPY-1 concentration under two separate conditions (variable dCTP concentrations). We obtained an average IC50 value of 6.5 ± 4 of three independent experiments at concentrations of 0.15 μm dCTP. IC50 values cannot be accurately determined for higher substrate concentrations because the reaction is not efficiently blocked under these conditions. Saturating concentrations of ∼20 μm of INDOPY-1 can further limit the accuracy of these measurements.
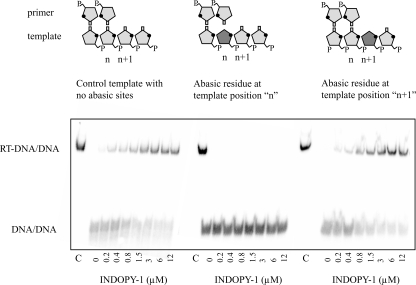
Effect of Abasic Sites in the Template Strand on Binding of INDOPY-1—The resistance profile and the combined biochemical data that point to a competitive mode of inhibition suggest that INDOPY-1 may bind in close proximity to the nucleotide binding site. In an attempt to further define the binding site for INDOPY-1, we employed chemically modified template strands with abasic sites at positions n and n+1 (Fig. 5A). Under this nomenclature, template position n lies opposite the 3′-end of the primer, and position n+1 pairs with the incoming dNTP (Fig. 5B). The planar structure of the inhibitor (Fig. 1) points to possible stacking interactions with the template strand, and the loss of the nucleobase at these strategic positions may therefore cause deficiencies in binding. We found that an abasic residue at position n fully prevents formation of a stable ternary complex with INDOPY-1, while an abasic site at position n+1 did not show any significant effect when compared with the unmodified template. The clear difference suggests that interaction with the ultimate base pair represents an important structural requirement for inhibitor binding, while the nucleobase in the template overhang appears to be dispensable in this regard.
FIGURE 5.
Effect of the template base on binding of INDOPY-1 to the RT-DNA/DNA complex. Band shift experiments were performed on templates containing an abasic residue in the presence of increasing concentrations of INDOPY-1. The left panel shows complex formation with no abasic sites while the middle panel shows the abasic residue at position n. The right panel shows complex formation when the abasic residue is at position n+1. The control C lane represents the complex formed in the absence of heparin trap.
DISCUSSION
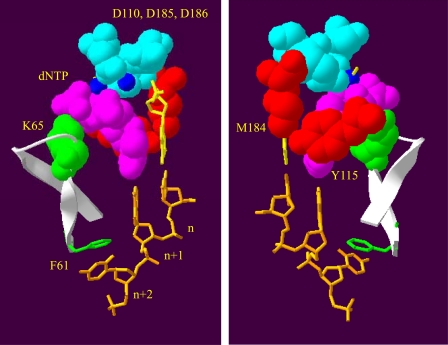
INDOPY-1 represents a family of compounds that inhibit HIV-1 RT through a novel mechanism of action. Previous biochemical studies by our group have shown that INDOPY-1 binds preferentially following incorporation of pyrimidines, stabilizes the RT-DNA/DNA complex in its post-translocated state, and blocks DNA synthesis in a reversible manner (5). While the resistance profile for this inhibitor is unique, changes in susceptibility to INDOPY-1 have been associated with a number of mutations that likewise change susceptibility to NRTIs (5, 6). Here we studied the biochemical phenotypes associated with these changes to provide novel insight into both mechanisms of resistance, and the binding properties of INDOPY-1. Key results are visualized against the structure of HIV-1 RT (Fig. 6).
FIGURE 6.
Structural features around the active site of HIV-1 RT that affect binding of INDOPY-1. The view of the polymerase active site is depicted from two opposing angles. In both panels, the catalytic aspartic acid residues (Asp-110, Asp-185, and Asp-186) are represented in cyan in contact with the two magnesium ions present in the active site (blue circles). The nucleic acid template shows positions n, n+1, and n+2 (orange), while the primer 3′-end is shown in yellow. The magenta residues indicate the incoming dNTP molecule present at the active site. Residues conferring resistance to INDOPY-1 (Met-184 and Tyr-115) are represented in red, while hypersusceptible residues Lys-65 and Phe-61 are represented in green. The β3-β4 loop is indicated in white. Upon nucleotide binding, the fingers close down allowing for Lys-65 to come in close contact with the γ-phosphate of dNTP. The aromatic side-chain of Phe-61 shows stacking interactions with the template at position n+2.
Decreased Susceptibility to INDOPY-1—M184V and Y115F have been associated with decreased susceptibility to INDOPY-1. Met-184 lies in close proximity to the 3′-end of the primer, while the aromatic side chain of Tyr-115 is in close proximity of the sugar moiety of the bound dNTP substrate ((20), visualized in Fig. 6). The combination of the two mutations amplifies the subtle effects of the individual mutations in cell-based and cell-free assays. Thus, these mutations in HIV-1 RT can be directly linked to the resistant phenotype. However, our data suggest that the contribution of the two mutations is based on different, yet complementary mechanisms. M184V diminishes complex formation with the inhibitor, while Y115F facilitates complex formation with the natural nucleotide substrate. These interlinked mechanisms help to explain the synergistic effects of the two mutations. The data provide strong evidence to suggest that the mechanism of resistance is based on substrate discrimination.
The band shift experiments used here to study complex formation provide an indirect measure for ligand binding. The differences in Kd(app) values are in good agreement with enzyme kinetic measurements that point to decreases in Kd and Km values with the Y115F mutant (21, 22). The Y115F mutation is seen, infrequently, in conjunction with M184V in isolates of patients treated with abacavir (9, 23). However, Y115F does not appear to further diminish the efficiency of incorporation of the NRTI (21).
Increased Susceptibility to INDOPY-1—K65R, which emerges under the selective pressure of tenofovir (10), confers hypersusceptibility to INDOPY-1. It is interesting to note that M184V can increase susceptibility to tenofovir (24); thus, the two compounds show inverse effects with regards to K65R and M184V and combining tenofovir and INDOPY-1 in cell culture affects the selection of resistance. Under these conditions, tenofovir selects for K70E (25). The properties of the mutant enzyme are reminiscent of K65R; however, K70E is severely compromised in regard to viral replication capacity (26).
We found that K65R shows subtle increases in complex formation with INDOPY-1, and subtle decreases in complex formation with the nucleotide substrate. In essence, this is the opposite of the results described for the combination of M184V and Y115F. Thus, K65R discriminates against the dNTP substrate, which is consistent with an INDOPY-1 hypersusceptible phenotype. Complex formation with the nucleotide appears to be an important parameter that affects susceptibility to INDOPY-1. The F61A mutation provides additional evidence in support of this notion. This mutation discriminates against dNTP substrate binding while the INDOPY-1 binding remains unaffected resulting in a heavily hypersusceptible enzyme.
Ternary Complexes with the Nucleotide Substrate versus INDOPY-1—The crystal structure of a ternary complex with a bound dNTP shows the polymerase domain in a closed structure (20, 27, 28). Prior to nucleotide binding, the β3-β4 hairpin loop in the fingers subdomain is open to facilitate this step. The closure of the fingers appears to mediate interactions between Lys-65 and the γ-phosphate of the bound dNTP (29), while the aromatic side chain of Phe-61 stacks against the nucleobase at template position n+2 (Ref. 20 and Fig. 6). The loss of these interactions, in particular the stacking between Phe-61 and template, appears to selectively destabilize the ternary complex with the nucleotide and not with INDOPY-1. Our studies with the F61A mutant suggest that the aromatic side chain of this residue is dispensable for formation of a stable ternary complex with the inhibitor. These data suggest that alternative contacts between the inhibitor and the DNA/DNA substrate provide an anchor to stabilize the complex with RT. The instability of a ternary complex with a template containing an abasic lesion opposite the primer terminus implicates important interactions between INDOPY-1 and the ultimate base pair. The specificity for pyrimidine residues at the 3′-end of the primer provides independent support for this notion; however, structural studies are required to address this issue in more detail.
Despite these differences in ternary complex formation, binding of INDOPY-1 and the nucleotide substrate are competitive in nature. Previous studies under steady-state conditions have pointed to competitive or mixed-type inhibition, respectively (5, 6). In theory, mixed-type inhibition may allow binding of the inhibitor to both binary RT-primer/template complexes and ternary complexes with the incoming dNTP. However, binding parameters suggested that the affinity of the inhibitor to ternary complexes is markedly reduced. In addition, INDOPY-1 stabilizes the product complex following nucleotide incorporation, which, in turn, prevents enzyme dissociation under multiple turnover (steady-state) conditions. Enzyme kinetics measured under these conditions can translate in mixed-type or non-competitive inhibition, although simultaneous binding of substrate and inhibitor is mutually exclusive (30). To avoid these complications we studied the inhibitory effects of INDOPY-1 under single turnover conditions, which indicates competitive binding, consistent with our previous observations (5).
Together, the data suggest that INDOPY-1 binds in close proximity to the nucleotide binding site of HIV-1 RT. Mechanisms associated with resistance and hypersusceptibility are both based on ligand discrimination. These findings along with complementary biochemical data show that the inhibitor competes with the natural dNTP substrates. In light of the collective data, we suggest to refer to members of this family of compounds as “nucleotide-competing RT inhibitors” (NcRTIs) (25, 31, 32). This work demonstrates that small molecules that are structurally distinct from classic nucleotides or nucleotide analogues can bind in close proximity to the active site of viral polymerases. This concept may be further explored in drug discovery and development efforts.
Supplementary Material
Acknowledgments
We thank Suzanne McCormick for excellent technical assistance.
This work was supported, in whole or in part, by the intramural research program of the Center for Cancer Research, NCI, National Institutes of Health (to S. L. G.). This work was also funded in part by Tibotec and a grant from the Canadian Institutes of Health Research (CIHR) (to M. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RT, reverse transcriptase; HIV-1, human immunodeficiency virus type 1; NRTIs, nucleoside-analogue RT inhibitors; NNRTIs, non-nucleoside analogue RT inhibitors; WT, wild type.
References
- 1.Basavapathruni, A., and Anderson, K. S. (2007) Faseb J. 21 3795-3808 [DOI] [PubMed] [Google Scholar]
- 2.Goldschmidt, V., and Marquet, R. (2004) Int. J. Biochem. Cell Biol. 36 1687-1705 [DOI] [PubMed] [Google Scholar]
- 3.Menendez-Arias, L. (2008) Virus Res. 134 124-146 [DOI] [PubMed] [Google Scholar]
- 4.Sluis-Cremer, N., and Tachedjian, G. (2008) Virus Res. 134 147-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jochmans, D., Deval, J., Kesteleyn, B., Van Marck, H., Bettens, E., De Baere, I., Dehertogh, P., Ivens, T., Van Ginderen, M., Van Schoubroeck, B., Ehteshami, M., Wigerinck, P., Gotte, M., and Hertogs, K. (2006) J. Virol. 80 12283-12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, Z., Walker, M., Xu, W., Shim, J. H., Girardet, J. L., Hamatake, R. K., and Hong, Z. (2006) Antimicrob. Agents Chemother. 50 2772-2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheaff, R., Ilsley, D., and Kuchta, R. (1991) Biochemistry 30 8590-8597 [DOI] [PubMed] [Google Scholar]
- 8.Cheng, C. H., and Kuchta, R. D. (1993) Biochemistry 32 8568-8574 [DOI] [PubMed] [Google Scholar]
- 9.Miller, V., Ait-Khaled, M., Stone, C., Griffin, P., Mesogiti, D., Cutrell, A., Harrigan, R., Staszewski, S., Katlama, C., Pearce, G., and Tisdale, M. (2000) AIDS 14 163-171 [DOI] [PubMed] [Google Scholar]
- 10.Miller, M. D. (2004) AIDS Rev. 6 22-33 [PubMed] [Google Scholar]
- 11.Jochmans, D., Van Ginderen, M., Ehteshami, M., Van Schoubroeck, B., Dehertogh, P., Hallenberger, S., Götte, M., and Hertogs, K. (2007) Conference on Retroviruses and Opportunistic Infections, Abstract 597, Los Angeles, CA
- 12.Le Grice, S. F., and Gruninger-Leitch, F. (1990) Eur. J. Biochem. 187 307-314 [DOI] [PubMed] [Google Scholar]
- 13.Arts, E. J., Li, X., Gu, Z., Kleiman, L., Parniak, M. A., and Wainberg, M. A. (1994) J. Biol. Chem. 269 14672-14680 [PubMed] [Google Scholar]
- 14.Marchand, B., and Gotte, M. (2003) J. Biol. Chem. 278 35362-35372 [DOI] [PubMed] [Google Scholar]
- 15.Tong, W., Lu, C. D., Sharma, S. K., Matsuura, S., So, A. G., and Scott, W. A. (1997) Biochemistry 36 5749-5757 [DOI] [PubMed] [Google Scholar]
- 16.Fisher, T. S., Darden, T., and Prasad, V. R. (2003) J. Mol. Biol. 325 443-459 [DOI] [PubMed] [Google Scholar]
- 17.Fisher, T. S., and Prasad, V. R. (2002) J. Biol. Chem. 277 22345-22352 [DOI] [PubMed] [Google Scholar]
- 18.Mandal, D., Dash, C., Le Grice, S. F., and Prasad, V. R. (2006) Nucleic Acids Res. 34 2853-2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman, A. P., Garforth, S. J., Prasad, V. R., and Kool, E. T. (2008) Biochemistry 47 4800-4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, H., Chopra, R., Verdine, G. L., and Harrison, S. C. (1998) Science 282 1669-1675 [DOI] [PubMed] [Google Scholar]
- 21.Ray, A. S., Basavapathruni, A., and Anderson, K. S. (2002) J. Biol. Chem. 277 40479-40490 [DOI] [PubMed] [Google Scholar]
- 22.Martin-Hernandez, A. M., Domingo, E., and Menendez-Arias, L. (1996) EMBO J. 15 4434-4442 [PMC free article] [PubMed] [Google Scholar]
- 23.Lanier, E. R., Givens, N., Stone, C., Griffin, P., Gibb, D., Walker, S., Tisdale, M., Irlbeck, D., Underwood, M., St Clair, M., and Ait-Khaled, M. (2004) HIV Med. 5 394-399 [DOI] [PubMed] [Google Scholar]
- 24.Wainberg, M. A., Miller, M. D., Quan, Y., Salomon, H., Mulato, A. S., Lamy, P. D., Margot, N. A., Anton, K. E., and Cherrington, J. M. (1999) Antivir. Ther. 4 87-94 [DOI] [PubMed] [Google Scholar]
- 25.Jochmans, D., Van Marck, H., Van Ginderen, M., De Baere, I., Dehertogh, P., Peeters, A., Kesteleyn, B., Pattery, T., McKenna, P., and Hertogs, K. (2006) Conference on Retroviruses and Opportunistic Infections Abstract 500, Denver, CO
- 26.Sluis-Cremer, N., Sheen, C. W., Zelina, S., Torres, P. S., Parikh, U. M., and Mellors, J. W. (2007) Antimicrob. Agents Chemother. 51 48-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarafianos, S. G., Clark, A. D., Jr., Das, K., Tuske, S., Birktoft, J. J., Ilankumaran, P., Ramesha, A. R., Sayer, J. M., Jerina, D. M., Boyer, P. L., Hughes, S. H., and Arnold, E. (2002) EMBO J. 21 6614-6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuske, S., Sarafianos, S. G., Clark, A. D., Jr., Ding, J., Naeger, L. K., White, K. L., Miller, M. D., Gibbs, C. S., Boyer, P. L., Clark, P., Wang, G., Gaffney, B. L., Jones, R. A., Jerina, D. M., Hughes, S. H., and Arnold, E. (2004) Nat. Struct. Mol. Biol. 11 469-474 [DOI] [PubMed] [Google Scholar]
- 29.Sluis-Cremer, N., Arion, D., Kaushik, N., Lim, H., and Parniak, M. A. (2000) Biochem. J. 348 77-82 [PMC free article] [PubMed] [Google Scholar]
- 30.Marchand, B., Tchesnokov, E. P., and Gotte, M. (2007) J. Biol. Chem. 282 3337-3346 [DOI] [PubMed] [Google Scholar]
- 31.Jochmans, D., Kesteleyn, B., Marchand, B., Gotte, M., Ivens, T., Dehertogh, P., Peeters, A., Pauwels, R., Wigerinck, P., and Hertogs, K. (2005) Conference on Retroviruses and Opportunistic Infections Abstract 156, Boston, MA
- 32.Jochmans, D. (2008) Virus Res. 134 171-185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.