Abstract
Appropriate glycoprotein O-glycosylation is essential for normal development and tissue function in multicellular organisms. To comprehensively assess the developmental and functional impact of altered O-glycosylation, we have extensively analyzed the non-glycosaminoglycan, O-linked glycans expressed in Drosophila embryos. Through multidimensional mass spectrometric analysis of glycans released from glycoproteins by β-elimination, we detected novel as well as previously reported O-glycans that exhibit developmentally modulated expression. The core 1 mucin-type disaccharide (Galβ1-3GalNAc) is the predominant glycan in the total profile. HexNAcitol, hexitol, xylosylated hexitol, and branching extension of core 1 with HexNAc (to generate core 2 glycans) were also evident following release and reduction. After Galβ1-3GalNAc, the next most prevalent glycans were a mixture of novel, isobaric, linear, and branched forms of a glucuronyl core 1 disaccharide. Other less prevalent structures were also extended with HexA, including an O-fucose glycan. Although the expected disaccharide product of the Fringe glycosyltransferase, (GlcNAcβ1-3)fucitol, was not detectable in whole embryos, mass spectrometry fragmentation and exoglycosidase sensitivity defined a novel glucuronyl trisaccharide as GlcNAcβ1-3(GlcAβ1-4)fucitol. Consistent with the spatial distribution of the Fringe function, the GlcA-extended form of the Fringe product was enriched in the dorsal portion of the wing imaginal disc. Furthermore, loss of Fringe activity reduced the prevalence of the O-Fuc trisaccharide. Therefore, O-Fuc glycans necessary for the modulation of important signaling events in Drosophila are, as in vertebrates, substrates for extension beyond the addition of a single HexNAc.
The O-linked glycans of metazoan organisms include any single monosaccharide or oligosaccharide glycosidically linked to the side chain hydroxyl of Ser/Thr residues. The diversity of this family of glycans begins with the first monosaccharide residue linked to the protein, which may be O-Man, O-Fuc, O-GlcNAc, O-Glc, O-GalNAc, or O-Xyl. In the case of O-Xyl, subsequent elaboration generates the highly diverse family of glycosaminoglycans, which play crucial roles in the modulation of morphogen and growth factor signaling and contribute specific structural characteristics to the extracellular matrix (1-3). In the case of O-GalNAc, elongation results in the generation of mucin type O-glycan cores, which can be linear or branched (core types 1-8) depending on the portfolio of glycosyltransferases expressed by a given cell or tissue (4, 5). Specific enzymes also extend O-Man, O-Fuc, and O-Glc, generating structures with unique functions and tissue distributions (6). Genetic, biochemical, and developmental studies in vertebrate and nonvertebrate species have repeatedly demonstrated the importance of O-linked glycans for normal tissue development and maintenance of mature tissue function (7-13).
O-Linked glycans function as recognition markers, as adhesive ligands, and as modulators of cell-cell signaling. In some cases, precise glycan structures have been assigned to specific functions. For example, a set of sulfated, fucosylated, branched glycans initiated by O-GalNAc, collectively known as 6-sulfo-sLex structures, serve as recognition and adhesive molecules for immune cells (14). The regulated expression of a specific branching enzyme (core 2 GlcNAc transferase) drives the synthesis of 6-sulfo-sLex glycans on appropriate protein backbones in response to inflammatory signals, imparting new adhesive specificities to endothelial and leukocyte surfaces (15, 16). The clinical importance of appropriate protein O-glycosylation is further demonstrated by the severity of a family of human congenital muscular dystrophies (17, 18). These disorders affect discrete steps in the biosynthesis of extended O-Man glycans, which modulate adhesive interactions between muscle α-dystroglycan and laminin in the extracellular matrix (19-21). Impaired α-dystroglycan O-glycosylation impacts muscle stability, leading to weakening and progressive degeneration.
In addition to their direct participation in adhesion and cell recognition, O-glycans function as both permissive and restrictive modulators of cell signaling during normal development and tissue morphogenesis. Members of the polypeptide GalNAc transferase family (ppGalNAcTs)2 initiate the synthesis of mucin-type, O-linked glycans in all animal species. Loss of a single ppGalNAcT in Drosophila (pgant35A) results in late embryonic, larval, and pupal lethality accompanied by defective epithelial polarization (12, 22). The epithelial phenotype is characterized by insufficient delivery of apical determinants, leading to the loss of tracheal integrity and indicating a role for O-glycans in directing or stabilizing polarized protein distributions. Cellular polarization is essential for morphogenesis and is driven in response to various environmental cues, including morphogens and cell-cell signaling. Cell fate choices are among the earliest developmental decisions driven by these cues and frequently depend on signaling that is initiated by the interaction of Notch receptors with ligands of the Delta or Serrate/Jagged family (23, 24). Appropriate O-glycosylation of Notch by the addition of O-Fuc to epidermal growth factor repeats in the Notch extracellular domain is permissive for signaling. Loss of the fucosylating enzyme in Drosophila (OFUT-1) phenocopies Notch with respect to aberrant wing development and neurogenesis (25, 26). O-Glycosylation can also restrict Notch signaling. Elongation of O-Fuc residues by the addition of β3-linked GlcNAc, catalyzed by the Fringe glycosyltransferase (Fng), generates a Notch glycoform that displays ligand preference for Delta over Serrate/Jagged (27-30). By modifying the ligand preference of the Notch receptor, distinct cell fates are induced within tissue compartments through differential ligand expression and spatially restricted receptor glycosylation.
In mammals, the disaccharide produced by Fringe is extended to a tetrasaccharide, NeuAcα2-3/6Galβ1-4GlcNAcβ1-3Fuc, in which the Gal residue is necessary for Fringe-dependent modulation of Notch signaling (30). Similarly extended O-Fuc structures have not been identified in Drosophila tissues or in tissue culture cells derived from insects. More generally, the glycosylation status of endogenously expressed Notch protein has not been directly determined in wild-type or in any mutant Drosophila tissues, nor has the full complement of O-linked glycans been characterized for any Drosophila tissue or for any single glycoprotein extracted from a Drosophila tissue. The absence of such an essential data base complicates efforts to assess the relative importance of O-glycan structures for signaling, development, or normal tissue function.
Previous characterizations of O-linked glycans expressed in Drosophila tissues have detected the presence of O-linked GalNAc (Tn-antigen) and the core 1 disaccharide (T-antigen) by direct chemical identification and by specific lectin and antibody probes (31-39). Various dipteran and lepidopteran cell lines have also yielded valuable clues regarding the capacity of insects to synthesize more diverse O-linked glycans (40-45). Here, we report in-depth characterization of the O-linked glycome of the Drosophila embryo, coupled with analysis of tissue- and stage-specific expression in wild-type and relevant mutant backgrounds, to provide an enriched context for assessing specific glycan functions.
EXPERIMENTAL PROCEDURES
Materials—Sodium hydroxide (50%) was obtained from Fisher Scientific and trifluoroacetic acid from Pierce. AG-50W-X8 cation exchange resin (H+) was purchased from Bio-Rad. Monosaccharide, monosaccharide alditol, and core 1 disaccharide standards were purchased from Ferro Pfanstiehl Laboratories, Inc. (Waukegan, WI) and from Sigma-Aldrich. Exoglycosidases and all other reagents and chemicals were purchased from Sigma-Aldrich. Standard media for propagation and growth of Drosophila were obtained from Genesee Scientific (San Diego, CA) or were prepared from commercially available agar and juices (apple or grape). Drosophila stocks were obtained from the Bloomington Drosophila Stock Center at Indiana University.
Preparation of Fly Embryo Powder—Fly embryo powder was prepared as described previously (46). Briefly, embryos were harvested from standard apple juice agar plates, dechorionated in 2.5% (w/v) sodium hypochlorite solution, transferred to a collection sieve and rinsed extensively with distilled water. Dechorionated, washed embryos were stored frozen until further use. To prepare fly embryo powder, frozen embryos were homogenized in ice-cold 50% methanol and delipidated by adjusting the solvent composition to 4:8:3 (chloroform:methanol:water (v/v/v)). Proteins were precipitated by centrifugation, and the resulting pellet was re-extracted with fresh solvent. The first lipid extraction and the pellet re-extraction were combined for each sample, dried under nitrogen, and stored for subsequent fatty acid analysis. Precipitated protein pellets were washed with ice-cold 20% aqueous acetone (v/v), which was found to reduce the prevalence of contaminating hexose oligomers, and the washed pellet was dried at 37 °C under a gentle nitrogen stream.
Release of Oligosaccharides from Fly Embryo Powder by β-Elimination—Conditions for reductive β-elimination were optimized for time, temperature, sodium hydroxide, and reducing agent concentrations using fly embryo powder as the starting material (47). Maximal recovery of the four major O-linked glycans of the Drosophila embryo was achieved by incubating with 100 mm sodium hydroxide, 1 m sodium borohydride for 18 h at 45 °C (supplemental Fig. 1). Other tested concentrations of sodium hydroxide and sodium borohydride gave similar glycan profiles but lower total glycan yield. For all of the reductive β-eliminations reported here, fly embryo powder (3-4 mg dry weight) was resuspended in 500 μl of 100 mm sodium hydroxide containing 1 m sodium borohydride and incubated for 18 h at 45 °C in a glass tube sealed with a Teflon-lined screw top. After incubation, the reaction mixture was neutralized with 10% acetic acid on ice and desalted by loading onto an AG-50W-X8 (H+ form) column (1 ml bed volume). Released oligosaccharide alditols were then eluted with 3 bed volumes of 5% acetic acid and lyophilized to dryness. Borate was removed as an azeotrope with methanol by adding 0.5 ml of 10% acetic acid in methanol, drying under a nitrogen stream at 37 °C, and repeating four additional times. Reduced recovery of a subset of glycans was noted upon excessive drying from methanol/acetic acid. Therefore, attention to the drying steps is required so that the drying time is limited to the minimum necessary to remove solvent for each wash. To remove residual peptide and reagent contaminants, the dried material was resuspended in 500 μl of 5% acetic acid and loaded onto a previously washed and equilibrated C18 cartridge column (100 mg, J. T. Baker Co., catalog No. 7020-1). Prior to use, C18 cartridges were washed with at least 3 ml of 100% acetonitrile and pre-equilibrated with at least 3 ml of 5% acetic acid. Released oligosaccharide alditols were recovered by collecting the column run-through and an additional 2 ml of wash with 5% acetic acid. The run-through and wash were combined and evaporated to dryness.
O-Linked glycans were also released from fly embryo powder by β-elimination under nonreductive conditions based on the method of Novotny (48). For release of O-linked glycans with an intact reducing terminal, fly embryo powder was reconstituted in 500 μl of 28% aqueous ammonium hydroxide saturated with ammonium carbonate. An additional 100 mg of solid ammonium carbonate was added, and the release reaction was incubated for 40 h at 60 °C in a glass tube sealed with a Teflon-lined screw top. At the end of the incubation, volatile reagent and salts were removed by vacuum centrifugation, and the dried material was washed 3-5 times by resuspension in 500 μl of water and redrying. Released oligosaccharides were purified on a C18 cartridge column as described for reductive β-elimination. After drying the combined C18 run-through and wash fractions, glycosylamines were reduced to free reducing termini by incubation in 50 μl of 0.5 m boric acid at 37 °C for 1 h. The reaction mixture was dried, and borate was removed by adding 0.5 ml of 10% acetic acid in methanol, drying under a nitrogen stream at 37 °C, and repeating four additional times.
Permethylation and Analysis of Glycans by Nanospray Ionization Mass Spectrometry (NSI-MSn)—Released oligosaccharide mixtures were permethylated to enhance sensitivity and structural determination by mass spectrometry (49). For mass analysis by NSI-MSn, permethylated glycans were dissolved in 1 mm sodium hydroxide in 50% methanol and infused directly into a linear ion trap mass spectrometer (LTQ, Thermo Finnigan) using a nanoelectrospray source at a syringe flow rate of 0.40 to 0.60 μl/min. The capillary temperature was set to between 210 and 240 °C as dictated by daily tuning to oligosaccharide standards. MS analysis was performed in positive ion mode. Most permethylated oligosaccharides were identified as singly charged species by NSI-MS. For fragmentation by CID in MS/MS and MSn modes, 25-35% collision energy was applied. Spectra generated in support of the reported structures are available upon request from the corresponding author. The systematic nomenclature of Domon and Costello was used to guide the depiction of fragmentation derived from MSn (50). Glycan nomenclature and the representation of oligosaccharides are in accordance with the guidelines proposed by the Consortium for Functional Glycomics.
As described previously, the total ion mapping (TIM) functionality of the Xcalibur software package (version 2.0) was utilized to detect and quantify the prevalence of individual glycans in the total glycan profile.3 Through TIM, automated MS and MS/MS spectra (at 25-35% collision energy) were obtained in collection windows that were 2.8 mass units in width. Five scans, each 150 ms in duration, were averaged for each collection window. The m/z range from 200 to 2000 was scanned in successive 2.8 mass unit windows with a window-to-window overlap of 0.8 mass units. Although it is possible to obtain MS spectra up to m/z = 4000 on the LTQ, software limitations currently disallow TIM beyond m/z = 2000. However, manual MS/MS on ions detected in the range of m/z = 2000-4000 did not detect additional O-linked glycans. Peaks in TIM scans were quantified if they were 2-fold or greater above background. Glycan prevalence was calculated as “% total profile” where the total profile was taken as the sum of the peak intensities for all quantified glycans. A polyhexose detected in the starting material gave rise to a ladder of repeating Δm/z = 204 fragments that overlapped with some O-linked glycan peaks detected by TIM. For peaks containing this contaminant, the MS signal intensity was apportioned to the relevant glycan based on the ratio of the first product ion of the glycan to the first contaminant ion measured in MS/MS.
Exoglycosidase Digestion—The O-linked glycans prepared from 3 mg of fly embryo powder by reductive β-elimination were incubated at 37 °C with 10 units of β-glucuronidase (Helix pomatia) in 100 μl of 50 mm citrate buffer, pH 4.5 (51). After 20 h of incubation, the reaction mixture was evaporated to dryness, resuspended in 250 μl of 5% acetic acid, and loaded onto a C18 cartridge that had been washed previously with 100% acetonitrile and pre-equilibrated with 5% acetic acid. Released GlcA and O-linked oligosaccharide alditols were recovered by collecting the column run-through and an additional 3 column volumes of wash with 5% acetic acid, which were combined and evaporated to dryness. GlcA was detected in the C18-cleaned and concentrated enzyme reaction by high pH anion exchange chromatography-PAD with in-line amino and borate traps (52, 53). Briefly, aliquots were injected onto a Dionex CarboPac column (PA-10 for GlcA and MA-1 for alditols) and eluted with a linear gradient of sodium acetate (in 100 mm sodium hydroxide) from 100 to 400 mm over 25 min at a flow rate of 0.5 ml/min. Following permethylation, O-linked oligosaccharides in the enzyme reaction were analyzed by NSI-MSn.
Characterization of O-Linked Monosaccharides by High Pressure Liquid Chromatography—Monosaccharides released from fly embryo powder as monosaccharide alditols by reductive β-elimination were separated on a Dionex CarboPac MA-1 column (250 × 4 mm) and detected by HPEAC-PAD. Monosaccharide alditols were eluted at a flow rate of 0.4 ml/min with 200 mm sodium hydroxide for 5 min followed by a linear gradient to 800 mm sodium hydroxide from 5.1 to 40 min. Peaks were identified and quantified relative to monosaccharide alditol standards.
Identification of Reducing Terminal Monosaccharides and Monosaccharide Composition of O-Linked Glycans—Following release from fly embryo powder by reductive β-elimination, O-linked oligosaccharide alditols were hydrolyzed for 4 h with 100 μl of 2 m trifluoroacetic acid at 100 °C (9). After removing the acid by evaporation under a nitrogen stream, the hydrolyzates, containing both monosaccharide alditols (original reducing terminus) and monosaccharide aldoses (internal and nonreducing terminal residues), were acetylated with 0.5 ml of pyridine:acetic anhydride (1:1 (v/v)) at 100 °C for 10 min (54). The resulting monosaccharide aldose and alditol acetates were analyzed by gas chromatographymass spectrometry (GC-MS; see below for conditions). Alternatively, for characterizing total monosaccharide composition, the mixture of aldoses and alditols resulting from acid hydrolysis were reduced with sodium borohydride before acetylation and GC-MS analysis. Peaks were identified and quantified relative to acetylated standards.
Monosaccharide Linkage Analysis—To determine monosaccharide linkage positions, O-linked glycans released by nonreductive β-elimination were partially methylated with sodium hydroxide and methyl iodide in dimethyl sulfoxide (54). The permethylated O-glycans were then hydrolyzed in 200 μl of HCl: water:acetic acid (0.5:1.5:8 (v/v/v)) at 80 °C for 18 h. The partially methylated monosaccharides were reduced with 0.5 ml of 2% NaBD4 in 5 mm sodium hydroxide at room temperature for 18 h. Following borate removal by drying from methanol, the partially methylated alditols were acetylated by adding 0.5 ml of pyridine:acetic anhydride (1:1 (v/v)) and incubation at 100 °C for 10 min. The resulting partially methylated alditol acetates were analyzed by GC-MS in comparison with established standards (see below for GC-MS conditions). To confirm linkage for the putative core 1 disaccharide and substitution assignments for other Drosophila O-glycans, intact oligosaccharide alditols released from 20 mg of fly embryo powder were permethylated and analyzed by GC-MS (55, 56).
Total Protein and Fatty Acid Analysis—Sufficient fly embryo powder could be obtained from wild-type embryos to allow total protein measurement by the BCA assay.3 For wing discs and hand-sorted mutant embryos (fng13 genotypes), fly powder was limiting. Therefore, total fatty acids were quantified in these samples to provide a basis for comparing relative glycan prevalences. Aliquots of the lipid extracts prepared during tissue delipidation were dried in a thick glass test tube, methanolyzed by resuspension in 200 μl of 1 m anhydrous methanolic HCl, and incubated at 80 °C for 18 h (57). The resulting fatty acid methyl esters were extracted three times with 200 μl of n-hexane and evaporated to dryness. Fatty acid components were identified and quantified by GC-MS (see below for GC-MS conditions).
Gas Chromatography-Mass Spectrometry Conditions—Electron impact ionization mass spectra were obtained with a Hewlett Packard 5890 gas chromatograph interfaced to a 5970 mass detector. Separations were performed on a 0.32 mm × 30 m capillary column (EC1, Alltech Associates, Deerfield, IL) under the following conditions: initial oven temperature 80 °C (hold 2.0 min), then increase temperature to 180 °C (at 20 °C/min), then increase temperature to 240 °C (at 4 °C/min); interface temperature, 250 °C (54). The same conditions were used for fatty acid, oligosaccharide, monosaccharide, alditol, and monosaccharide linkage analysis.
RESULTS
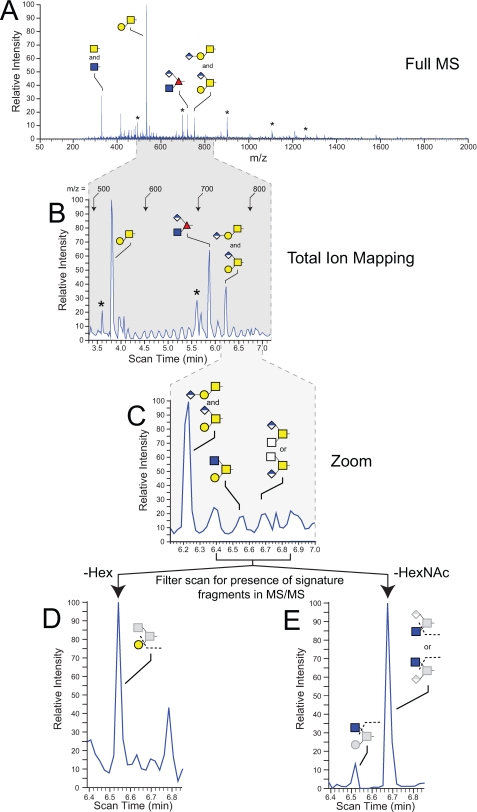
O-Linked Glycan Profile of the Drosophila Embryo Is Dominated by a Single Disaccharide—O-Linked glycans were released from embryo glycoproteins by β-elimination and analyzed by NSI-MSn following permethylation. Full MS scans (from m/z = 50-2000) detected four sodiated parent ions (Fig. 1A). These four major ions were assigned to specific glycan structures by subsequent MSn fragmentation, exoglycosidase sensitivity, HPAEC-PAD, and GC-MS analysis of monosaccharides, monosaccharide alditols, and oligosaccharide alditols. The least complex glycan was detected at m/z = 330 as a single N-acetylhexosamine, a mixture of both GalNAc-O- (known also as Tn-antigen) and GlcNAc-O-. HPAEC-PAD analysis of β-eliminated, reduced monosaccharide alditols indicates that GalNAc-ol3 is 20-fold higher than GlcNAc-ol (supplemental Fig. 2). The glycan of highest prevalence in the Drosophila embryo is recognized in many animal species as the core 1 disaccharide (T-antigen, Galβ1-3GalNAc-O-) at m/z = 534 (supplemental Fig. 3). Two other ion peaks are easily detectable in full MS scans at m/z = 722 and 752 (Fig. 1, A and B). Each of these peaks and their associated MS/MS profiles indicate the presence of glycans bearing hexuronic acid and are discussed in further detail below.
FIGURE 1.
Detection and identification of Drosophila embryo O-linked glycans by NSI-MS and TIM. Glycans released from fly embryo powder by reductive β-elimination were permethylated and analyzed. A, full MS mode demonstrates the predominance of O-linked HexNAc and four more complex structures. Glycans are detected as singly charged species, and structural assignments are based on fragmentation (MSn), exoglycosidase digestion, GC-MS linkage analysis, and monosaccharide composition. In this and other figures (Figs. 5 and 6), asterisks denote ions assigned to an oligomeric hexose ladder detected as a common, but variable, contaminant of fly embryo powder. B, automated acquisition of MS and MS/MS spectra by TIM enhances detection and quantification of minor glycans. In this TIM scan, the mass range covered by scan times from 3.3 to 7.0 min contains the most prevalent O-linked glycans. C, expanded view of a subregion of the TIM scan shows relatively unimpressive evidence for the presence of the indicated minor glycans, a core 2 trisaccharide (Galβ1-3(GlcNAcβ1-4)GalNAc) and a hexuronylated di-HexNAc. In the bottommost panels (D and E), the indicated region of the TIM profile was filtered for the presence of predictive fragment ions in the MS/MS spectra. For the region expanded in D, filtering for the loss of a nonreducing terminal Hex residue supports the presence of the core 2 trisaccharide at m/z = 780 (scan time 6.52 min). For the region expanded in E, filtering for the loss of a nonreducing terminal HexNAc detects both the core 2 trisaccharide revealed in D and also a signal of greater relative intensity arising from the hexuronylated di-HexNAc at m/z = 794 (scan time 6.67 min).
To characterize the diversity of O-linked glycans in the Drosophila embryo, we utilized the TIM functionality of the Xcalibur software (version 2.0) as an unbiased approach to detect minor structures. Unlike conventional data-dependent MS/MS acquisition, in which predefined losses trigger further fragmentation to identify interesting parent ions, MS/MS spectra are acquired continuously across a selected mass range in TIM scans. Subsequent inspection of all MS/MS spectra for any signature of glycan fragmentation provides confidence that unexpected structures are not ignored. The application of TIM analysis to Drosophila glycans has been described previously.3 Briefly, the peaks in TIM scans (Fig. 1B) provide landmarks behind which lie MS/MS spectra that indicate whether one-dimensional MS signals arise from glycan or from non-glycan contaminants. TIM also generates MS/MS spectra in regions where the MS signal is insufficient to generate a discernible peak (Fig. 1C). Examination of these spectra frequently reveals fragment ions diagnostic for glycan structures that fall below the quantification threshold (Fig. 1D). The MS/MS fragmentation profiles provide significant clues for assigning structures and also focus subsequent MSn analyses (usually MS3-MS5) by manual acquisition to m/z values of interest.
Recovery of O-Linked Glycans from the Drosophila Embryo—The recovery of O-linked glycans from complex protein mixtures by reductive β-elimination reflects the competition between efficient release from glycoprotein and the need for rapid reduction before base-catalyzed peeling reactions drive glycan degradation (47). Empirically optimized conditions (see “Experimental Procedures” and supplemental Fig. 1) were employed to assess overall recovery of glycan from Drosophila embryo powder. Previous analysis demonstrated that GalNAc is essentially undetectable as a component of Drosophila N-linked glycans (36).3 Therefore, we compared GalN-ol prepared directly from delipidated fly embryo powder by acid hydrolysis (2 m trifluoroacetic acid, 100 °C, 4 h) and subsequent reduction (sodium borohydride) to GalN-ol prepared by acid hydrolysis of oligosaccharides released by reductive β-elimination. Quantification by HPAEC-PAD relative to standards demonstrated that 72% of the expected GalN-ol, based on the reduced acid hydrolysates of fly embryo powder, was recovered in O-linked glycans released by reductive β-elimination.
We also assessed analytic recovery and glycan quantity by adding external spikes of core 1 disaccharide standard into Drosophila embryo powder before β-elimination or into O-linked glycan preparations prior to MS analysis. Spike recovery was 115% of that expected when the standard disaccharide was added to embryo powder. Recovery through permethylation and MS analysis was assessed in two ways. In one series of experiments, recovery was determined by adding a known amount of core 1 disaccharide to Drosophila O-linked glycans before permethylation. In another series of experiments, a known amount of core 1 disaccharide previously permethylated with [13C]methyl iodide was mixed together with Drosophila O-linked glycans previously permethylated with [12C]methyl iodide. Peak heights at m/z = 534 (12C-methylated core 1, with or without spike) and at 543 (13C-methylated core 1) were compared with calculate spike recovery, which was determined to be 100 ± 9% (n = 3 independent determinations). Therefore, neither ion suppression nor sample matrix contaminants compromised the sensitivity of the detection method. By referencing the signal intensities for endogenous core 1 disaccharide to standard spikes of core 1 disaccharide and correcting for recovery, the amount of core 1 disaccharide in fly embryo powder is calculated to be 74 pmol/mg dry weight (6.4 pmol/mg wet weight of embryos). Adjusting for the prevalence of the core 1 disaccharide in the wild-type embryo (55% of the total profile in OreR), the content of O-linked glycan is 134 pmol/mg dry weight of fly embryo powder (11.6 pmol/mg wet weight of embryos).
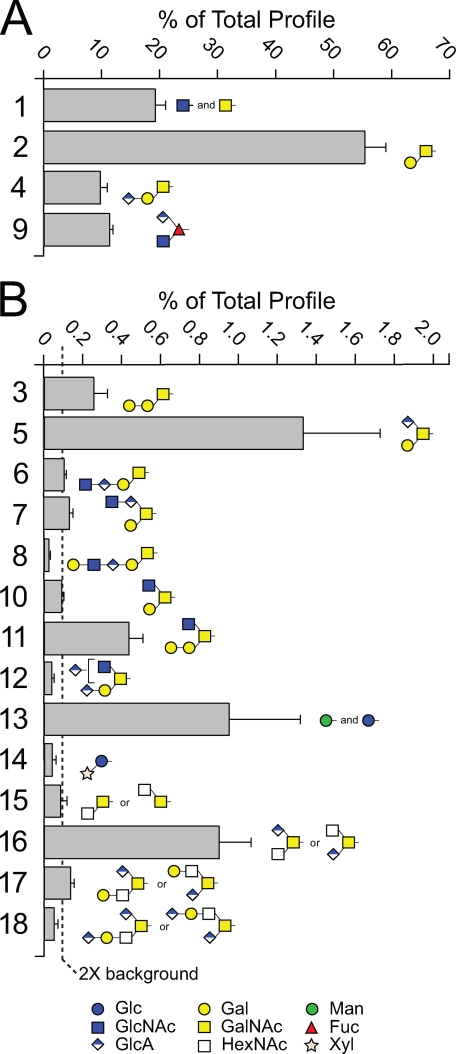
The Less Prevalent O-Linked Glycans of the Drosophila Embryo Include Extended Core 1, Core 2, and Other Structures—TIM scans were examined for MS/MS fragmentation consistent with the presence of glycans within each of the overlapping 2.8-mass unit windows comprising the mass range between m/z = 200 and 2000. The primary fragmentation signatures that we detected as diagnostic for glycan were loss of Hex-ol or HexNAc-ol from the reducing terminus and loss of unsubstituted Hex or HexNAc from a nonreducing terminus. Further fragmentation analyses (MS3-MS4) were performed as needed to determine structures of candidate glycans (supplemental Figs. 3-6). The full diversity of O-linked glycans detected in the Drosophila embryo includes a predominance of core 1-related glycans (Fig. 2, Structures 1-8, and supplemental Table 1), a single O-Fuc glycan (Structure 9), a small pool of core 2 structures (Structures 10-12), and other glycans possessing HexNAc-HexNAc cores (Structures 15-18). Representatives of each core type were detected with HexA extensions, as terminal, internal, or branching residues. Among the less complex O-linked glycans, the single monosaccharides GalNAc-O-/GlcNAc-O-(Structure 1), and Glc-O-/Man-O-(Structure 13), and the Xyl-Glc-O-disaccharide (Structure 14) were also detected (58, 59).
FIGURE 2.
The O-linked glycan profile of wild-type Drosophila embryos. Glycans were released from fly embryo powder by β-elimination. Following permethylation, glycans were identified and quantified by NSI-MSn and TIM. The prevalence of each major glycan (A) and minor glycan (B) is expressed as a percent of the total pool of detected glycans. The dashed line in B indicates the threshold for quantification (0.1% of total profile, which corresponds to 2× background). The structure numbers next to each bar are used consistently throughout all figures and tables to facilitate rapid comparison and identification of the same glycan structure. The data represent the mean ± S.D. for determinations performed on three independent glycan preparations (see also supplemental Table 1). Glycan structures are assigned based on combinations of parent ion mass, MSn fragmentation, exoglycosidase digestion, and GC-MS analysis of permethylated oligosaccharide alditols. The unshaded squares depicted for Structures 15-18 indicate that the identity of the HexNAc residue is not currently assigned.
Structural assignments presented here are based on multiple lines of evidence, including extensive NSI-MSn, composition and linkage analysis by GC-MS of permethylated oligosaccharide alditols or of partially methylated alditol acetates, and exoglycosidase sensitivity. The core 1 disaccharide, Galβ1-3GalNAc, has been identified previously in Drosophila tissues and cultured Drosophila cells; it is released from Drosophila proteins by the enzyme O-glycanase, which has a strict anomeric specificity for Galβ1-3GalNAc, and is recognized by structurally specific lectin and antibody probes (34, 35, 37, 40, 60). Combined with the fragmentation and composition data presented here, the core 1 disaccharide structure can be assigned with confidence as expected. Other less prevalent core 1 type glycans are built from this defined disaccharide based on fragmentation data, compositional analysis, and exoglycosidase digestion. For the single O-Fuc structure detected in Drosophila tissues, fragmentation and exoglycosidase digestion were sufficient to define the order and substitution positions of the monosaccharide constituents (see below).
For glycans of other core types, previously described specificities for animal O-glycan biosynthetic enzymes place important constraints on structural diversity. A core 2 GlcNAc transferase has not yet been identified in Drosophila; the best gene candidate actually possesses Xyl transferase activity (61). Therefore, the glycans depicted as core 2 structures (Structures 10-12) are assigned based on the detection of both Hex and HexNAc residues branching from the reducing terminal GalNAc-ol, consistent with the activity expected for the yet to be identified core 2 GlcNAc transferase. The glycans grouped as other core structures (Structures 15-18) also contain a HexNAc-HexNAc-ol disaccharide but lack the branching hexose and cannot, therefore, be classified as core 2. However, we cannot currently distinguish between other core possibilities (core 3, GlcNAcβ1-3GalNAc; core 5, GalNAcα1-3GalNAc; core 6, GlcNAcβ1-6GalNAc; core 7, GalNAcα1-6GalNAc) that have been described previously in animal tissues (4).
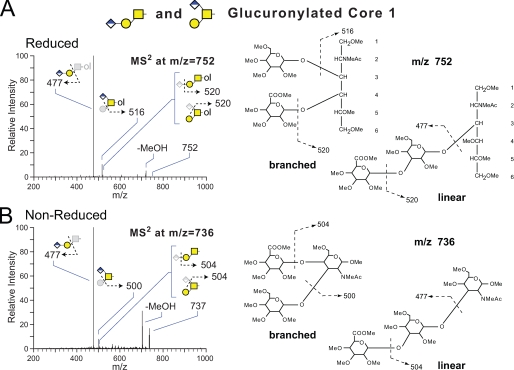
Hexuronic Acid Extension Results in Novel O-Linked Glycans on Multiple Cores—A broad range of major and minor O-linked glycans are detected as both neutral and acidic forms in which hexuronic acid residues impart charge. Two isobaric forms of the core 1 disaccharide extended with hexuronic acid are resolved by NSI-MS2 (Fig. 3). One form is a linear trisaccharide capped with a nonreducing terminal HexA. GC-MS analysis of the permethylated oligosaccharide alditol verified HexA-Hex at the nonreducing terminal as well as a singly substituted HexNAc-ol at the reducing terminal. The other, less prevalent form of the core 1 trisaccharide is branched by the addition of HexA to the 4-position of the reducing terminal GalNAc (62, 63). Although the linear form of the trisaccharide has recently been described on proteins expressed by the Drosophila S2 cell line in culture, the branched core 1 trisaccharide is novel, and neither glycan has been detected previously in embryos (40).
FIGURE 3.
NSI-MSn fragmentation of a mixture of two permethylated, isobaric, core 1 trisaccharides bearing glucuronic acid, extracted from Drosophila embryos. Following release from fly embryo powder by reductive (A) or nonreductive (B) β-elimination, glycans were permethylated and characterized by NSI-MSn. Fragmentation of the parent ions corresponding to permethylated HexA1Hex1HexNAc-ol (m/z = 752) or permethylated HexA1Hex1HexNAc (m/z = 737) demonstrate that both are a mixture of two isobaric O-glycans, one branched and one linear. A, the reduced trisaccharide yields a strong C-ion at m/z = 477, which corresponds to loss of the reducing terminal HexNAc-ol from the linear structure. The Z-ion at m/z = 516 arises from loss of a nonreducing terminal Hex and assigns the HexA residue to a branching position on a minor isomer. B, fragmentation of the nonreduced trisaccharide mixture reveals the same pattern of ions, supporting the predominance of the linear structure, C-ion at m/z = 477, and the presence of a minor branched isomer, Z-ion at m/z = 500 (62, 63). Further fragmentation of the ions at m/z = 477, 500, 504, 516, and 520 by MS3-MS4 are consistent with the indicated structures.
The only O-Fuc glycan detected in the Drosophila embryo (Structure 9) also carries a single HexA residue as a branch from the reducing terminal Fuc (Fig. 4). O-Linked Fuc is known to be modified with β3GlcNAc by Fringe, a specific GlcNAc transferase, but the addition of uronic acid has not previously been described on any O-Fuc glycan. Therefore, this novel glycan structure was extensively characterized to fully assign monosaccharide sequence and substitution positions. GC-MS analysis of the O-Fuc trisaccharide as its permethylated oligosaccharide alditol verified terminal HexNAc and HexA as well as di-substituted deoxy-Hex-ol. Cross-ring and glycosidic fragmentation by NSI-MSn placed the HexNAc substitution at the 3-position and the HexA substitution at the 4-position of the Fuc ring (supplemental Fig. 6). Multiple lines of evidence support the structural assignments for the O-Fuc trisaccahride. The identity of the reducing terminal deoxy-Hex-ol as Fuc is based on the detected levels of Fuc-ol as its alditol acetate by GC-MS following acid hydrolysis (see below). The β3GlcNAc is assigned based on MSn fragmentation and the known substrate specificity of the Fringe enzyme (29, 30). Finally, the novel branching HexA is assigned as β4GlcA based on fragmentation, composition, and exoglycosidase digestion (see below).
FIGURE 4.
NSI-MSn fragmentation of a permethylated, glucuronylated O-Fuc glycan extracted from Drosophila embryos. Following release from fly embryo powder by reductive (A) or nonreductive (B) β-elimination, glycans were permethylated and characterized by NSI-MSn. Fragmentation of the parent ions corresponding to permethylated HexA1HexNAc1deoxy-Hex-ol (m/z = 722) or permethylated HexA1HexNAc1deoxy-Hex (m/z = 706) reveal a branched trisaccharide with a reducing terminal Fuc. A, fragmentation of the reduced trisaccharide yields strong Z-ions at m/z = 445 and 472, which correspond to loss of nonreducing terminal HexNAc and HexA branches, respectively. The major MS/MS ion at m/z = 576 arises from an incompletely defined cross-ring cleavage through the HexA residue (broken line arrow). The same cross-ring cleavage is evident as a very minor fragment in MS2 of the glucuronylated core 1 disaccharide (see Fig. 3) in both reduced (m/z = 606) and nonreduced (m/z = 593) forms, suggesting that glycosidic linkage to a deoxy-Hex imparts stability. Although the cleavage and possible rearrangements associated with this cross-ring event are not resolved, the resulting X-ion fragments in MS3 to produce the expected losses for the predicted structure: a Z-ion at m/z = 472 from loss of the residual HexA, a 0,4X-ion at m/z = 398 from cross-ring cleavage through the HexNAc, and a C-ion at m/z = 298 from liberation of the nonreducing terminal HexNAc branch. B, fragmentation of the nonreduced trisaccharide reveals the same pattern of supporting ions as well as the Y-ion at m/z = 301 in MS3, corresponding to the reducing terminal deoxy-Hex. Further fragmentation (MS4-MS5) identified substitution positions for the HexA and HexNAc termini (supplemental Fig. 6).
Although the O-Fuc trisaccharide accounts for 11.3% of the total O-linked glycan profile in wild-type embryos, its expected biosynthetic precursors, Fuc-ol and GlcNAcβ1-3Fuc-ol, are not present at detectable levels in fly embryo powder. To verify the major contribution of Structure 9 to the total glycan profile, O-linked glycans released from fly embryo powder by reductive β-elimination were subjected to acid hydrolysis. The resulting mixture of free monosaccharides and alditols were treated with borohydride, acetylated, and analyzed by GC-MS. The ratio between GalNAc-ol and Fuc-ol detected by GC-MS (GalNAc-ol:Fuc-ol (4.3:1)) is in reasonable agreement with the ratio between the prevalences of the major core 1 glycans and the O-Fuc trisaccharide determined by NSI-MS (Structures 2 and 4:Structure 9 (65.0:11.3% = 5.8:1)). Therefore, two independent analytic techniques report a significant level of O-Fuc trisaccharide in Drosophila embryos.
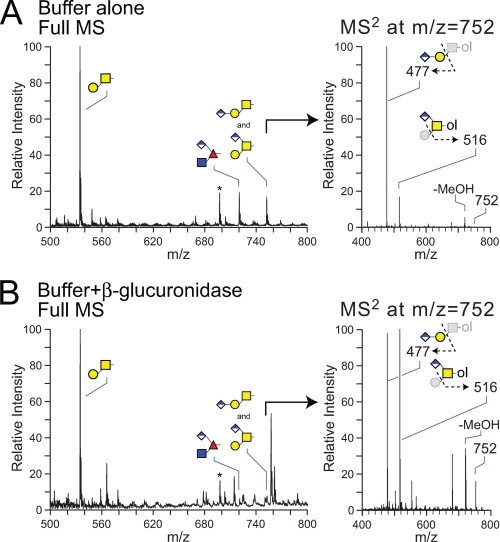
The Hexuronic Acid on Drosophila O-Linked Glycans Is β-Linked Glucuronic Acid—The O-linked glycans released from fly embryo powder by reductive β-elimination were treated with the β-glucuronidase from H. pomatia, and the resulting changes in the prevalences of the acidic core 1 and O-Fuc trisaccharides were monitored by NSI-MS (51). The dramatic reductions of ion signals at m/z = 722 and 752 indicate the susceptibility of the uronic acid residues on these two structures to removal by this β-specific glucuronidase (Fig. 5). Focusing on the isobaric mixture of the two core 1 trisaccharides at m/z = 752 (Fig. 5, right panels), the linear structure is reduced to a proportionally greater extent than the branched structure see (compare MS2 ions at m/z = 477 and 516), suggesting that steric factors hinder the removal of the branching GlcA residue on this core.
FIGURE 5.
The major hexuronylated O-linked glycans of the Drosophila embryo are sensitive to digestion with β-glucuronidase from H. pomatia. O-Glycans released from fly embryo powder by β-elimination were treated with buffer alone (A) or with β-glucuronidase from H. pomatia (B) before permethylation and analysis by NSI-MS. The core 1 disaccharide (m/z = 534) predominates the MS profile with or without enzymatic digestion. Incubation with the enzyme reduces detected parent ion signals in full MS for both the O-Fuc trisaccharide at m/z = 722 and the core 1 trisaccharide isomers at m/z = 752. A, right panel, without enzyme digestion, the MS/MS spectra for the permethylated HexA1Hex1HexNAc-ol at m/z = 752 exhibits an intense signal at m/z = 477, corresponding to the loss of monosubstituted HexNAc-ol (linear structure) and a characteristic ion at m/z = 516, corresponding to the loss of terminal Hex (branched structure). B, right panel, incubation with β-glucuronidase attenuates the intensity of the MS/MS ion at m/z = 477 (linear trisaccharide) to near equivalence with the m/z = 516 ion (branched trisaccharide). Comparison of the relative signal intensities for the MS/MS ions at m/z = 516 and 752 in panels A and B indicates that the branched core 1 trisaccharide is also reduced by enzyme digestion, although to a lesser extent than the linear trisaccharide (>80% versus ∼30%), indicating partial resistance of the branched structure.
The Glucuronylated O-Fuc Trisaccharide Is Enriched along the Dorsal/Ventral Axis of the Wing Imaginal Disc—The addition and elongation of O-Fuc on Ser/Thr residues of the Notch receptor protein drives differential ligand activation across a broad range of developing tissues in many animal species. In the imaginal wing disc of the third instar Drosophila larvae, differential Notch signaling is established by asymmetric expression of the Fringe glycosyltransferase. Elevated Fng expression in the dorsal half of the wing disc results in the spatially restricted production of Notch glycoforms that are preferentially activated by the ligand Delta. Spatial distributions of Delta and Fng expression cooperate to establish a dorsal/ventral boundary across the single-cell epithelial layer of the imago at a meridian that divides the disc pouch roughly into two halves (Fig. 6A). If the O-Fuc trisaccharide identified in whole embryos is relevant for Notch signaling, it should be distributed across the wing disc with an asymmetry that parallels the Fng enzyme.
FIGURE 6.
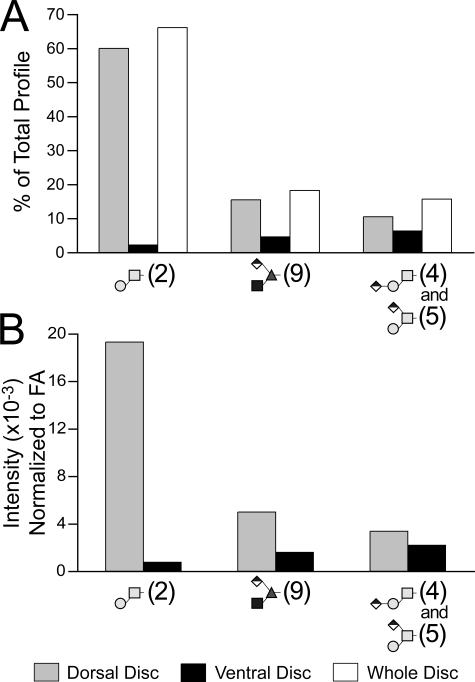
Major O-linked glycans are enriched in the dorsal portion of the larval wing disc. Imaginal wing discs were harvested from wandering third instar larvae. At this stage of development, the wing disc is differentiated into distinct spatial compartments based on the expression of cellular differentiation markers. A, cells at the boundary between the dorsal and ventral compartments express the Wingless protein in response to activation of the Notch signaling pathway (DIC, differential interference contrast; anti-Wingless immunofluorescence). B, a total of 269 freshly harvested wing discs, including the associated peripodial membrane, were microdissected into dorsal and ventral halves roughly along the meridian defined by Wingless expression (pseudocolored for illustrative purposes). Following tissue delipidation, O-linked glycans were released by reductive β-elimination. Equal portions of the dorsal and ventral preparations were analyzed by NSI-MS and TIM. C, O-linked glycans are enriched in the dorsal portion of the wing disc. D, filtered TIM scans reveal that the O-Fuc trisaccharide (m/z = 722) is enriched by almost 4-fold in the dorsal disc compared with the ventral disc. Similar dorsal enrichment is detected for the glucuronylated linear core 1 trisaccharide (signature fragment at m/z = 477 from MS/MS at 752), but the branched isomer (signature fragment at m/z = 516 from MS/MS at 752) is enriched in the ventral disc.
A total of 269 wing discs were microdissected into dorsal and ventral halves, and their O-linked glycans were prepared by reductive β-elimination from delipidated wing disc powders (Fig. 6B). NSI-MS analysis of equivalent portions of the total permethylated glycans isolated from each half-disc preparation reveals a significant enrichment of the O-Fuc trisaccharide (m/z = 722) in the dorsal disc, consistent with Fng distribution (Fig. 6C). The core 1 disaccharide (m/z = 534), as well as the isobaric mixture of the linear and branched forms of the glucuronylated core 1 disaccharide (m/z = 752), are also enriched in the dorsal wing disc (Fig. 6C). The dorsal enrichment of the O-Fuc trisaccharide, evident in full MS spectra, is verified in TIM scans filtered for the detection of signature fragment ions (Fig. 6D). Filtered MS/MS data also revealed that expression of the linear form of the glucuronylated core 1 trisaccharide parallels the O-Fuc trisaccharide. In contrast, the branched form of the glucuronylated core 1 structure is enriched in the ventral portion of the disc.
To assess relative glycan recovery from microdissected wing discs, the profile of the major O-glycans isolated from disc halves was compared with the profile determined for intact isolated discs. For this comparison, the total glycan profile for disc halves was calculated by summing together the ion signals for both the dorsal and ventral glycans, providing a virtual reassembly of the disc (Fig. 7A). For each of the indicated glycan structures, the summed contribution of the dorsal and ventral profile closely approximates the independently determined profile for whole discs, demonstrating that the detected asymmetries are not an artifact of the microdissection. The ion intensities resulting from NSI-MS analysis of equal proportions of the total disc halves indicate that the O-Fuc trisaccharide is enriched 3.8-fold in the dorsal disc (Fig. 6). However, this relative enrichment factor does not take into account potential differences in tissue mass. Normalization by protein content is not possible because all of the protein powder was carried forward into the β-elimination to maximize glycan detection. The delipidation performed during sample preparation provides a ready source of fatty acid for normalization between tissue samples. Fatty acid methyl ester profiles of the larval imaginal wing disc halves are very similar to each other but somewhat different than the profiles detected for whole embryos (supplemental Table 2). In particular, disc fatty acid profiles are generally lower in their prevalence of unsaturated species than are embryos. The profile of fatty acid methyl esters detected in embryos is in good agreement with previous characterizations of lipid diversity in Drosophila and other insects (64, 65). Total fatty acid content of the dorsal disc was determined to be 126% of the ventral disc. Therefore, normalized for tissue mass by fatty acid content, the O-Fuc trisaccharide is enriched 3.1-fold and the core 1 disaccharide is enriched 25-fold in the dorsal relative to the ventral wing disc (Fig. 7B).
FIGURE 7.
Quantification of the asymmetric distribution of major O-linked glycans across the dorsal/ventral axis of the wing disc. O-Linked glycans prepared from dorsal or ventral wing disc halves or from intact wing discs were quantified by NSI-MS. A, to assess glycan recovery from microdissected tissues, signal intensities for all of the indicated major glycans detected in dorsal and ventral disc halves were summed to give a total signal for the combined profiles. Structure numbers are shown in parentheses. The percentage of the total combined profile contributed by each individual glycan in each half-disc was then calculated (gray box, dorsal glycans; black box, ventral glycans). An independent preparation and analysis of the O-linked glycans isolated from nondissected wing discs generated the whole disc profile, which is shown for comparison (white box, whole disc). For each major glycan, the summed contribution of the dorsal and ventral disc closely approximates the independently determined prevalence in the whole disc, verifying the significant asymmetry in O-glycan expression across the imaginal tissue. B, the determination of relative prevalence in A and in Fig. 6 is based on analysis of equal proportions of the whole preparation. To control for differences in tissue mass between dorsal and ventral disc halves, the MS signal intensities were normalized to total fatty acids (supplemental Table 2). The dorsal disc preparation yielded 26% greater fatty acid content than the ventral disc preparation. Correcting for this tissue difference, the core 1 disaccharide (Structure 2) is enriched 25-fold and the O-Fuc trisaccharide (Structure 9) is enriched 3.1-fold in the dorsal disc.
Expression of the Glucuronylated O-Fuc Trisaccharide Is Partially Dependent on Fringe—The spatial enrichment of the O-Fuc trisaccharide in the dorsal wing disc is consistent with an expectation that the GlcNAc transferase activity of the Fng enzyme participates in its biosynthesis. However, it is not possible to obtain wing discs that are entirely mutant for fng to directly test this assumption. Although clones of fng mutant cells can be generated within otherwise wild-type tissue, the small amount of material and the resulting heterogeneity of cell types across the whole disc preclude meaningful glycan analysis. Therefore, we assessed the O-glycan profile of fng13 mutant embryos, which can be obtained as homozygotes by hand-sorting in late embryogenesis (supplemental Table 3 and supplemental Fig. 7, A-C). The fng13 mutation is genetically an amorphic allele, a loss-of-function mutation in which a premature stop codon severely truncates the catalytic domain; no enzymatic activity has been reported (66). Staged embryos were collected from balanced fng13/TM3-GFP parents and sorted based on age and fluorescence. Nonfluorescent fng13/fng13 embryos (884 total) were separated from their fluorescent siblings (fng13/TM3-GFP and TM3-GFP/TM3-GFP) who were accumulated together as a mixed control pool. O-Linked glycans were harvested from fng13 homozygotes, from their sibling controls, and from w1118 embryos, the genetic background in which the fng13/TM3-GFP stock is maintained.
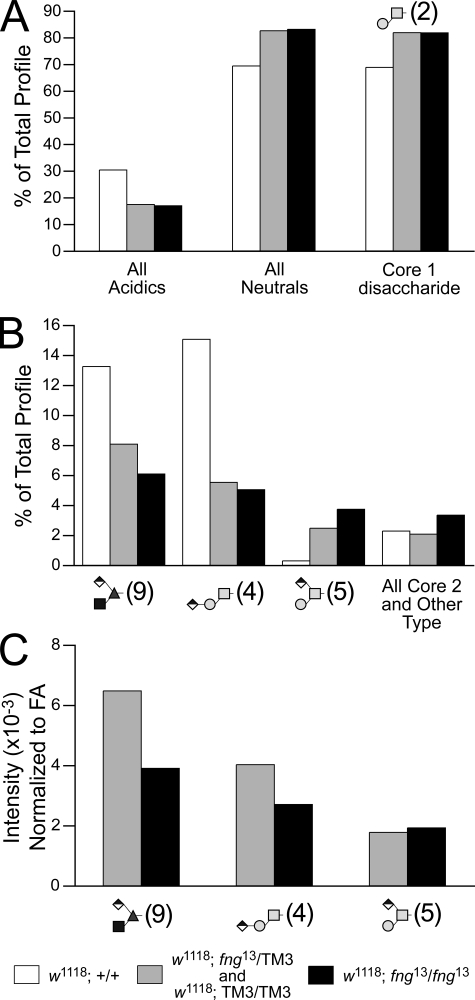
The prevalence of acidic glycans, as a class, is reduced with the introduction of even a single copy of the fng13 mutation (Fig. 8A). A slight increase in neutral glycans offsets the decreased acidics and is entirely attributable to the predominant neutral structure, the core 1 disaccharide. Filtered TIM scans detected decreased prevalences for two of the three major glucuronylated O-linked glycans (Fig. 8B and supplemental Fig. 7, D and E). Compared with w1118 embryos, the O-Fuc trisaccharide was decreased by 41 and 56%, respectively, in sibling controls (which includes fng13 heterozygotes) and fng13 homozygotes. Similar to wing disc halves, expression of the linear glucuronylated core 1 glycan parallels expression of the O-Fuc trisaccharide in the fng mutation. As a percent of the total profile, an increase in the branched, glucuronylated core 1 glycan offsets the decrease in the linear structure. However, when normalized to fatty acid content, the branched core 1 trisaccharide is unchanged, whereas both the linear core 1 trisaccharide and the O-Fuc trisaccharide exhibit decreases in prevalence that track with the dosage of the mutant gene (Fig. 8C).
FIGURE 8.
A mutation in the fringe gene decreases the prevalence of the O-Fuc trisaccharide and the linear core 1 structure. Embryos collected from adults heterozygous for the fng13 mutation carried over a GFP balancer chromosome (TM3-GFP) were manually sorted by fluorescence (supplemental Fig. 7), yielding 884 homozygous fng13 late-stage embryos and a pool of age-matched control siblings (mixed fng13/TM3-GFP heterozygotes and TM3-GFP homozygotes). O-Linked glycans were prepared from the sorted embryos and from w1118 embryos, the genetic background in which the fng13 mutation is maintained. A, the prevalence of acidic glycans is decreased in fng mutant embryos. A corresponding increase in neutral glycans is driven almost entirely by a higher relative prevalence of the core 1 disaccharide (Structure 2). B, two of the three major glucuronylated O-linked glycans are reduced in fng mutant embryos as a percent of the total profile, including the O-Fuc trisaccharide (Structure 9), which is predicted to require Fng enzyme activity for its biosynthesis. C, normalized to total fatty acid content, the MS signal intensities for the O-Fuc trisaccharide and for the linear core 1 trisaccharide (Structure 4) decrease in proportion to the dose of the mutant fng gene. The amount of the branched, glucuronylated core 1 trisaccharide (Structure 5) is unchanged by loss of Fng function.
DISCUSSION
Previous analyses of the O-linked glycans expressed in Drosophila melanogaster tissues have verified the existence of O-linked GalNAc (Tn-antigen) and the core 1 disaccharide (T-antigen) by direct chemical identification and by indirect binding of specific lectin and antibody probes (31-39). Characterization of the O-linked glycans synthesized by cultured insect cell lines (dipteran and lepidopteran) have also indicated the predominance of these two glycans, and a recent report describes a linear isomer of a glucuronylated core 1 disaccharide (designated here as Structure 4) in Drosophila S2 cells (40-42, 44, 45). Our analysis of O-linked glycans of the Drosophila embryo detected these three glycans and added another novel structure (the O-Fuc trisaccharide, Structure 9) to the complement of major glycans that together account for 96% of the total O-linked profile of wild-type embryos (OreR). The addition of a novel, less prevalent, branched form of the glucuronylated core 1 glycan (Structure 5) increases this portfolio to 98% of the total profile. We also detected a group of expected, minor structures (O-Man, O-Glc, monoxylosylated O-Glc, O-GlcNAc) that have been predicted to exist in embryonic tissues based on phenotypes associated with mutations in the proposed biosynthetic enzymes or on the in vitro identification of appropriate enzymatic activities: protein O-mannosyltransferases, rotated/POMT1 and twisted/POMT2; protein O-glucosyltranferase, rumi/Poglut; and O-GlcNAc transferase (OGT) (8, 58, 59, 67-71).
We searched for the fragmentation signatures of glycans in MS/MS spectra collected by automated incremental scanning over a broad mass range (TIM), and our unbiased detection methods revealed additional minor components within the total profile of Drosophila O-linked glycans. These least prevalent structures include neutral and acidic extensions of the major O-linked glycans, of core 2 type structures, and of other glycans built on a HexNAc-HexNAc core. Two striking properties of the minor glycans are worth emphasizing. First, HexA is found on almost every type of core, with some cores carrying more than one HexA. Second, HexA is found both at nonreducing terminal positions (Structures 12 and 16-18) and at internal positions (Structures 6-8). The terminal and internal HexNAc-HexA disaccharides are evocative of glycosaminoglycan repeat units. Whether these proto-glycosaminoglycan structures, which have also been described in Caenorhabditis elegans, are substrates for sulfation or are capable of mediating the developmental functions of extended glycosaminoglycan chains remains to be determined (72). To date, however, the glycosyltransferase activities required to build the minor Drosophila O-linked glycans have not been assigned to any of the putative glycosyltransferase genes annotated in the Drosophila genome. Therefore, a well characterized O-linked glycome offers a scaffold for linking enzymes to biosynthetic pathways, providing new targets for identifying glycan functions.
In comparing the O-linked glycan profile of the Drosophila embryo with the O-linked glycans of other organisms, notable differences are apparent. Two post-synthetic core modifications were not detectable on Drosophila glycans. Phosphoethanolamine is a common addition to GlcNAc residues on Drosophila glycosphingolipids and on the glycans of other invertebrates, but it was not detected on the O-linked glycans of fly embryo powder (73-75). Sulfation of Hex, HexA, and HexNAc residues on glycosphingolipids, glycosaminoglycans, and N-linked and O-linked glycans frequently modifies oligosaccharide function in developing and mature animal tissues. Sulfated, non-glycosaminoglycan O-linked oligosaccharides were not detected in fly embryo powder. Although the high pH of the elimination and permethylation steps may place this modification at risk for cleavage prior to MS analysis, reasonable recovery of a sulfated standard (sulfatide, 3-O-sulfated galactosylceramide) was achieved under our analytic conditions (data not shown). The possible existence of O-sulfate on the non-glycosaminoglycan glycans of the Drosophila embryo will require the development of analytic techniques specifically optimized for the identification of post-synthetic glycan modifications.
Other comparative differences are evident in the monosaccharide composition of the O-linked glycans. Despite the ability of Drosophila embryos to sialylate N-linked glycans, we were unable to detect sialic acid on O-linked structures, a common terminal modification in vertebrates (76).3 Instead, it is tempting to speculate that GlcA serves as the sialic acid equivalent in Drosophila. The occurrence of terminal GlcA on Drosophila glycosphingolipids and of terminal sialic acid on the vertebrate ganglioside family of glycosphingolipids is also consistent with functional equivalence between these two acidic monosaccharides (74, 75, 77). However, distinct differences indicate that this hypothesis is probably an oversimplification. For instance, we detected GlcA in Drosophila as an internal residue, capped with HexNAc, whereas sialic acid is almost exclusively a nonreducing terminal residue in vertebrate oligosaccharides. The only occurrence of vertebrate sialic acid residues in other than terminal positions is when the sialic acid is covered in α2-8 linkage by another sialic acid or by a polymer of sialic acid (42, 78, 79). We did not detect analogous GlcA-GlcA dimers or extended GlcA homopolymers in our O-linked preparations.
Another set of vertebrate O-glycans that appeared to be absent, or at least below our detection limit, in Drosophila embryos is the family of extended O-Man structures, most clearly described as modifications to the α-dystroglycan protein (80). Although normal muscle development in Drosophila requires appropriate mannosylation of the Drosophila dystroglycan homologue by POMT-1/POMT-2, the addition of the single hexose appears to be sufficient (81). Consistent with the O-linked glycan profile, a homologue of POMGnT-1, the enzyme that initiates extension of O-Man glycans in vertebrates, has not been identified in the Drosophila genome. Lewis-type structures, carrying Fuc as branching residues off of nonreducing terminal N-acetyllactosamine disaccharides, are also not detected in the Drosophila O-linked glycome. In vertebrates and in some blood-dwelling pathogens, these glycans mediate essential cell-cell interactions at the interface between circulating leukocytes, platelets, and the vascular endothelium, a tissue context without analogy in Drosophila embryos (82).
Although not detected as a nonreducing terminal modification, Fuc is at the core of one of the four major O-linked glycans expressed in the Drosophila embryo. The O-Fuc trisaccharide, GlcNAcβ1-3(GlcAβ1-4)Fucitol (Structure 9) is the only detectable O-Fuc glycan in the embryo. The addition of β3-linked GlcNAc onto Fuc in this structure is consistent with the known specificity of the Fringe glycosyltransferase, but neither the expected disaccharide product nor the unsubstituted O-Fuc precursor was detected in our embryo glycan preparations. Both Fuc-O- and GlcNAcβ1-3Fuc-O- have been detected on Notch protein forms expressed in cultured insect cells, where they modulate ligand specificity in vitro (10, 28). However, the efficient processing of O-Fuc into the trisaccharide, coupled with the surprisingly high prevalence of the glycan (11-14% of the total profile) in the embryo, indicates that the O-Fuc structure capable of modulating Notch signaling in vivo may not be faithfully replicated in cell culture. The ability of the O-Fuc trisaccharide to modulate ligand binding specificity should now be assessed, and such studies would be greatly facilitated by identifying the relevant glucuronyl transferase(s).
It is formally possible that a small pool of Fuc-O- and GlcNAcβ1-3Fuc-O-glycan is entirely responsible for modulating Notch signaling and that these modifications exist at levels below our detection threshold. Thus, the O-Fuc trisaccharide that we detected might be an inactive glycan or might constitute a previously unidentified, broadly distributed O-glycan expressed on many proteins, perhaps largely irrelevant to Notch signaling. Two results indicate that this is not the case. First, the O-Fuc trisaccharide is enriched in the dorsal half of the imaginal wing disc, a tissue in which spatially restricted Fringe expression modulates Notch signaling. Second, the O-Fuc trisaccharide is reduced in embryos homozygous for a loss-of-function fng mutation that exhibits ligand-dependent, Notch-like phenotypes (25, 26). Thus, the O-Fuc trisaccharide increases where Notch signaling is modulated by Fringe activity and decreases in the absence of Fringe. In neither case, however, is expression of the O-Fuc trisaccharide completely dependent on Fringe activity; the O-Fuc trisaccharide is present in the ventral disc and is not entirely eliminated in the fng mutant.
It is unlikely that the residual O-Fuc trisaccharide detected in fng mutant embryos reflects maternal contribution of fng mRNA because the message is not detected by in situ hybridization until embryonic stage 10, well after the initiation of zygotic transcription (83, 84). For this study, glycans were harvested from stage 15-16 mutant embryos. Furthermore, although maternal fng contributes to ovarian follicle development through the somatic lineage, it is not required in germ line cells for the generation of viable oocytes (85, 86). Currently, there are no genetic data to support a maternal contribution to fng embryonic phenotypes (87-89). Although the O-Fuc trisaccharide might be maternally deposited into eggs on yolk proteins during oogenesis, we detected increasing levels of O-Fuc trisaccharide as development proceeds (supplemental Table 3) rather than decreasing levels, as would be expected for the consumption of yolk during embryogenesis. Drosophila genes with weak similarity to fng have been identified molecularly, and residual O-Fuc trisaccharide in the fng mutant may reflect the activity of these enzymes on Notch or on other, as yet unidentified, target proteins (66). Although the task is immensely challenging, it will ultimately be essential to place specific glycan structures at defined sites on endogenous protein isolated from developing tissues in order to resolve the functional relevance of Notch glycoform heterogeneity.
A particularly striking and completely unexpected differential distribution of glycan was detected for the prevalence of the core 1 disaccharide across the wing disc. The significant enrichment of core 1 disaccharide in the dorsal disc could not have been predicted by previously published patterns of lectin binding (Jacalin), antibody probe (anti-Tn antigen for precursor), or ppGalNAcT mRNA localization (39, 60). Multiple ppGalNAcTs are expressed in the larval wing disc, some are uniformly distributed, and some are expressed in spatially enriched patterns (39). To date, however, the expression of a relevant core 1 galactosyltransferase, capable of extending O-GalNAc residues in the wing disc, has not been described. Furthermore, many of the lectin probes that reveal potential polypeptide O-linked glycan distributions possess significant potential cross-reactivity with identical terminal glycan moieties found on glycosphingolipids (74). Therefore, the relation between the described lectin binding patterns and measured glycan expression profiles is currently too indirect to yield useful comparisons.
Evidence for coordinated expression of a subset of O-glycans arises from our characterization of the total O-glycan profile in larval imaginal disc halves and in wild-type and mutant embryos. Comparing wild-type (w1118) with fng mutant embryos, decreased expression of the O-Fuc trisaccharide (Structure 9) was accompanied by an even greater decrease in the linear isomer of the glucuronylated core 1 disaccharide (Structure 4). However, the branched form of the glucuronylated core 1 disaccharide (Structure 5) was increased in the same mutant as a percent of the total profile (Figs. 8B and 9A). Likewise, Structures 9 and 4 are greatly reduced in the ventral wing disc in comparison with the dorsal disc, but Structure 5 is enriched. Therefore, the ventral wing disc, which genetically and developmentally behaves as a tissue that lacks Fng, presents a portfolio of O-linked glycans that resembles the O-glycan profile of the fng mutant embryo. Furthermore, a comparison of the O-glycan profiles of early and late wild-type embryos also supports coordinated expression of this glycan subset. The prevalence of Structures 9 and 4 increases by 40% between early and late embryonic stages, whereas Structure 5 increases by more than 500% (supplemental Table 3). Given the predicted substitution positions and the overall structural dissimilarity between the core 1 and O-Fuc disaccharide acceptors, unique glucuronyltransferases are likely responsible for generating each of these three trisaccharides. Our results indicate that the enzymes that synthesize Structures 9 and 4 are separately regulated from the enzyme that makes Structure 5.
FIGURE 9.
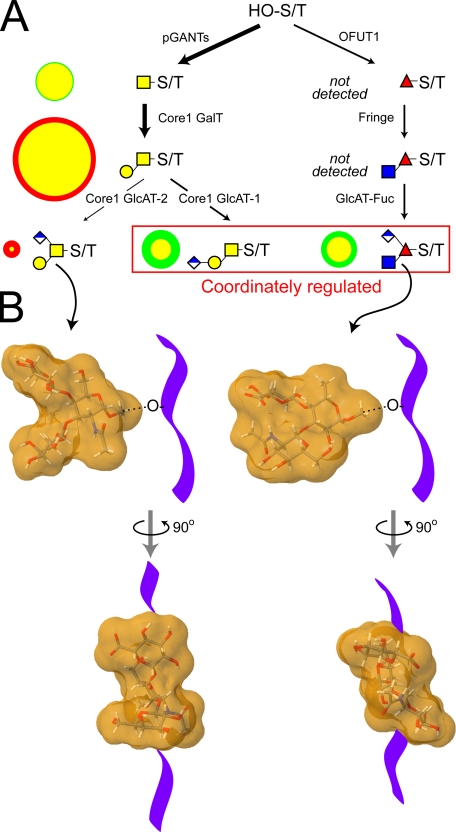
Differential fluxes through the biosynthetic pathways that produce major O-linked glycans yield distinctive structural profiles in wild-type and fringe mutant embryos. Only a portion of the full complement of glycosyltransferases that are necessary for synthesizing Drosophila O-linked glycans have been identified. A, among the activities already identified in Drosophila are the pGANTs (ppGalNAcT) and the OFUTs (O-fucosyltransferases 1 and 2), which transfer monosaccharide directly to serine/threonine residues (HO-S/T) of the polypeptide backbone (9, 28, 90). To date, a single core 1 galactosyltransferase candidate has been characterized that can generate core 1 disaccharide, the predominant O-linked glycan of the embryo (96). The Fringe glycosyltransferase adds β3GlcNAc to O-Fuc on epidermal growth factor repeats (26, 30). The putative glucuronyltransferases necessary for generating the characterized glucuronylated O-glycans have yet to be identified (denoted here as core 1 GlcAT-1/2 and GlcAT-Fuc). In A, the arrow densities reflect predicted flux through pathways based on the relative prevalence of glycan products in wild-type embryos. To compare biosynthesis in fng mutant and wild-type embryos, circles to the left of each structure are drawn such that the area of the circle (green for wild-type and red for fng13 mutant) is directly proportional to the prevalence of that glycan (expressed as % total profile). For the two circles that describe the prevalence of an individual glycan in each background, the smaller is centered on top of the larger, and the area of overlap is coded in yellow. Therefore, a yellow circle rimmed in green indicates a glycan that is more prevalent in the wild type, and a yellow circle rimmed in red indicates a glycan that is more prevalent in fng13 mutant embryos. In the fringe mutant, the profile of the major acidic glycans shifts such that the relative prevalence of the branched core 1 trisaccharide increases in relation to both the linear core 1 trisaccharide and the O-Fuc trisaccharide. Coordinate regulation of these latter two glucuronylated structures, built on entirely different cores, is also apparent in the wing disc (see Fig. 6D). B, coordinate reduction of the branched O-Fuc trisaccharide and the linear core 1 trisaccharide, coupled to increased relative prevalence of the branched core 1 trisaccharide, shifts the distribution of glycan shapes on glycoprotein polypeptide backbones (diagrammed as a purple ribbon). Energy minimized models (supplemental Fig. 8) of the molecular shapes of the branched core 1 trisaccharide and the O-Fuc trisaccharide predict significantly different dispositions for their 3- and 4-linked substitutions (91, 92).5 The clustering of multiple O-glycans with distinct structural characteristics on mucins or on other types of polypeptide backbones may impart significant functional constraints to glycoproteins expressed in different cell types or in mutant backgrounds.
The coordinate regulation of glucuronylating enzyme activities may have important consequences for the structure of the glycoproteins targeted for this modification. An energy-minimized model of the branched core 1 trisaccharide (Structure 5) produces a molecular shape in which the two antennae are separated by a distinct groove (Fig. 9B and supplemental Fig. 8). The gaping-jawed appearance of Structure 5 is dramatically different from the predicted structure of the O-Fuc trisaccharide (Structure 9), in which the branches are closely packed into a smoother, continuous molecular shape (91, 92).4 These distinct molecular shapes may be expected to impart unique structural constraints on the peptide backbone to which they are attached or on the potency of protein-ligand interactions. Thus, induction of the enzyme that generates Structure 5 on core 1 glycans of a particular protein, perhaps Notch, may antagonize or attenuate the structural influence of the enzyme activity that generates Structure 9. For any particular protein, it will be essential to define the full distribution of glycoforms to assess the structural impact of glycan diversity.
Perhaps Fringe enzymes in vertebrates and in Drosophila function primarily to provide substrate for subsequent elaboration, either glucuronylation in Drosophila or extension with sialylgalactose in vertebrates. Recent genetic analysis in Drosophila demonstrates that a subset of OFUT-1 phenotypes are identical to Fng loss-of-function phenotypes, consistent with the proposal that O-Fuc addition by itself is not as essential as elaboration of the O-Fuc (93). The fully extended O-Fuc tetrasaccharide on vertebrate Notch (NeuAcα2-3/6Galβ1-4GlcNAcβ1-3Fuc-O-) possesses little primary sequence similarity to the O-Fuc trisaccharide of Drosophila. However, a subtle structural congruence results from the close packing of the two antennae in the Drosophila trisaccharide. The proximity of the GlcA carboxyl and the acetamido group of the GlcNAc in the O-Fuc trisaccharide is similar to the cross-ring separation between the C-1 carboxyl and the acetamido group of sialic acid (supplemental Fig. 8). This minimal similarity involves structural components on two separate branches of the trisaccharide and, therefore, could easily be disrupted by relatively unrestricted, independent bond rotations. Furthermore, the functional significance of this congruence is unclear because sialylation, but not galactosylation, seems to be dispensable for vertebrate Notch ligand binding, at least in vitro (94). However, it is now evident that Drosophila, like vertebrates, possesses the biosynthetic capacity to extend O-Fuc by more than a single HexNAc residue.
The portfolio of O-linked glycans that we detected in the Drosophila embryo predicts the existence of previously unappreciated glycosyltransferase activities. Among these activities are the addition of GlcA to Gal, GalNAc, and Fuc, the addition of HexNAc to GalNAc, and the addition of GlcNAc to GalNAc and GlcA. Some of these activities have been described in other species or are evident in the biosynthesis of Drosophila glycosphingolipids and glycosaminoglycans (77, 95). However, in the context of O-linked core 1 or core 2 or O-Fuc, these modifications are novel, and identification of the relevant biosynthetic enzymes will provide new opportunities for dissecting the functions of O-linked glycans.
Supplementary Material
Acknowledgments
We acknowledge the advice and technical assistance of Mayumi Ishihara, the generosity and expert guidance of Lance Wells and Jae-Min Lim, and the access to instrumentation provided through the National Center for Research Resource's support of the Integrated Technology Resource for Biomedical Glycomics (Complex Carbohydrate Research Center, University of Georgia).
This work was supported, in whole or in part, by National Institutes of Health Grants 1-R01-GM072839 from NIGMS and 1-U01-CA128454 from NCI (both to M. T.). This work was also supported by a Toyobo Biotechnology Foundation long-term research grant (to K. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-8 and Tables 1-3.
Footnotes
The abbreviations used are: ppGalNAcT, polypeptide N-acetlygalactosaminyltransferase; Hex, undetermined hexose; HexNAc, undeterminedN-acetylhexosamine; HexA, undetermined hexuronic acid; CID, collision-induced dissociation; NSI-MS, nanospray ionization mass spectrometry; MS/MS, tandem mass spectrometry; GC-MS, gas chromatography-mass spectrometry; TIM, total ion mapping; OFUT, O-linked fucosyltransferase; GlcAT, glucuronosyltransferase; GFP, green fluorescent protein; HPAEC, High pH Anion Exchange Chromatography.
The reduced (alditol) forms of monosaccharides are designated by the suffix “-ol” appended to the abbreviation. Glycan nomenclature and the representation of oligosaccharides are in accordance with the guidelines proposed by the Consortium for Functional Glycomics.
The Woods Group, GLYCAM Web, March 15, 2008, Complex Carbohydrate Research Center, The University of Georgia, Athens, GA.
References
- 1.Bishop, J. R., Schuksz, M., and Esko, J. D. (2007) Nature 446 1030-1037 [DOI] [PubMed] [Google Scholar]
- 2.Esko, J. D., and Selleck, S. B. (2002) Annu. Rev. Biochem. 71 435-471 [DOI] [PubMed] [Google Scholar]
- 3.Lin, X. (2004) Development (Camb.) 131 6009-6021 [DOI] [PubMed] [Google Scholar]
- 4.Hanisch, F.-G., Chai, W., Rosankiewicz, J. R., Lawson, A. M., Stoll, M. S., and Feizi, T. (1993) Eur. J. Biochem. 217 645-655 [DOI] [PubMed] [Google Scholar]
- 5.Schachter, H. (1986) Biochem. Cell Biol. 64 163-181 [DOI] [PubMed] [Google Scholar]
- 6.Haltiwanger, R. S., and Lowe, J. B. (2004) Annu. Rev. Biochem. 73 491-537 [DOI] [PubMed] [Google Scholar]
- 7.Ju, T., and Cummings, R. D. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16613-16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly, W. G., and Hart, G. W. (1989) Cell 57 243-251 [DOI] [PubMed] [Google Scholar]
- 9.Moloney, D. J., Shair, L. H., Lu, F. M., Xia, J., Locke, R., Matta, K. L., and Haltiwanger, R. S. (2000) J. Biol. Chem. 275 9604-9611 [DOI] [PubMed] [Google Scholar]
- 10.Okajima, T., Xu, A., and Irvine, K. D. (2003) J. Biol. Chem. 278 42340-42345 [DOI] [PubMed] [Google Scholar]
- 11.Ten Hagen, K. G., Fritz, T. A., and Tabak, L. A. (2003) Glycobiology 13 1R-16R [DOI] [PubMed] [Google Scholar]
- 12.Ten Hagen, K. G., and Tran, D. T. (2002) J. Biol. Chem. 277 22616-22622 [DOI] [PubMed] [Google Scholar]
- 13.Van den Steen, P., Rudd, P. M., Dwek, R. A., and Opdenakker, G. (1998) Crit. Rev. Biochem. Mol. Biol. 33 151-208 [DOI] [PubMed] [Google Scholar]
- 14.Rosen, S. D. (2004) Annu. Rev. Immunol. 22 129-156 [DOI] [PubMed] [Google Scholar]
- 15.Snapp, K. R., Heitzig, C. E., Ellies, L. G., Marth, J. D., and Kansas, G. S. (2001) Blood 97 3806-3811 [DOI] [PubMed] [Google Scholar]
- 16.White, S. J., Underhill, G. H., Kaplan, M. H., and Kansas, G. S. (2001) J. Immunol. 167 628-631 [DOI] [PubMed] [Google Scholar]
- 17.Barresi, R., and Campbell, K. P. (2006) J. Cell Sci. 119 199-207 [DOI] [PubMed] [Google Scholar]
- 18.Martin, P. T., and Freeze, H. H. (2003) Glycobiology 13 67R-75R [DOI] [PubMed] [Google Scholar]
- 19.Ervasti, J. M., Burwell, A. L., and Geissler, A. L. (1997) J. Biol. Chem. 272 22315-22321 [DOI] [PubMed] [Google Scholar]
- 20.Ervasti, J. M., and Campbell, K. P. (1991) Cell 66 1121-1131 [DOI] [PubMed] [Google Scholar]
- 21.Moore, S. A., Saito, F., Chen, J., Michele, D. E., Henry, M. D., Messing, A., Cohn, R. D., Ross-Barta, S. E., Westra, S., Williamson, R. A., Hoshi, T., and Campbell, K. P. (2002) Nature 418 422-425 [DOI] [PubMed] [Google Scholar]
- 22.Tian, E., and Ten Hagen, K. G. (2007) J. Biol. Chem. 282 606-614 [DOI] [PubMed] [Google Scholar]
- 23.Stanley, P. (2007) Curr. Opin. Struct. Biol. 17 530-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Artavanis-Tsakonas, S., Rand, M. D., and Lake, R. J. (1999) Science 284 770-776 [DOI] [PubMed] [Google Scholar]
- 25.Okajima, T., and Irvine, K. D. (2002) Cell 111 893-904 [DOI] [PubMed] [Google Scholar]
- 26.Panin, V. M., Shao, L., Lei, L., Moloney, D. J., Irvine, K. D., and Haltiwanger, R. S. (2002) J. Biol. Chem. 277 29945-29952 [DOI] [PubMed] [Google Scholar]
- 27.Haines, N., and Irvine, K. D. (2003) Nat. Rev. Mol. Cell Biol. 4 786-797 [DOI] [PubMed] [Google Scholar]
- 28.Xu, A., Haines, N., Dlugosz, M., Rana, N. A., Takeuchi, H., Haltiwanger, R. S., and Irvine, K. D. (2007) J. Biol. Chem. 282 35153-35162 [DOI] [PubMed] [Google Scholar]
- 29.Haltiwanger, R. S., and Stanley, P. (2002) Biochim. Biophys. Acta 1573 328-335 [DOI] [PubMed] [Google Scholar]
- 30.Moloney, D. J., Panin, V. M., Johnston, S. H., Chen, J., Shao, L., Wilson, R., Wang, Y., Stanley, P., Irvine, K. D., Haltiwanger, R. S., and Vogt, T. F. (2000) Nature 406 357-358 [DOI] [PubMed] [Google Scholar]
- 31.Callaerts, P., Vulsteke, V., Peumans, W., and De Loof, A. (1995) Roux's Arch. Dev. Biol. 204 229-243 [DOI] [PubMed] [Google Scholar]
- 32.D'Amico, P., and Jacobs, J. R. (1995) Tissue Cell 27 23-30 [DOI] [PubMed] [Google Scholar]
- 33.Fredieu, J. R., and Mahowald, A. P. (1994) Acta Anat. (Basel) 149 89-99 [DOI] [PubMed] [Google Scholar]
- 34.Kramerov, A. A., Arbatsky, N. P., Rozovsky, R. M., Mickhaleva, E. A., Polesskaya, O. O., Gvozdev, V. A., and Shibaev, F. N. (1996) FEBS Lett. 378 213-218 [DOI] [PubMed] [Google Scholar]
- 35.Kramerov, A. A., Mikhaleva, E. A., Rozovsky, Y. M., Pochechueva, T. V., Baikova, N. A., Arsenjeva, E. L., and Gvozdev, V. A. (1997) Insect Biochem. Mol. Biol. 27 513-521 [DOI] [PubMed] [Google Scholar]
- 36.North, S. J., Koles, K., Hembd, C., Morris, H. R., Dell, A., Panin, V. M., and Haslam, S. M. (2006) Glycoconj. J. 23 345-354 [DOI] [PubMed] [Google Scholar]
- 37.Schwientek, T., Mandel, U., Roth, U., Muller, S., and Hanisch, F.-G. (2007) Proteomics 7 3264-3277 [DOI] [PubMed] [Google Scholar]
- 38.Theopold, U., Dorian, C., and Schmidt, O. (2001) Insect Biochem. Mol. Biol. 31 189-197 [DOI] [PubMed] [Google Scholar]
- 39.Tian, E., and Ten Hagen, K. (2006) Glycobiology 16 83-95 [DOI] [PubMed] [Google Scholar]
- 40.Breloy, I., Schwientek, T., Lehr, S., and Hanisch, F.-G. (2008) FEBS Lett. 582 1593-1598 [DOI] [PubMed] [Google Scholar]
- 41.Lopez, M., Tetaert, D., Juliant, S., Gazon, M., Cerutti, M., Verbert, A., and Delanoy, P. (1999) Biochim. Biophys. Acta 1427 49-61 [DOI] [PubMed] [Google Scholar]
- 42.Mourad, R., Morelle, W., Neveu, A., and Strecker, G. (2001) Eur. J. Biochem. 268 1990-2003 [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, N., Nakagawa, T., Fujikawa, K., Kawamura, Y., and Tomiya, N. (1995) Anal. Biochem. 226 139-146 [DOI] [PubMed] [Google Scholar]
- 44.Thomsen, D. R., Post, L. E., and Elhammer, A. P. (1990) J. Cell. Biochem. 43 67-79 [DOI] [PubMed] [Google Scholar]
- 45.Uttenweiler-Joseph, S., Moniatte, M., Lambert, J., Van Dorsselaer, A., and Bulet, P. (1997) Anal. Biochem. 247 366-375 [DOI] [PubMed] [Google Scholar]
- 46.Aoki, K., Perlman, M., Lim, J.-M., Cantu, R., Wells, L., and Tiemeyer, M. (2007) J. Biol. Chem. 282 9127-9142 [DOI] [PubMed] [Google Scholar]
- 47.Greis, K. D., Hayes, B. K., Comer, F. I., Kirk, M., Barnes, S., Lowary, T. L., and Hart, G. W. (1996) Anal. Biochem. 234 38-49 [DOI] [PubMed] [Google Scholar]
- 48.Huang, Y., Konse, T., Mechref, Y., and Novotny, M. V. (2002) Rapid Commun. Mass Spectrom. 16 1199-1204 [DOI] [PubMed] [Google Scholar]
- 49.Anumula, K. R., and Taylor, P. B. (1992) Anal. Biochem. 203 101-108 [DOI] [PubMed] [Google Scholar]
- 50.Domon, B., and Costello, C. E. (1988) Glycoconj. J. 5 397-409 [Google Scholar]
- 51.Sugita, M., Itonori, S., Inagaki, F., and Hori, T. (1989) J. Biol. Chem. 264 15028-15033 [PubMed] [Google Scholar]
- 52.Hardy, M. R., Townsend, R. R., and Lee, Y. C. (1988) Anal. Biochem. 170 54-62 [DOI] [PubMed] [Google Scholar]
- 53.Weitzhandler, M., Pohl, C., Rohrer, J., Narayanan, L., Slingsby, R., and Avdalovic, N. (1996) Anal. Biochem. 241 128-134 [DOI] [PubMed] [Google Scholar]
- 54.Aoki, K., Uchiyama, R., Yamauchi, S., Katayama, T., Itonori, S., Sugita, M., Hada, N., Yamada-Hada, J., Takeda, T., Kumagai, H., and Yamamoto, K. (2004) J. Biol. Chem. 279 32028-32034 [DOI] [PubMed] [Google Scholar]
- 55.Hansson, G. C., Li, Y. T., and Karlsson, H. (1989) Biochemistry 28 6672-6678 [DOI] [PubMed] [Google Scholar]
- 56.Karlsson, H., Carlstedt, I., and Hansson, G. C. (1989) Anal. Biochem. 182 438-446 [DOI] [PubMed] [Google Scholar]
- 57.Graver, R. C., and Sweeley, C. C. (1965) J. Am. Oil Chem. Soc. 42 294-298 [DOI] [PubMed] [Google Scholar]
- 58.Acar, M., Jafar-Nejad, H., Takeuchi, H., Rajan, A., Ibrani, D., Rana, N. A., Pan, H., Haltiwanger, R. S., and Bellen, H. J. (2008) Cell 132 247-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shao, L., Luo, Y., Moloney, D. J., and Haltiwanger, R. S. (2002) Glycobiology 12 763-770 [DOI] [PubMed] [Google Scholar]
- 60.Tian, E., and Ten Hagen, G. (2007) Glycobiology 17 820-827 [DOI] [PubMed] [Google Scholar]
- 61.Wilson, I. B. (2002) J. Biol. Chem. 277 21207-21212 [DOI] [PubMed] [Google Scholar]
- 62.Ashline, D. J., Lapadula, A. J., Liu, Y. H., Lin, M., Grace, M., Pramanik, B., and Reinhold, V. N. (2007) Anal. Chem. 79 3830-3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prien, J. M., Huysentruyt, L. C., Ashline, D. J., Lapadula, A. J., Seyfried, T. N., and Reinhold, V. N. (2008) Glycobiology 18 353-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cmelik, S. H. W., Hurrell, D. P., and Lunat, M. (1969) Comp. Biochem. Physiol. 31 65-78 [DOI] [PubMed] [Google Scholar]
- 65.Overgaard, J., Tomcala, A., Sorensen, J. G., Holmstrup, M., Krogh, P. H., Simek, P., and Kostal, V. (2008) J. Insect Physiol. 54 619-629 [DOI] [PubMed] [Google Scholar]
- 66.Correia, T., Popayannopoulos, V., Panin, V., Woronoff, P., Jiang, J., Vogt, T. F., and Irvine, K. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6404-6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ishimizu, T., Sano, K., Uchida, T., Teshima, H., Omichi, K., Hojo, H., Nakahara, Y., and Hase, S. (2007) J. Biochem. 141 593-600 [DOI] [PubMed] [Google Scholar]
- 68.Kreppel, L. K., Bloomberg, M. A., and Hart, G. W. (1997) J. Biol. Chem. 272 9308-9315 [DOI] [PubMed] [Google Scholar]
- 69.Lyalin, D., Koles, K., Roosendaal, S. D., Repnikova, E., Van Wechel, L., and Panin, V. M. (2006) Genetics 172 343-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ichimiya, T., Manya, H., Ohmae, Y., Yoshida, H., Takahashi, K., Ueda, R., Endo, T., and Nishihara, S. (2004) J. Biol. Chem. 279 42638-42647 [DOI] [PubMed] [Google Scholar]
- 71.Martin-Blanco, E., and Garcia-Bellido, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 6048-6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guerardel, Y., Balanzino, L., Maes, E., Leroy, Y., Coddeville, B., Oriol, R., and Strecker, G. (2001) Biochem. J. 357 167-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maes, E., Garenaux, E., Strecker, G., Leroy, Y., Wieruszeski, J.-M., Brassart, C., and Guerardel, Y. (2005) Carbohydr. Res. 340 1852-1858 [DOI] [PubMed] [Google Scholar]
- 74.Seppo, A., Moreland, M., Schweingruber, H., and Tiemeyer, M. (2000) Eur. J. Biochem. 267 3549-3558 [DOI] [PubMed] [Google Scholar]
- 75.Wiegandt, H. (1992) Bochim. Biophys. Acta 1123 117-126 [DOI] [PubMed] [Google Scholar]
- 76.Koles, K., Irvine, K. D., and Panin, V. M. (2004) J. Biol. Chem. 279 4346-4357 [DOI] [PubMed] [Google Scholar]
- 77.Kim, B. T., Tsuchida, K., Lincecum, J., Kitagawa, H., Bernfield, M., and Sugahara, K. (2003) J. Biol. Chem. 278 9116-9124 [DOI] [PubMed] [Google Scholar]
- 78.Acheson, A., Sunshine, J. L., and Rutishauser, U. (1991) J. Cell Biol. 114 143-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saito, M., Sugano, K., and Nagai, Y. (1979) J. Biol. Chem. 254 7845-7854 [PubMed] [Google Scholar]
- 80.Michele, D. E., and Campbell, K. P. (2003) J. Biol. Chem. 278 15457-15460 [DOI] [PubMed] [Google Scholar]
- 81.Haines, N., Seabrooke, S., and Stewart, B. A. (2007) Mol. Biol. Cell 18 4721-4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang, H.-H., Tsai, P.-L., and Khoo, K.-H. (2001) Glycobiology 11 395-406 [DOI] [PubMed] [Google Scholar]
- 83.Irvine, K. D., and Wieschaus, E. (1994) Cell 79 595-606 [DOI] [PubMed] [Google Scholar]
- 84.Tomancak, P., Beaton, A., Weiszmann, R., Kwan, E., Shu, S., Lewis, S. E., Richards, S., Ashburner, M., Hartenstein, V., Celniker, S. E., and Rubin, G. M. (2002) Genome Biol. 2002 3(12):research0088 [DOI] [PMC free article] [PubMed]
- 85.Grammont, M., and Irvine, K. D. (2001) Development (Camb.) 128 2243-2253 [DOI] [PubMed] [Google Scholar]
- 86.Zhao, D., Clyde, D., and Bownes, M. (2000) J. Cell Sci. 113 3781-3794 [DOI] [PubMed] [Google Scholar]
- 87.Iwaki, D. D., and Lengyel, J. A. (2002) Mech. Dev. 114 71-84 [DOI] [PubMed] [Google Scholar]
- 88.Thomas, G. B., and van Meyel, D. J. (2007) Development (Camb.) 134 591-600 [DOI] [PubMed] [Google Scholar]
- 89.Walters, J. W., Dilks, S. A., and DiNardo, S. (2006) Dev. Biol. 297 323-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ten Hagen, K. G., Tran, D. T., Gerken, T. A., Stein, D. S., and Zhang, Z. (2003) J. Biol. Chem. 278 35039-35048 [DOI] [PubMed] [Google Scholar]
- 91.Kirschner, K. N., Yongye, A. B., Tschampel, S. M., Gonzalez-Outeirino, J., Daniels, C. R., Foley, B. L., and Woods, R. J. (2007) J. Comput. Chem. 29 622-655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Woods, R. J., Dwek, R. A., Edge, C. J., and Fraser-Reid, B. (1995) J. Phys. Chem. 99 3832-3846 [Google Scholar]
- 93.Okajima, T., Reddy, B., Matsuda, T., and Irvine, K. D. (2008) BMC Biol. 2008 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen, J., Moloney, D. J., and Stanley, P. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13716-13721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bellaiche, Y., The, I., and Perrimon, N. (1998) Nature 394 85-88 [DOI] [PubMed] [Google Scholar]
- 96.Muller, R., Hulsmeier, A. J., Altmann, F., Ten Hagen, K., Tiemeyer, M., and Hennet, T. (2005) FEBS J. 272 4295-4305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.