Abstract
Two-component signaling systems allow bacteria to adapt to changing environments. Typically, a chemical or other stimulus is detected by the periplasmic sensor domain of a transmembrane histidine kinase sensor, which in turn relays a signal through a phosphotransfer cascade to the cognate cytoplasmic response regulator. Such systems lead ultimately to changes in gene expression or cell motility. Mechanisms of ligand binding and signal transduction through the cell membrane in histidine kinases are not fully understood. In an effort to further understand such processes, we have solved the crystal structures of the periplasmic sensor domains of Escherichia coli DcuS and of Vibrio cholerae DctB in complex with the respective cognate ligands, malate and succinate. Both proteins are involved in the regulation of the transport and metabolism of C4-dicarboxylates, but they are not highly related by sequence similarity. Our work reveals that despite disparate sizes, both structures contain a similar characteristic α/β PDC (PhoQ-DcuS-CitA) sensor-domain fold and display similar modes of ligand binding, suggesting similar mechanisms of function.
The ability of bacteria to monitor and adapt to their environment is crucial to their survival, and two-component signal transduction systems mediate most of these adaptive responses. One component is a histidine kinase sensor, most commonly part of a homodimeric transmembrane sensor protein, and the second component is a cytoplasmic response regulator. The two components interact in tandem through a phosphotransfer cascade (1-4). A typical transmembrane sensor protein contains a periplasmic sensor domain and a cytoplasmic histidine kinase domain. Upon binding of a ligand to the periplasmic sensor domain, this signal is transduced across the membrane to the cytoplasmic domain where an ATP-dependent autophosphorylation of a conserved histidine residue occurs (1). The phosphate is subsequently transferred to a conserved aspartate residue in the response regulator protein by an auto-catalyzed reaction (2), ultimately leading to adaptive modulation of gene expression. In some circumstances, sensor stimulation leads to dephosphorylation. Within the large family of protein histidine kinases, the sequence of the sensor domain is considered to be modular, whereas that of the histidine kinase domain is more conserved. Over the past decade, especially with the determination of numerous bacterial genome sequences, hundreds of new histidine kinase proteins have been identified but remain unstudied.
The DcuS-DcuR two component system is involved in the regulation of the anaerobic fumarate respiratory pathway in Escherichia coli (5, 6). The sensor kinase DcuS is a member of the CitA family of histidine kinases, and it responds to C4-dicarboxylates such as fumarate, succinate, malate, and tartrate (5, 7). Upon detection of its cognate ligand, the DcuS-DcuR system up-regulates the synthesis of both fumarate reductase (frdABCD) and also the anaerobic fumarate-succinate anti-porter DcuB. DcuS is predicted to contain an ∼140-residue periplasmic domain flanked by two hydrophobic transmembrane helices and a cytoplasmic portion comprising a coiled-coil domain, a cytoplasmic PAS domain, and a histidine kinase domain (6). It has been shown in vitro that the sensing of C4-dicarboxylates by DcuS is through direct binding (8). Although the solution structure of the periplasmic domain of DcuS has been solved (9), the mechanism of ligand binding is still unclear.
The DctB-DctD two-component system is involved in the regulation of C4-dicarboxylate uptake in rhizobia as part of nitrogen fixation (10). DctB, like DcuS, is a transmembrane sensor kinase that has been shown to be involved in the direct sensing of C4-dicarboxylates such as succinate (11). But its periplasmic sensor domain is predicted to contain ∼270 amino acid residues, almost twice that of the DcuS sensor domain (12). The response regulator DctD controls the expression of the dicarboxylate transporter DctA, another integral membrane protein (10). Studies have shown that either the ligand specificity or the signaling state of DctB may be modified by DctA, through a possible direct interaction between DctA and DctB (7, 11). The DctB ortholog in Vibrio cholerae is the product of gene VC1925, a protein annotated in the Swiss-PROT/TrEMBL (13) data base as a putative histidine kinase sensor under accession number Q9KQS3. It shows high sequence similarity to the previously studied rhizobial DctB sensors, and it has been grouped into the NtrB C4-dicarboxylate sensor histidine kinase family (7). The exact function of DctB in V. cholerae has yet to be determined.
In this study we present high resolution crystal structures of the periplasmic domains of both E. coli DcuS and V. cholerae DctB in complexes with their cognate ligands malate and succinate, respectively. These two sensor domains show similarities in structure and in ligand binding that provide possible clues toward the understanding of signal transduction in two-component histidine kinase proteins.
EXPERIMENTAL PROCEDURES
Cloning—For the generation of the DcuS construct, a DNA fragment corresponding to the periplasmic domain of DcuS from residues 42 to 181 was PCR-amplified from genomic E. coli K12 DNA (ATCC Bioproducts) using the appropriate 5′ and 3′ primers. The primers were engineered to produce a DNA fragment flanked by a BamHI restriction site at the 5′ end and an XhoI restriction site at the 3′ end followed by a stop codon. The amplified DNA was subsequently ligated into the ampicillin-selectable pGEX-4T-2 (Amersham Biosciences) expression vector at the BamHI/XhoI sites. The resulting construct allowed for the isopropyl β-d-thiogalactopyranoside (IPTG)2-inducible expression of a soluble N-terminal glutathione S-transferase fusion protein containing an internal thrombin cleavage site that would leave only an additional glycine and serine residue on the N-terminal end of the protein upon cleavage.
Generation of the periplasmic DctB construct entailed PCR amplification of a DNA fragment corresponding to residues 28-286 of DctB from genomic V. cholerae MO45 DNA (ATCC Bioproducts). The 5′ primer used in the PCR contained an NdeI restriction site, and the 3′ primer contained a stop codon and an XhoI restriction site. The resulting DNA fragment could be ligated into the ampicillin-resistant Novagen pET22b+ expression vector at the NdeI/XhoI polylinker site for IPTG-inducible expression of the protein. The product of this expression retains the N-terminal methionine residue from the vector ahead of Arg-28.
Expression and Purification—Native DcuS-(42-181) protein was expressed as a glutathione S-transferase fusion protein from a 4-liter culture of Novagen E. coli BL21 (DE3) cells grown in Luria-Bertani (LB) media containing 100 μg/ml ampicillin. The culture was originally started as a 1:100 inoculation from an overnight culture grown in LB (100 μg/ml ampicillin) at 37 °C. Induction began with the addition of IPTG to 1 mm for 2 h at 37 °C upon reaching an optical density (OD) of 0.6. Selenomethionyl (SeMet) DcuS-(42-181) was produced in the same manner except that 4 liters of minimal media containing SeMet was used instead of LB, and the overnight culture was grown in minimal media containing methionine instead of SeMet. The cells were harvested by centrifugation and resuspended in 40 ml of phosphate-buffered saline, pH 7.4, and 5 mm dithiothreitol. Cell supernatant was prepared by sonication and cleared by centrifugation, passed through an 8-ml glutathione-Sepharose™ 4B (GE Healthcare) column, and washed with phosphate-buffered saline, pH 7.4. The fusion protein was eluted from the column with 0.1 m Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, and 20 mm glutathione. The protein was further purified by gel filtration on a Superdex 200 26/60 (GE Healthcare) column previously equilibrated with 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol and then cleaved for 4 h at 20 °C by the addition of thrombin to a concentration of 1 unit of thrombin/mg of fusion protein. The cleaved DcuS-(42-181) was then purified by gel filtration on a Superdex 75 26/60 (GE Healthcare) column previously equilibrated with 20 mm HEPES, pH 8.0, 50 mm NaCl, 1 mm EDTA, and 5 mml-malate. The protein appeared to be monomeric by gel filtration chromatography and was homogeneous when analyzed by SDS-PAGE and native PAGE.
SeMet DctB-(28-286) was expressed in the same manner as SeMet DcuS-(42-181) except that only a 2-liter culture of cells was used instead with induction at 30 °C for 3 h. The cells were harvested by centrifugation and resuspended in 20 ml of Q-buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, and 1 mm EDTA). Cell supernatant was prepared by sonication and cleared by centrifugation. DctB-(28-286) was purified by passing the supernatant through a 5-ml Hi-Trap Q™ column (GE Healthcare) previously equilibrated with Q-buffer, washing with 20 volumes of Q-buffer, and elution using a 50-400 mm NaCl gradient over a total volume of 120 ml. DctB-(28-286) eluted from the column at around 150 mm NaCl and was further purified by gel filtration on a Superdex 75 26/60 (GE Healthcare) column previously equilibrated with Q-buffer, in which it appeared monomeric. A final purification step was performed by ion exchange chromatography on a MonoQ 10/10 (GE Healthcare) column, using a 50-300 mm NaCl gradient over a total volume of 120 ml. The purified fractions were pooled and dialyzed against 1 liter of 20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm EDTA, and appeared homogeneous by SDS-PAGE and native PAGE.
Crystallization—Native DcuS-(42-181) crystals were grown by hanging-drop vapor diffusion against a reservoir buffer containing 24% polyethylene glycol monoethyl ether (PEG MME) 2000, 15% isopropyl alcohol, 0.2 m ammonium citrate, and 0.1 m sodium acetate, pH 4.5 at 4 °C. A protein concentration of 18.5 mg/ml and a protein to buffer ratio of 1:1 (1 + 1 μl) was used. SeMet DcuS-(42-181) crystals were grown by the same method with the same buffer, except the concentration of PEG MME 2000 was reduced to 20%, and a protein concentration of 16 mg/ml was used. Long hexagonal rod-shaped crystals typically appeared overnight and grew to a reasonable size within a week. Crystals were briefly soaked in cryoprotectant containing 25% PEG MME 2000, 15% isopropyl alcohol, 0.2 m ammonium citrate, 0.1 m sodium acetate, pH 4.5, and 7.5% glycerol prior to freezing in liquid nitrogen.
SeMet DctB-(28-286) crystals were grown by hanging-drop vapor diffusion at 4 °C against a buffer containing 6% isopropyl alcohol, 0.2 m calcium acetate, and 0.1 m Tris-HCl, pH 7.5. A protein concentration of 8.5 mg/ml was used with a protein to buffer ratio of 1:1 (2 μl + 2 μl). Crystals typically appeared overnight and grew to an optimal size within 2 weeks. The crystals were initially soaked in crystallization buffer supplemented with glycerol and ethylene glycol at 7.5 and 5%, respectively; and crystals were then subsequently transferred to a comparable buffer in which the glycerol and ethylene glycol was raised to 15 and 10%, respectively, prior to freezing in liquid nitrogen.
Structure Determination of DcuS-(42-181) in Complex with Malate—A four-wavelength MAD experiment at the selenium K-edge was collected on a single frozen SeMet crystal at the X4A beamline of the National Synchrotron Light Source at Brookhaven National Laboratory. Diffraction data to Bragg spacings of 2.0 Å (165 mm detector distance) were collected with 10-s exposure times and 1.2° oscillations at each of the four wavelengths, using inverse beam measurements for the accurate determination of Bijvoet differences. Data were also collected from a single native crystal in two parts, first at a 125-mm detector distance for 15-s exposure times at 1° oscillations and second at a detector distance of 225 mm for 3-s exposure times at 4° oscillations. Two passes were required because very strong reflections at low angles overloaded the detector at the longer exposure times required for the collection of high angle data. The native crystal diffracted to a limit of 1.4-Å spacings. Denzo and Scalepack of the HKL program package (14) were used to process the datasets, which showed the symmetry compatible with space groups P3121 and P3221. Solve (15) was used to determine the positions of two sites, which were verified by analysis of Patterson maps using RSPS (16) of the CCP4 program suite (17) and subsequently refined in SHARP (18). One site refined to a high B-factor, indicative of disorder, and the other refined to reasonable values. Phases were calculated to 2.0-Å spacings in the two possible space groups, but after solvent flipping using Solomon (19) interpretable electron density maps showing clear secondary structure only resulted in the P3121 enantiomer. The calculated figure of merit in the correct space group was 0.62. Phases were extended from 2.2 to 1.4 Å against the native dataset using DM (20), and Arp/wArp 5.1 (21) was used for automatic model building. Refinement of the structure against the native dataset to 1.45 Å was accomplished by iterative cycles of simulated annealing, conjugate-gradient minimization, temperature-factor refinement, and manual rebuilding using CNS (22) and O (23). An extra feature of electron density was fitted by l-malate, which was included in the protein purification, and this model was refined. Data collection and refinement statistics are listed in Table 1 and Table 3. The atomic coordinates and structure factors for DcuS-(42-181) have been deposited with the Protein Data Bank with accession code 3BY8.
TABLE 1.
DcuS-(42-181) diffraction data
| Dataset | dmin | Wavelength | No. of reflections | Average redundancy | <I>/(δ) | Completenessa | Rmergea,b |
|---|---|---|---|---|---|---|---|
| Å | Å | % | % | ||||
| Native | 1.4 | 0.9678 | 29,824 | 8.6 | 17.0 | 99.9 (98.9) | 4.6 (23.8) |
| SeMet λ1 | 2.0 | 0.9918 (low) | 19,879 | 5.9 | 10.5 | 99.8 (99.8) | 6.4 (26.2) |
| SeMet λ2 | 2.0 | 0.9793 (edge) | 19,839 | 5.9 | 10.9 | 99.8 (100.0) | 6.2 (24.7) |
| SeMet λ3 | 2.0 | 0.9787 (peak) | 19,857 | 5.9 | 10.6 | 99.8 (100.0) | 6.5 (26.5) |
| SeMet λ4 | 2.0 | 0.9678 (high) | 19,869 | 5.9 | 9.6 | 99.8 (100.0) | 7.6 (34.1) |
Values in outermost shell are given in parentheses.
Rmerge = (∑|Ii - <Ii>|)/∑|Ii|, where Ii is the integrated intensity of a given reflection.
TABLE 3.
DcuS and DctB refinement statistics
| Parameter | DcuS-(42-181) (with malate) | SeMet DctB-(28-286) (with succinate) |
|---|---|---|
| Bragg spacings (Å) | 30 to 1.45 | 20 to 1.7 |
| Space group | P3121 | P212121 |
| Cell parameters: a, b, c (Å) | 86.11, 86.11, 35.21 | 56.31, 90.98, 114.20 |
| Za/solvent content (%) | 1/48.7 | 2/49.9 |
| Ra/Rfreeb (%) | 21.9/23.2 | 16.2/19.9 |
| No. of unique reflections | 26,860 | 64,940 |
| No. of total atoms (non-hydrogen) | 1312 | 5139 |
| No. of protein atoms | 1151 | 4400 |
| No. of ligand atoms | 9 (malate) | 16 (succinate) |
| No. of waters | 152 | 721 |
| Average B factor (Å2) | 25.1 | 20.5 |
| Root mean square bond ideality (Å) | 0.010 | 0.015 |
| Root mean square angle ideality (°) | 1.6 | 1.7 |
| Protein Data Bank accession code | 3BY8 | 3BY9 |
R = (∑||Fo| - |Fc||)/∑| Fo|, where Fo and Fc denote observed and calculated structure factors, respectively.
Rfree was calculated using 5% of data excluded from refinement.
Structure Determination of DctB-(28-286) in Complex with Succinate—MAD data were collected on a single frozen SeMet crystal at National Synchrotron Light Source beamline X4A. Phasing data were collected to a limit of 2.2-Å spacings (220 mm detector distance) using 5-s exposure times and 1° oscillations with inverse beam at four wavelengths. An additional high angle dataset extending to 1.6-Å spacings (135 mm detector distance) was collected from the same crystal at the low remote wavelength using 12-s exposure times and 0.75° oscillations. The data were processed in space group P212121 using Denzo and Scalepack. Four ordered selenium sites belonging to two molecules in the asymmetric unit were found in Solve (15), which calculated phases to 2.2 Å with a mean figure of merit of 0.61 for one of the enantiomers. The programs Arp/wArp 6.0 (21) and GUISIDE (21) were used for model building, and CNS (22) and O (23) were used for refinement of the structure to a resolution of 1.7 Å against the high angle data, using iterative cycles of simulated annealing, conjugate gradient minimization, temperature-factor refinement, and manual rebuilding. Adventitious electron density found in the structure could be best fitted with succinate, and this structure was included in the model and refined. Data collection and refinement statistics are listed in Table 2 and Table 3. The atomic coordinates and structure factors for DctB-(28-286) have been deposited with the Protein Data Bank with accession code 3BY9.
TABLE 2.
DctB-(28-286) diffraction data
| Dataset | dmin | Wavelength | No. of reflections | Average redundancy | <I>/(δ) | Completenessa | Rmergea,b |
|---|---|---|---|---|---|---|---|
| Å | Å | % | % | ||||
| SeMet (high angle) | 1.6 | 0.9946 | 76,821 | 3.6 | 11.5 | 98.1 (91.8) | 5.7 (39.5) |
| SeMet λ1 | 2.2 | 0.9946 (low) | 56,296 | 4.7 | 16.8 | 97.6 (84.9) | 5.6 (10.7) |
| SeMet λ2 | 2.2 | 0.9791 (edge) | 56,596 | 4.7 | 19.2 | 98.1 (87.3) | 5.0 (8.5) |
| SeMet λ3 | 2.2 | 0.9788 (peak) | 56,581 | 4.7 | 18.8 | 98.1 (87.6) | 5.1 (8.6) |
| SeMet λ4 | 2.2 | 0.9637 (high) | 56,978 | 5.0 | 15.4 | 98.8 (91.2) | 6.1 (12.9) |
Values in outermost shell are given in parentheses.
Rmerge = (∑|Ii - <Ii>|)/∑|Ii|, where Ii is the integrated intensity of a given reflection.
Analytical Ultracentrifugation of DcuS-(42-181)—A Beckman/Coulter XLI analytical ultracentrifuge with absorbance optics was used for sedimentation equilibrium experiments. Purified DcuS-(42-181) was concentrated to 45 mg/ml and loaded into one channel of a two-channel cell of very low path length (24) (∼0.04 mm) with sapphire windows for absorption measurements at 280 nm. Because of the extremely thin path length of the cell, an air blank was used in the empty channel. The sample was sedimented to equilibrium in an AN-50Ti rotor spun at 17,000, 21,000, and 26,000 rpm at 4 °C, and absorbance scans were taken at 1-h intervals. The program WinMatch (written by J. Lary and D. A. Yphantis, available from the SPIN6 FTP site) was used to verify the attainment of equilibrium, and equilibrium data from all three speeds were fit as a group using the nonlinear least squares program Win-NONLIN (25). The partial specific volume and the solution density were calculated to be 0.7359 ml/g and 1.0081 g/ml, respectively, from the protein sequence and buffer composition using SEDNTERP (26).
RESULTS
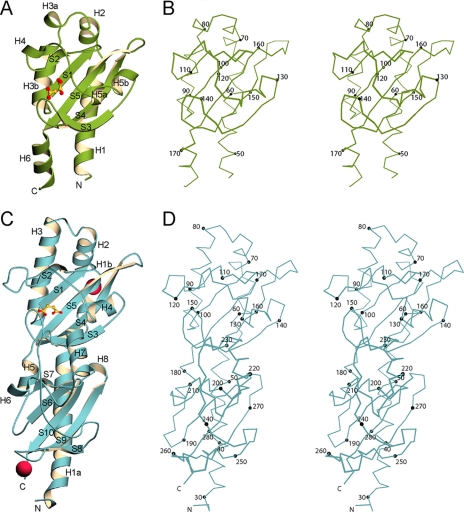
Overall Structure of DcuS—The structure of DcuS-(42-181) in complex with malate is shown in Fig. 1, A and B. The asymmetric unit contains one molecule, with clear electron density for residues 46-178 in the initial maps. A total of 133 ordered residues (46-178), 152 water molecules, and one l-malate molecule were refined against native data to a resolution of 1.45 Å with an R and Rfree of 21.9 and 23.2%, respectively (Tables 1 and 3).
FIGURE 1.
Structures of the malate-DcuS-(42-181) and succinate-DctB-(28-286) ligand complexes. A, overall structure of DcuS-(42-181) is shown as a ribbon diagram with secondary structure elements labeled in black. Bound malate is shown in ball-and-stick representation with carbon atoms in yellow and oxygen atoms in red. B, stereo plot of a Cα trace of DcuS-(42-181). Every 10th Cα atom is depicted as a black sphere and labeled accordingly. Malate was omitted for greater clarity. C, overall structure of DctB-(28-286) is shown as a ribbon diagram with secondary structure elements labeled in black. Bound succinate is shown in ball-and-stick representation with carbon atoms in yellow and oxygen atoms in red. Calcium ions are shown as magenta spheres. D, stereo plot of a Cα trace of DctB-(28-286). Every 10th Cα atom is depicted as a black sphere and labeled accordingly. Succinate was left out for greater clarity. The diagrams were created using MolScript (41) and BobScript (42).
The structure of DcuS-(42-181) consists of a mixed α/β-structure containing a central β-sheet flanked on either side by α-helices. The central β-sheet is composed of antiparallel strands S1 to S5. N- and C-terminal helices H1 and H6 lie on one side of the sheet; helices H2, H3a, H3b, H4, H5a, and H5b lie on the other side of the sheet, connected to the central sheet through loop regions. Extents of structural elements are shown in Fig. 2. Electron density for malate, a component of the purification buffer, was found in a concave pocket located on the front side of the sheet.
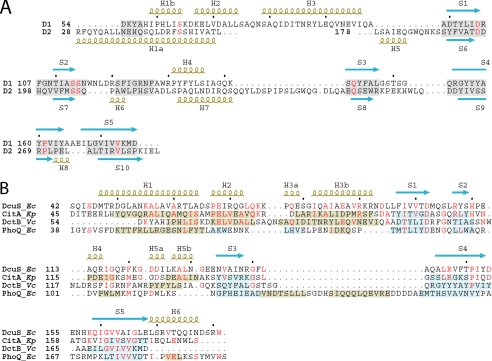
FIGURE 2.
Structure-based sequenced alignments. A, structure-based sequence alignment between the two subdomains of the V. cholerae DctB periplasmic sensor domain. Secondary structure elements for the distal subdomain 1 (D1) and the proximal subdomain 2 (D2) are labeled above and below the sequences, respectively, and are colored olive (helices) and light blue (strands). Conserved residues are shown in red. The shaded regions represent aligned regions. B, structure-based sequence alignment of DcuS-(42-181) and DctB-(28-286) distal subdomain 1 (D1) to each other and to the sensor domains of CitA and PhoQ. Conserved residues are shown in red. The secondary structure elements of DcuS-(42-181) are labeled above the alignment, whereas that for the other structures are shown as shaded regions that overlay the sequences themselves. Secondary structure elements are colored olive (helices) and light blue (strands). Organism names are abbreviated in italics (Ec for Escherichia coli, Kp for Klebsiella pneumoniae, and Vc for Vibrio cholerae).
Overall Structure of DctB—The crystal structure of SeMet DctB-(28-286) in complex with succinate is shown in Fig. 1, C and D. Two nearly identical molecules were found in the asymmetric unit, with residues 28-285 ordered in one unit, including the N-terminal Met, and residues 29-286 ordered in the other. A total of 517 residues, two succinate molecules, two calcium ions, and 722 water molecules were refined to a resolution of 1.7 Å to a final R and Rfree of 16.2 and 19.9%, respectively (Tables 2 and 3).
The structure of DctB-(28-286) is a mixed α/β-structure containing two subdomains of similar folds, each consisting of a five-stranded antiparallel β-sheet flanked by helices on either side. The overall architecture (Fig. 1, C and D) is such that the distal (relative to the membrane surface) upper subdomain (residues 54-177) is inserted between the first two helices (H1a and H5) of the proximal lower subdomain (residues 28-53 and 178-286). Strands S1 to S5 form the central core of the distal subdomain, and strands S6 to S10 form the core of the proximal subdomain. Helices H1a, H5, H6, H7, and H8 are in the proximal subdomain, whereas helices H1b, H2, H3, and H4 are in the distal subdomain (Fig. 2A). Helices H1-H3 and H5 lie on one side of the central sheet in each subdomain, and the other helices lie on the opposite side. An unusual solvent-exposed Trp in position 259 is found near the apex of the S8 to S9 loop of the proximal subdomain.
Four features of non-protein electron density were found in the structure. Equivalent distinctive features, best fit as succinate, were located in concave pockets along the front side of each distal β-sheet. Succinate is an adventitious ligand, presumably from the E. coli cytosol, and was not a crystallization additive. Two additional spherical features were modeled in as calcium ions with well defined coordination by carbonyl, carboxyl, and water oxygens. Both, one near Asp-66 and the other near Asp-263, mediate lattice contacts between protein molecules. Calcium acetate was a required crystallization ingredient.
Structural Similarities between DcuS-(42-181) and DctB-(28-286)—The DcuS and DctB sensor domains contain similar protein folds, where the two subdomains of DctB-(28-286) form a molecule of twice the size and a more elongated shape than DcuS-(42-181). The two DctB subdomains can be superimposed with a root-mean-square deviation (r.m.s.d.) of 1.37 Å for 56 structurally aligned Cα positions (Fig. 2A) where each segment of correspondence contains at least three contiguous Cα positions all within 3.0 Å of one another.
DcuS-(42-181) is similar in fold to each DctB-(28-286) subdomain. Using criteria specified above, the DcuS structure superimposes onto the distal DctB subdomain with an r.m.s.d. of 1.54 Å from 115 structurally aligned Cα positions (Fig. 2B) and onto the proximal DctB subdomain with an r.m.s.d. of 1.68 Å from 67 corresponding Cα positions. The malate-binding DcuS domain relates most closely to the distal, succinate-binding DctB subdomain.
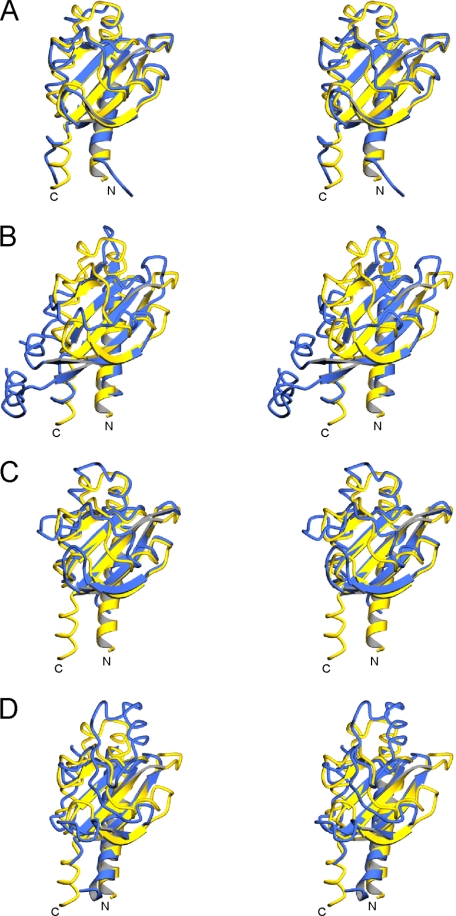
Structural Similarities of DcuS-(42-181) and DctB-(28-286) with Other Proteins—Other crystal structures of periplasmic histidine kinase sensor domains having similar folds include the citrate sensor CitA (27) and the magnesium sensor PhoQ. In our previous analysis of the E. coli PhoQ crystal structure (28), we found that these sensor domain structures are distinct from the PAS domain structures with which they share a common β-sheet topology. We introduced the term PDC (PhoQ-DcuS-CitA) sensor domain to describe such folds. Accordingly, a DALI (29) search for structural similarity to our DcuS structure yields highest Z-scores of 15.7 and 8.2 for the PDC sensor domains of CitA and PhoQ, respectively. Although the sequence similarity between DcuS and CitA sensor domains is modest (23.4% identity), they superimpose well with an r.m.s.d. of 1.38 Å over 117 corresponding Cα positions (Fig. 2B and Fig. 3A). Somewhat surprisingly, E. coli DcuS-(42-181) and the E. coli DcuS solution structure (9) match appreciably less well (r.m.s.d. of 1.68 Å over only 78 Cα positions). The DcuS and PhoQ sensor domains are dissimilar in sequence similarity (4.5% identity), but their structures superimpose with an r.m.s.d. of 1.50 Å over 63 corresponding Cα atoms (Fig. 2B and Fig. 3B and supplemental Fig. 1).
FIGURE 3.
Superimpositions of PDC sensor domains. PDC sensor domains are shown superimposed upon DcuS in stereo. Only the first N-terminal helix and all β-strands for each structure are shown in ribbon representation. All other segments are shown in worm representation. In each panel, DcuS is shown in yellow, and the superimposed PDC sensor is shown in blue. A, CitA is superimposed with DcuS, with an r.m.s.d. of 1.38Å over 117 Cα residues. B, PhoQ is superimposed with DcuS, with an r.m.s.d. of 1.50Å over 63 Cα residues. C, distal domain D1 of DctB is superimposed with DcuS, with an r.m.s.d. of 1.54Å over 115 Cα residues. D, proximal domain D2 of DctB is superimposed with DcuS, with an r.m.s.d. of 1.68Å over 67 Cα residues. The diagrams were created using MolScript (41).
As each DctB subdomain is structurally similar to DcuS (Fig. 2B and Fig. 3, C and D), both are consequently also similar to CitA and PhoQ, but the dual arrangement of two PDC subdomains is unique. Shortly after our structure determinations, it was shown that the sensor domain of Vibrio harveyi LuxQ also displays a similar quasi-tandem subdomain topology (30, 31).
It should be noted that although we have used the program DSSP (33) as a guide for assigning secondary structure elements (Fig. 2), by exception we have assigned DcuS segment S2 as a β-strand despite failing DSSP identification for having only one hydrogen bond. DcuS strand S2 is β-like in conformation, however, and it is similar enough to the S2 segments of CitA, PhoQ, and DctB to be in structural alignment with all of them. The tenuous β-nature of S2 is also evident in the DcuS solution structure (9).
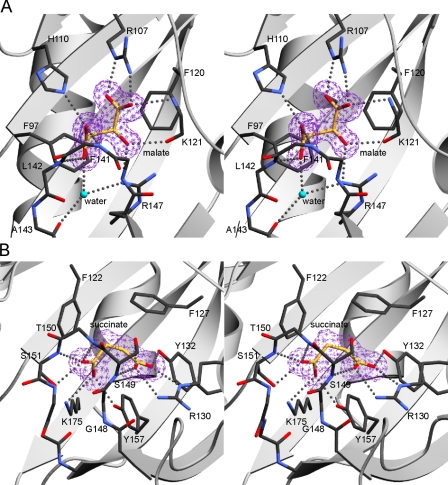
Ligand-binding Sites in DcuS and DctB—In DcuS-(42-181), the ligand binding pocket is formed by residues in the β-sheet and in the S2 to H4, H4 to H5a, and S3 to S4 loops (Fig. 4A). The majority of the protein-ligand interactions involve direct hydrogen bonding between the carboxyl and hydroxyl oxygen atoms of malate to main chain and side chain atoms of the protein. Residues Lys-121, Gly-140, Phe-141, and Leu-142 engage the ligand through main chain hydrogen bonding, whereas residues Arg-107, His-110, and Arg-147 contribute hydrogen bonding through their side chains. Water-mediated hydrogen bonds connect malate to the side chain of Arg-147 and the main chain carbonyl of Ala-143. There also are hydrophobic contacts between malate and the phenyl groups of Phe-97 and Phe-120. Stereospecificity for l-malate is conferred by the side chain of Phe-120, which packs against it in such a manner that would sterically clash with the hydroxyl group of malate in the other enantiomer. All of the residues in the binding pocket appear to be highly conserved between DcuS sequences from different organisms; however, the conservation of these residues with other members of the CitA family of histidine kinases is lower.
FIGURE 4.
Stereodrawings of ligand-binding sites of DcuS and DctB. A, malate-binding site of DcuS. The protein backbone is shown in a ribbon representation, except that loop residues 139-144 are shown in full stick representation. An Fo - Fc omit map contoured at the 4σ contour level, colored purple, is shown around malate. Malate and its contacting protein side chain and main chain atoms are shown in stick representation and labeled accordingly. Hydrogen bonds are depicted as gray dots. Carbon backbone atoms of the ligand are colored yellow for greater clarity. All other atoms are colored by atom type: carbon, black; oxygen, red; nitrogen, blue. Water is depicted in turquoise. B, succinate-binding site of DctB. The protein backbone is shown in a ribbon representation, except that loop residues 147-154 are shown in full stick representation. An Fo - Fc omit map at the 4σ contour level, colored purple, is shown around succinate. Succinate and its contacting protein side chain and main chain atoms are shown labeled in stick representation, with hydrogen bonds depicted as gray dots. The same coloring scheme for atom types in A is used. The diagrams were created using MolScript (41).
The ligand binding pocket in DctB-(28-286) is formed by residues in the first β-sheet, H4, and in the S2 to S3 and S3 to S4 loops (Fig. 4B). Residues Arg-130, Tyr-132, Ser-149, Ser-151, Tyr-157, and Lys-175 make hydrogen bonds via their side chains to the carboxylate oxygens of succinate. Residues Ser-149, Thr-150, and Ser-151 are also involved with hydrogen bonding interactions to succinate via their main chain nitrogens. There are also hydrophobic interactions between succinate and the phenyl rings of Phe-122 and Phe-127, as well as with Gly-148. The majority of these residues appear to be conserved within the DctB sequences of other organisms.
The relative locations of bound l-malate in DcuS-(42-181) and succinate in DctB-(28-286) are similar, and there is partial overlap of the residues involved in ligand binding in terms of similarities in either relative position and/or composition. The only two residues that are strictly conserved between the two sensors are Phe-120 and Gly-140 of DcuS-(42-181) corresponding to Phe-127 and Gly-148 in DctB-(28-286).
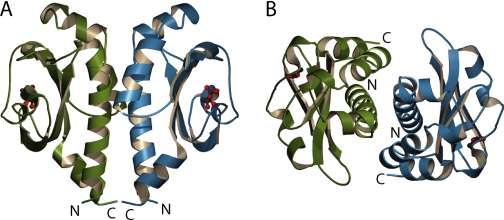
Dimerization of DcuS-(42-181)—In the crystal structure of DcuS-(42-181), an association observed about a crystallographic 2-fold axis is suggestive of a biologically relevant dimer (Fig. 5, A and B). The relative orientation of the two molecules is such that the N- and C-terminal ends are facing the same direction. The dimer interface consists primarily of residues on corresponding facing surfaces of helix H1 and H3b of the two molecules and buries a combined total of 1684 Å2 of accessible surface from the two protomers. The dimerization interface is situated on the most hydrophobic surface of the structure. The calculated shape complementarity (34) statistic for the interface is also relatively high at 0.708.
FIGURE 5.
Ribbon diagrams of the DcuS-(42-181) crystallographic dimer. A, side view of the dimer showing relative orientations of the N and C termini. B, top view of the dimer showing packing between helices H1b and H3b of opposing protomers. Individual protomers are colored green and blue with malate shown in ball-and-stick representation. The figure was created using MolScript (41).
The relative orientation between the DcuS-(42-181) subunits within its putative dimer closely resembles the E. coli PhoQ dimer, which we showed to be physiologically relevant (28), and a recent described molybdate-free/citrate-bound CitA dimer (35), although it is markedly different from a putative molybdate-bound/citrate-bound CitA dimer (27) or a putative dimer of Salmonella typhimurium PhoQ (36). The entire DcuS-(42-181) dimer can be superimposed upon the PhoQ dimer such that there are reasonable structural alignments between conserved secondary structural elements, while allowing overlap between the N- and C-terminal regions of corresponding subunits of both dimers. A similar superimposition can be performed with the molybdate-free/citrate-bound CitA dimer (supplemental Fig. 2), revealing similar dimer interfaces along structurally conserved helices. The orientation of each subunit within each respective dimer differs by a rotation of 13.1 and 14.1° along an axis running roughly perpendicular to the plane of the dimer interface.
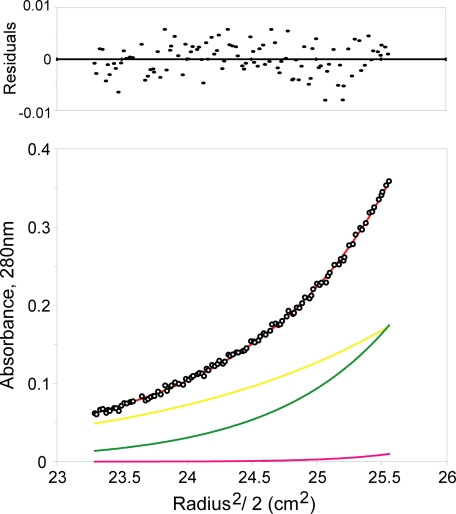
Analytical Ultracentrifugation of DcuS-(42-181)—DcuS-(42-181) appears to be monomeric by gel filtration chromatography at low concentration in the presence or absence of ligand, but a monomer-dimer equilibrium could be observed at millimolar protein concentrations by sedimentation equilibrium analytical ultracentrifugation at a physiological pH and ionic strength. Equilibrium data, measured at three different speeds from DcuS-(42-181) in the final purification buffer at 4 °C, indicated self-association. The molecular weight derived from fitting of the data with an only-monomer model is significantly greater than the theoretical monomer molecular weight of DcuS-(42-181). The data were best fit with a monomer-dimertetramer model with a dimerization Kd of 9.7 ± 3.7 mm, which yields a calculated monomer molecular weight within 5% of the theoretical (Fig. 6). The small tetramer component was included to improve the residuals slightly, yielding a more accurate monomer molecular weight. We found that the data could not be fit using only monomer, monomer-tetramer, or monomer-trimer model.
FIGURE 6.
Equilibrium centrifugation analysis of DcuS-(42-181) oligomerization. The black open circles represent absorbance (280 nm) data taken at 17,000 rpm, starting with a loading concentration of 45 mg/ml (2.8 mm). The smooth red curve through the circles represents the model for a monomer/dimer/tetramer reversible equilibrium. The distribution of residuals for a least squares fit to the model is shown above the plot. The yellow, green, and pink curves represent the absorbance calculated for the individual monomer, dimer, and tetramer species, respectively. The data can be fit with a Kd of dimerization of 9.7 ± 3.7 mm.
DISCUSSION
Canonical sensor histidine kinases detect specific chemicals from outside the cell and signal through the plasma membrane to downstream response regulators. Although there is substantial conservation in cytoplasmic portions of such two-component systems, the external sensor domains are diverse in sequence, and mechanisms for transmembrane signaling remain obscure.
Only a few sensor domain structures have been determined to date, but among these PhoQ, DcuS, CitA, and DctB all adopt the α/β PDC sensor fold. PDC domains are completely distinct from the four-helical bundles of sensor domains from Tar (37) and NarX.3 DcuS and CitA sensor domains belong to the same sequence family, and their structural similarity was expected, but the structural kinship of PhoQ, DcuS and DctB sensors could not be detected from sequences. The DctB family is most remarkable, having sensor domains approximately twice the size of DcuS/CitA sensors, apparently derived from an evolutionary duplication-insertion event that gave rise to similar subdomains arranged in tandem. It seems that the PDC fold might be prevalent among sensor domains of histidine kinases, despite large sequence variations within the superfamily.
Biochemical and genetic studies of DcuS and DctB have shown specificity to C4-dicarboxylates (7, 11), and activation of the DcuS histidine kinase sensor by l-malate has been characterized (5). We purified DcuS in the presence of 5 mm malate and crystallized the complex in 0.2 m citrate, but we find malate specifically bound in the structure to the exclusion of citrate. Although the physiological ligand for V. cholerae DctB was previously unclear, studies have shown that rhizobial DctB is responsive to succinate (5, 7, 11). The co-purification and subsequent co-crystallization of the V. cholerae DctB sensor domain with succinate is undeniable evidence for its specificity. Succinate is a natural metabolite in cells and was not added to any of the buffers used in protein purification or crystallization. It was preferentially selected from other C4-dicarboxylates such as fumarate, malate, or aspartate, which are also present in E. coli. Although the dissociation constant for succinate binding has never been reported, the binding must be reversible for proper biological function of the receptor. Succinate binds to the distal subdomain of DctB in a manner like that of malate binding to DcuS or citrate binding to CitA (35). Many of the succinate-binding residues in DctB are conserved within the DctB family, and a small subset of these are conserved in the DcuS/CitA family as well. The direct binding of small molecule ligands to the sensor domains of DcuS and DctB contrasts with that of the LuxPQ quorum-sensing complex (32), where the signaling ligand is bound to the periplasmic receptor LuxP, which in turn is bound to the LuxQ periplasmic sensor domain. Although we find calcium bound to two sites in the DctB structure, both mediate lattice contacts, and there is no indication of a requirement for calcium in DctB function.
Sensor histidine kinases exist as homodimers on the cell membrane (4), and signal transduction involves histidine phosphorylation in trans through these dimers upon ligand binding. Although intact sensor domains are in a dimeric environment, intrinsic associations of the isolated domains are often weak. Dimers of sensor domains have been characterized in crystal structures of Tar (37) and PhoQ (28), and we observe meaningful dimers in the DcuS structure as well; but we also show by analytical ultracentrifugation that this DcuS association is very weak (Kd = 9.7 ± 3.7 mm), albeit sufficient for appropriate dimerization in the crystal (44 mm protein concentration). We do not see relevant dimers in our DctB crystals, even though intramolecular complementation studies indicate that DctB is active as a dimer (12). Weak intrinsic affinity for self-association by isolated sensor domains is as expected for membrane-tethered protein domains, which can have up to a 106-fold lower likelihood for self-association when freed from membrane localization (38, 39). Ligand binding seems to occur independently of receptor dimerization in CitA (27), and this also seems likely for DcuS and DctB.
We believe that the dimer observed in the DcuS crystal lattice is representative of the physiological dimer. Just as in the PhoQ dimer, which we have shown by mutational analysis to be a functionally relevant interface (28), protomers of the DcuS dimer are properly oriented for connecting its N and C termini to the transmembrane four-helix bundle dimer. The DcuS dimer is also structurally similar to the molybdate-free CitA dimer (35), which is also believed to be in a physiologically relevant state. Other putative sensor domain dimers (27, 36) do not relate to this model. Because isolated PDC sensor domains have very low intrinsic affinity for dimerization (millimolar level Kd values; see Refs. 27, 40 and this work), it is not surprising that lattice contacts can compete effectively with dimerization interfaces.
Ligand binding to the sensor domain must in some manner elicit changes for activation of the sensor histidine kinase. How this might happen in the case of these C4-dicarboxylate sensors is not clear, but given the envelopment of ligands by protein loops in DcuS and DctB (Fig. 4), ligand-dependent conformational changes seem inevitable. We do find substantial conformational differences when we compare our malate-bound structure with the solution structure of DcuS solved in presumably the apo state (9). It is difficult, however, to distinguish changes that truly result from ligand binding from differences due to the technique used to solve the structure. Because ligand binding is centered over β-strand S5 which then leads to the membrane through helix H6 of DcuS, a mode for signal transmission is suggested. Whether conformational change caused by ligand binding induces a piston-sliding motion between the N- and C-terminal helices, as suggested in structural studies of Tar (37) and NarX,3 has yet to be determined.
In DctB the mode for communication from the ligand-binding site to the transmembrane domain is complicated by the presence of the proximal subdomain, which separates the distal ligand-binding site from the transmembrane helices. The function of this proximal subdomain is unknown, but it might be a site for direct interaction with DctA, which is thought to modulate substrate specificity and signaling characteristics of DctB (7, 11). The indole of Trp-259 projects out from the S8 to S9 loop roughly at the level where putative transmembrane helices enter the membrane, with which it may interact.
Supplementary Material
Acknowledgments
We thank Craig Ogata, Randy Abramowitz, and Alberto Marina for help with synchrotron data collection. We also thank Rich Olson of the Gouaux Lab at Columbia University and Qing Fan for aid in use of the Beckman XLI Analytical Ultracentrifuge (supported by a shared National Institutes of Health Grant S10 RR12848 (instrumentation)) and with use of data analysis software.
The atomic coordinates and structure factors (codes 3BY8 and 3BY9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant GM34102, and it made use of Beamline X4A of the National Synchrotron Light Source at Brookhaven National Laboratory (Department of Energy facility), which is supported by the New York Structural Biology Center. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: IPTG, isopropyl β-d-thiogalactopyranoside; r.m.s.d., root-mean-square deviation; SeMet, selenomethionyl; PEG MME, polyethylene glycol monoethyl ether.
J. Cheung and W. A. Hendrickson, unpublished data.
References
- 1.West, A. H., and Stock, A. M. (2001) Trends Biochem. Sci. 26 369-376 [DOI] [PubMed] [Google Scholar]
- 2.Robinson, V. L., Buckler, D. R., and Stock, A. M. (2000) Nat. Struct. Biol. 7 626-633 [DOI] [PubMed] [Google Scholar]
- 3.Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000) Annu. Rev. Biochem. 69 183-215 [DOI] [PubMed] [Google Scholar]
- 4.Wolanin, P. M., Thomason, P. A., and Stock, J. B. (2002) Genome Biology 3 3013.1-3018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zientz, E., Bongaerts, J., and Unden, G. (1998) J. Bacteriol. 180 5421-5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golby, P., Davies, S., Kelly, D. J., Guest, J. R., and Andrews, S. C. (1999) J. Bacteriol. 181 1238-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janausch, I. G., Zientz, E., Tran, Q. H., Kroger, A., and Unden, G. (2002) Biochim. Biophys. Acta 1553 39-56 [DOI] [PubMed] [Google Scholar]
- 8.Janausch, I. G., Garcia-Moreno, I., and Unden, G. (2002) J. Biol. Chem. 277 39809-39814 [DOI] [PubMed] [Google Scholar]
- 9.Pappalardo, L., Janausch, I. G., Vijayan, V., Zientz, E., Junker, J., Peti, W., Zweckstetter, M., Unden, G., and Griesinger, C. (2003) J. Biol. Chem. 278 39185-39188 [DOI] [PubMed] [Google Scholar]
- 10.Giblin, L., Boesten, B., Turk, S., Hooykaas, P., and O'Gara, F. (1995) FEMS Microbiol. Lett. 126 25-30 [DOI] [PubMed] [Google Scholar]
- 11.Reid, C. J., and Poole, P. S. (1998) J. Bacteriol. 180 2660-2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giblin, L., Archdeacon, J., and O'Gara, F. (1996) FEMS Microbiol. Lett. 139 19-25 [DOI] [PubMed] [Google Scholar]
- 13.Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., Martin, M. J., Michoud, K., O'Donovan, C., Phan, I., Pilbout, S., and Schneider, M. (2003) Nucleic Acids Res. 31 365-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight, S. D. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 42-47 [DOI] [PubMed] [Google Scholar]
- 17.Bailey, S. (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 18.de la Fortelle, E., and Bricogne, G. (1997) Methods Enzymol. 276 472-494 [DOI] [PubMed] [Google Scholar]
- 19.Abrahams, J. P., and Leslie, A. G. W. (1996) Acta Crystallogr. Sect. D Biol. Crystallogr. 52 30-42 [DOI] [PubMed] [Google Scholar]
- 20.Cowtan, K. (1994) CCP4/ESF-EACBM Newsletter on Protein Crystallography 31 34-38 [Google Scholar]
- 21.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 23.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 24.Minton, A. P., and Lewis, M. S. (1981) Biophys. Chem. 14 317-324 [DOI] [PubMed] [Google Scholar]
- 25.Johnson, M. L., Correia, J. J., Yphantis, D. A., and Halvorson, H. R. (1981) Biophys. J. 36 575-588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohn, E. J., and Edsall, J. T. (1943) in Proteins, Amino Acids, and Peptides (Cohn, E. J., and Edsall, J. T., eds) pp. 370-381, Reinhold Publishing Corp., New York
- 27.Reinelt, S., Hofmann, E., Gerharz, T., Bott, M., and Madden, D. R. (2003) J. Biol. Chem. 278 39189-39196 [DOI] [PubMed] [Google Scholar]
- 28.Cheung, J., Bingman, C. A., Reyngold, M., Hendrickson, W. A., and Waldburger, C. D. (2008) J. Biol. Chem. 283 13762-13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm, L., and Sander, C. (1993) J. Mol. Biol. 233 123-138 [DOI] [PubMed] [Google Scholar]
- 30.Cedergrenzeppezauer, E. S., Goonesekere, N. C. W., Rozycki, M. D., Myslik, J. C., Dauter, Z., Lindberg, U., and Schutt, C. E. (1994) J. Mol. Biol. 240 459-475 [DOI] [PubMed] [Google Scholar]
- 31.Neiditch, M. B., Federle, M. J., Miller, S. T., Bassler, B. L., and Hughson, F. M. (2005) Mol. Cell 18 507-518 [DOI] [PubMed] [Google Scholar]
- 32.Neiditch, M. B., Federle, M. J., Pompeani, A. J., Kelly, R. C., Swem, D. L., Jeffrey, P. D., Bassler, B. L., and Hughson, F. M. (2006) Cell 126 1095-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch, W., and Sander, C. (1983) Biopolymers 22 2577-2637 [DOI] [PubMed] [Google Scholar]
- 34.Lawrence, M. C., and Colman, P. M. (1993) J. Mol. Biol. 234 946-950 [DOI] [PubMed] [Google Scholar]
- 35.Sevvana, M., Vijayan, V., Zweckstetter, M., Reinelt, S., Madden, D. R., Herbst-Irmer, R., Sheldrick, G. M., Bott, M., Griesinger, C., and Becker, S. (2008) J. Mol. Biol. 377 512-523 [DOI] [PubMed] [Google Scholar]
- 36.Cho, U. S., Bader, M. W., Amaya, M. F., Daley, M. E., Klevit, R. E., Miller, S. I., and Xu, W. Q. (2006) J. Mol. Biol. 356 1193-1206 [DOI] [PubMed] [Google Scholar]
- 37.Yeh, J. I., Biemann, H. P., Prive, G. G., Pandit, J., Koshland, D. E., and Kim, S. H. (1996) J. Mol. Biol. 262 186-201 [DOI] [PubMed] [Google Scholar]
- 38.Metzger, H. (1992) J. Immunol. 149 1477-1487 [PubMed] [Google Scholar]
- 39.Grasberger, B., Minton, A. P., Delisi, C., and Metzger, H. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 6258-6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waldburger, C. D., and Sauer, R. T. (1996) J. Biol. Chem. 271 26630-26636 [DOI] [PubMed] [Google Scholar]
- 41.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24 946-950 [Google Scholar]
- 42.Esnouf, R. M. (1997) J. Mol. Graph. Model. 15 132-134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.