Abstract
Intracellular trafficking and proteolytic processing of amyloid precursor protein (APP) have been the focus of numerous investigations over the past two decades. APP is the precursor to the amyloid β-protein (Aβ), the 38–43-amino acid residue peptide that is at the heart of the amyloid cascade hypothesis of Alzheimer disease (AD). Tremendous progress has been made since the initial identification of Aβ as the principal component of brain senile plaques of individuals with AD. Specifically, molecular characterization of the secretases involved in Aβ production has facilitated cell biological investigations on APP processing and advanced efforts to model AD pathogenesis in animal models. This minireview summarizes salient features of APP trafficking and amyloidogenic processing and discusses the putative biological functions of APP.
APP Gene Family
The human APP3 gene, located on chromosome 21, was first identified in 1987 by several laboratories independently using partial protein sequence information obtained by the Glenner and Beyreuther/Masters laboratories several years earlier. More than 25 mutations in APP have been identified that are causative of the hereditary form of familial AD and a related condition of hereditary cerebral amyloid angiopathy. These mutations introduce amino acid substitutions within or flanking the Aβ domain (for a listing of the mutations, see the Alzheimer Disease & Frontotemporal Dementia Mutation Database at www.molgen.ua.ac.be/ADMutations/). Moreover, APP gene duplication alone also causes early-onset AD with cerebral amyloid angiopathy. The latter findings fit nicely with the consistent finding of AD changes in individuals with trisomy 21 (Down syndrome), in which the APP gene is triplicated. Nonetheless, although mutations in APP are found only in rare cases of AD, they are nevertheless important because they provided early and seminal evidence that APP and Aβ play a central role in AD pathogenesis.
APP is now known to be one of three members of a small gene family, which includes APLP1 and APLP2 (human), Appl (fly), and apl-1 (worm). All encode type I membrane proteins with a large extracellular domain and a short cytoplasmic region that undergo similar processing (see below). Notably, only APP, but not any of the other APP-related genes, contains sequence encoding the Aβ domain.
APP Processing
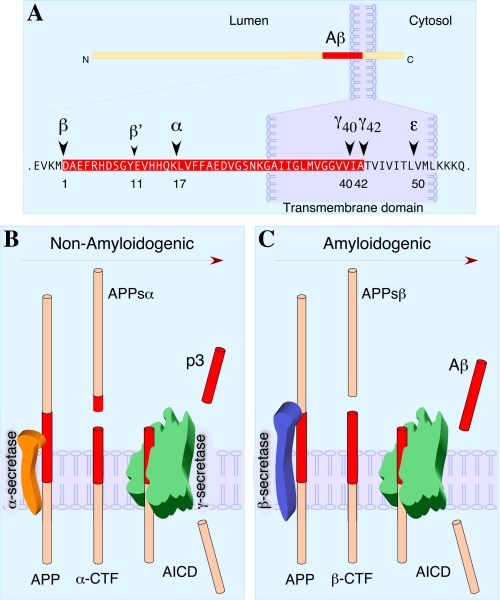
Two predicted cleavages, one in the extracellular domain (β-secretase cleavage) and the other in the transmembrane region (γ-secretase cleavage), are necessary to release Aβ from the precursor molecule (Fig. 1). APP is first cleaved within the lumenal domain by β-or α-secretase, resulting in the shedding of nearly the entire ectodomain and generation of membrane-tethered β-or α-C-terminal fragments, respectively. The β- and α-C-terminal fragments are subsequently cleaved within the transmembrane domain by γ-secretase to release Aβ (4 kDa) and p3 (3 kDa) peptides, respectively, into the extracellular milieu. In addition, γ-secretase cleavage generates a cytoplasmic polypeptide termed AICD.
FIGURE 1.
Proteolytic processing of APP. A, the schematic structure of APP is shown with the Aβ domain shaded in red and enlarged. The major sites of cleavage by α-, β-, and γ-secretases are indicated along with Aβ numbering from the N terminus of Aβ (Asp1). B, non-amyloidogenic processing of APP refers to sequential processing of APP by membrane-bound α- and γ-secretases. α-Secretase cleaves within the Aβ domain, thus precluding generation of intact Aβ peptide. The fates of N-terminally truncated Aβ (p3) and AICD are not fully resolved. C, amyloidogenic processing of APP is carried out by sequential action of membrane-bound β- and γ-secretases. CTF, C-terminal fragment.
APP Secretases—Several zinc metalloproteinases such as TACE/ADAM17, ADAM9, ADAM10 and MDC-9 and the aspartyl protease BACE2 can cleave APP at the α-secretase site, located within the Aβ domain between Lys16 and Leu17, essentially precluding the generation of intact Aβ (1). The major neuronal β-secretase is a transmembrane aspartyl protease termed BACE1 (β-site APP-cleaving enzyme 1) (2). Cleavage by BACE1 generates the N terminus of Aβ. In addition, BACE1 can also cleave within the Aβ domain between Tyr10 and Glu11 (β′-cleavage site). γ-Secretase is made of four essential subunits: presenilin-1 or -2, nicastrin, APH-1, and PEN-2 (3). γ-Secretase cleaves at multiple sites within the transmembrane domain of APP, generating Aβ peptides ranging in length from 38 to 43 residues (4). Nearly 90% of secreted Aβ ends in residue 40, whereas Aβ42 accounts for <10% of secreted Aβ. Moreover, minor amounts of shorter Aβ peptides such as Aβ38 and Aβ37 have also been detected. Familial AD-linked mutations in APP just beyond the C terminus of the Aβ domain increase Aβ42 production. Intriguingly, familial AD-linked mutations in presenilin-1 and -2 influence γ-secretase cleavage by elusive mechanisms that variably influence the cleavage site specificity, in general favoring cleavage at position 42 relative to that at position 40, thus increasing the Aβ42/40 ratio (4).
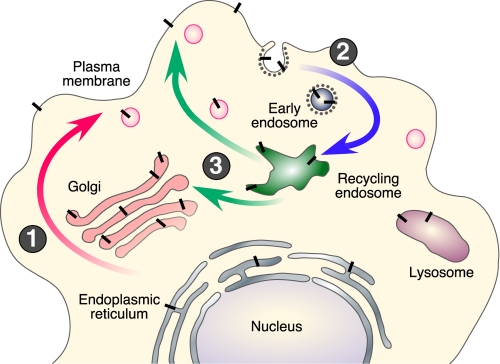
Intracellular Itinerary and Processing of APP—The pathways of APP trafficking are depicted in Fig. 2. During its transit from the ER to the plasma membrane, nascent APP is post-translationally modified by N- and O-glycosylation, ectodomain and cytoplasmic phosphorylation, and tyrosine sulfation. Only a small fraction of nascent APP molecules is present at the plasma membrane (estimated at ∼10% based on APP overexpression in cultured cells), whereas the majority of APP at steady state localizes to the Golgi and TGN. In non-neuronal cells, APP is internalized within minutes of arrival at the cell surface due to the presence of the YENPTY internalization motif near the C terminus of APP (residues 682–687 of the APP695 isoform). Following endocytosis, APP is delivered to endosomes, and a fraction of endocytosed molecules is recycled to the cell surface. Measurable amounts of internalized APP also undergo degradation in the lysosome.
FIGURE 2.
Intracellular trafficking of APP. Nascent APP molecules (black bars) mature through the constitutive secretory pathway (step 1). Once APP reaches the cell surface, it is rapidly internalized (step 2) and subsequently trafficked through endocytic and recycling compartments back to the cell surface (step 3) or degraded in the lysosome. Non-amyloidogenic processing occurs mainly at the cell surface, where α-secretases are present. Amyloidogenic processing involves transit through the endocytic organelles, where APP encounters β- and γ-secretases.
APP can be processed at the cell surface by α-secretase cleavage, resulting in the shedding of the APPsα ectodomain (5). Activation of protein kinase C increases APPsα secretion by mechanisms involving the formation and release of secretory vesicles from the TGN, thus enhancing APP (and possibly α-secretase) trafficking to the cell surface. BACE1 predominantly localizes to the late Golgi/TGN and endosomes, consistent with amyloidogenic cleavage of wild-type APP during endocytic/recycling steps (6). Available data indicate the presence of γ-secretase complex and enzyme activity in multiple compartments, including the ER, ER-Golgi intermediate compartment, Golgi, TGN, endosomes, and plasma membrane. Studies conducted in non-neuronal and neuroblastoma cell lines show that Aβ is generated mainly in the TGN and endosomes as APP is trafficked through the secretory and recycling pathways (Fig. 2) (7). Evidence converging from a number of studies also indicates that amyloidogenic processing occurs in cholesterol- and sphingolipid-enriched membrane raft microdomains of intracellular organelles (8–11). Further investigations are needed to clarify the mechanisms regulating raft association of APP and the secretases.
The high level neuronal expression of BACE1 preferentially channels APP through the amyloidogenic processing pathway in the brain. Moreover, in neurons, APP is trafficked anterogradely along peripheral and central axons and proteolytically processed during transit (12, 13). Axonal transport of APP is thought to be mediated by direct or indirect binding of APP to the kinesin light chain subunit of kinesin-1. It has also been proposed that APP may represent a kinesin cargo receptor, linking kinesin-1 to a unique subset of transport vesicles (14). However, this notion remains highly controversial (e.g. see Ref. 15). Nevertheless, the intracellular organelles/transport vesicles where Aβ is generated in neurons are not fully characterized.
Endocytic APP Sorting and Aβ Production—Mutations within the YENPTY endocytosis motif selectively inhibit APP internalization and decrease Aβ generation (16). This motif and the flanking region serve as the binding site for many cytosolic adaptors with phosphotyrosine-binding domains, including Fe65, Fe65L1, Fe65L2, Mint1 (also called X11α), Mint2, Mint3, Dab1, and JNK (c-Jun N-terminal kinase)-interacting protein family members. Overexpression of Mint1, Mint2, or Fe65 causes reduction in Aβ generation and deposition in the brains of transgenic mice, strongly suggesting a physiological role for these adaptors in regulating APP processing in the nervous tissue (17). Interestingly, Fe65 acts as a functional linker between APP and LRP (another type I membrane protein containing two NPXY endocytosis motifs) in modulating endocytic APP trafficking and Aβ production (18). A conformational change introduced by phosphorylation at Thr668 (14 amino acids proximal to the YENPTY motif) interferes with Fe65 binding to APP and facilitates BACE1 and γ-secretase cleavage of APP (19, 20). Moreover, Fe65 stabilizes the highly labile AICD, which may serve as a regulatory step in modulating the physiological function of AICD (see below). In addition to APP, Mint proteins can directly bind ADP-ribosylation factors; thus, Mint proteins can potentially regulate vesicular trafficking of APP by serving as coat proteins (21). Finally, the type I transmembrane protein SorLA/LR11 (a member of the VPS10p domain receptor family), which functionally interacts with cytosolic adaptors GGA and PACS-1, attenuates Aβ production by acting as a Golgi/TGN retention factor (22). SorLA/LR11 is also genetically associated with AD, thus further implicating this sorting molecule in APP biology (23).
APP Function
Trophic Properties—Since the discovery of APP, a number of physiological roles have been attributed to the molecule, some unique to certain isoforms, but its actual functions remain unclear. The literature covering APP function is extensive and cannot be reviewed comprehensively here (24). Suffice to say that a number of functional domains have since been mapped to the extra- and intracellular regions of APP. These include metal (copper and zinc) binding, extracellular matrix components (heparin, collagen, and laminin), neurotrophic and adhesion domains, and protease inhibition (the Kunitz protease inhibitor domain present in APP751 and APP770 isoforms). One of the earliest indications of APP function came from assessing the growth pattern of fibroblasts in which APP levels were decreased by expression of an antisense APP construct (25). These cells grew slowly, but the growth retardation could be restored by treatment with APPs. The active domain was subsequently mapped to a pentapeptide domain (RERMS) near the middle of the extracellular domain (positions 403–407) (26). The activity is not limited to fibroblasts, as infusion of this pentapeptide as well as APPs into brain resulted in increased synaptic density and improved memory retention in animals (27, 28). Because, as mentioned above, APPs is constitutively released from cells following α-secretase cleavage, these findings indicated that APP has autocrine and paracrine functions in growth regulation.
In all, a trophic role for APP has been perhaps the most consistently and arguably the best established function for the molecule. APP has been shown to stimulate neurite outgrowth from a variety of settings. This phenotype is compatible with the up-regulation of APP expression during neuronal maturation (29). The N-terminal heparin-binding domain of APP (residues 28–123) upstream from the RERMS sequence also stimulates neurite outgrowth and promotes synaptogenesis. Interestingly, the crystal structure of this domain shows similarities to known cysteine-rich growth factors (30). Conversely, injection of anti-APP antibodies directly into the brain led to impairment in behavioral tasks in adult rats (27). Not surprisingly, studies have also implicated a role for APPs in regulating stem cells. On one hand, APPsα induces the differentiation of neural stem cells into astrocytic lineage (31). On the other, in adult rodent brains, APPsα acts in concert with EGF to stimulate the proliferation of EGF-responsive neural stems cells in the subventricular zone (32). However, APPs is necessary but not sufficient for full activity, as it appears to act as a cofactor with EGF.
Cell Adhesion—An RHDS motif near the extralumenal portion of APP or at the C terminus of APPs lying within the Aβ region appears to promote cell adhesion. It is believed that this region acts in an integrin-like manner and can, accordingly, be blocked by RGDS peptide sequence derived from the fibronectin-binding domain (33). Similarly, APP colocalizes with integrins on the surface of axons and at sites of adhesion (34, 35). Evidence of interaction with laminin and collagen provides further evidence of adhesion-promoting properties. Interestingly, because the RHDS sequence is contained within the N terminus of Aβ, similar cell adhesion-promoting properties have also been attributed to the Aβ peptide itself. This latter property is, however, difficult to tease out in view of the cytotoxicity of Aβ peptide when tested in a variety of cell systems in vitro. Furthermore, it is difficult to separate the cell adhesion-from the neurite outgrowth-promoting roles of APP. Clearly, these are probably somewhat inseparable, as neuronal migration, neurite outgrowth, and even synaptogenesis would involve substrate adhesion. Consistent with this view, a recent study using short hairpin RNA silencing methodology in utero showed that APP is required for migration of neuronal precursors to the cortical plate; furthermore, this activity is mediated by Dab1 acting downstream of APP (36). The phenotypes of APP- and APLP-deficient animals are certainly in agreement with these proposed physiological activities of these molecules (see below).
Is APP a Receptor?—Although APP was initially proposed to act as a cell-surface receptor, the evidence supporting this idea has been unconvincing. Aside from interactions with extracellular matrix proteins, only recently has a candidate ligand been proposed. It was reported that F-spondin, a neuronally secreted signaling glycoprotein that may function in neuronal development and repair, binds to the extracellular domain of APP as well as APLP1 and APLP2 (37). This binding reduces β-secretase cleavage of APP and nuclear transactivation of AICD (see below), therefore suggesting that F-spondin may be a ligand that regulates APP processing.
As mentioned above, γ-secretase processing of APP also releases AICD (Fig. 1). This processing step is not unique for APP and indeed may be a rather generalized phenomenon whereby membrane-anchored proteins are cleaved either to release cytosolic fragments that participate in cell signaling, as in the Notch receptor, or for degradation (“proteasome of the membrane” as coined by Kopan and Ilagan (38)). Because APP undergoes the same γ-secretase membrane proteolysis as Notch, the analogy to Notch is simply too tempting or obvious, even though the evidence that APP is itself a cofactor for transcriptional activation within the nucleus remains to be firmly established. Using a heterologous signaling reporter system, AICD can form a transcriptionally active complex together with two other molecules, Fe65 and Tip60 (39). Although it was initially felt that AICD must enter the nucleus with Fe65, subsequent study showed that nuclear translocation of AICD is not required but may be indirect through Fe65 (40). An alternative approach to address this question is to look for AICD-activated candidate genes. In this regard, several genes have been proposed to date: KAI1 (a tumor suppressor gene), neprilysin (a neutral endopeptidase with Aβ-degrading activity), LRP1, and the EGF receptor (41–44). The activation of the EGF receptor and LRP is particularly interesting because it links APP to a number of in vivo phenotypes of presenilin activity and cholesterol metabolism, respectively. Finally, a recent study reported a surprising twist, viz. that APP in conjunction with TAG1, a molecule found in the outer plasma membrane, and presenilins act to suppress neurogenesis. Thus, both TAG1- and APP-deficient animals showed more neuroprogenitor cells than did wild-type animals. In this proposed pathway, it is the release of AICD that suppresses neurogenesis in a pathway that may be dependent on the binding to Fe65 because the NPTY motif of AICD is required. At present, it is unclear which proteins or genes are downstream of this pathway or whether this pathway is active in vivo (45).
APP-deficient Animals—In view of the above discussion, it is perhaps a little surprising then that with so many functions attributed to APP, the initial phenotype of APP-deficient mice obtained by gene targeting was rather unrevealing (46). These mice were lighter in body mass, and with age, there was weakness in the extremities. Examination of the brain revealed gliosis only, a rather nonspecific astrocytic reaction. Consistent with the trophic properties of APP described above, one report suggested that synaptic markers are reduced in APP-deficient mice (47), and this is correlated with deficits in learning and memory as well as in synaptic plasticity. Postnatal growth deficit was also noted in APLP1-deficient mice, but APLP2-deficient mice demonstrated no apparent phenotype. Interestingly, Aplp2–/–/Aplp1–/– and App–/–/Aplp2–/– double mutants, but not App–/–/Aplp1–/– animals, showed early postnatal lethality, indicating that members of the APP gene family are essential genes that exhibit partial overlapping functions (48). Curiously, the histopathological phenotype of the animals that displayed early lethality was also rather bland by initial descriptions. Similarly, neurons cultured from these animals were unaltered in their basal growth rates or response to excitotoxicity. However, in the peripheral nervous system, App–/–/ Aplp2–/– double knock-out animals exhibited poorly formed neuromuscular junction with reduced apposition of pre- and postsynaptic elements of the junctional synapses (49). The number of synaptic vesicles at the presynaptic terminals was also reduced, a finding confirmed by defective neurotransmitter release. With knowledge of the neuromuscular junction phenotypes of App–/–/Aplp2–/– mice in mind, examination of the parasympathetic submandibular ganglia of these animals also showed a reduction in active zone size, synaptic vesicle density, and number of docked vesicles per active zone (50). Furthermore, APP also regulates the presynaptic expression and activity of the high affinity choline transporter. In this setting, loss of APP leads to aberrant localization of the choline transporter at neuromuscular junctions (51). This function of APP/APLP in synapse formation is evolutionarily conserved, as evidenced by the decreased number of synaptic boutons in neuromuscular junctions of Drosophila larvae lacking Appl, and involves interaction of APPL with the cytosolic adaptor Mint and a transmembrane cell adhesion molecule named Fasciclin II (52).
In Caenorhabditis elegans, deficiency in the ortholog APL-1 disrupts molting and morphogenesis and results in larval lethality. Interestingly, the APL-1 lethality can be rescued by neuronal expression of only the extracellular domain of APL-1, analogous to the α-secretase-derived APPsα, suggesting that the predominant function of APL-1 originates from the secreted product. However, adding to this complexity, the human APP gene cannot substitute for the loss of APL-1, arguing that this function was not evolutionarily conserved (53).
Deficiency of all three App genes led to death shortly after birth. The majority of the animals showed cortical dysplasia suggestive of migrational abnormalities of the neuroblasts and partial loss of cortical Cajal-Retzius cells (54). Taken together, these findings presented a convincing picture that members of the APP gene family play essential roles in the development of the nervous system relating to synapse structure and function as well as in neuronal migration. Whether these abnormalities underlie the early postnatal survival of the animals remains to be established. Furthermore, whether these activities are due to mechanical properties or mediated by activating signaling pathways, or both, is an interesting question that remains to be elucidated.
Summary
This minireview has covered some of the salient aspects of APP biology, concentrating on the recent advances in processing, trafficking, and function of APP and related family members. The importance of APP in AD clearly lies in its role as a precursor to the Aβ peptide, which plays a central role in the amyloid hypothesis. However, APP has a number of additional biological activities, some of which impact neuronal development and function. Growing evidence suggests that perturbations of some of these activities may also contribute to AD pathogenesis and neurodegeneration. As such, it will be important to continue to investigate the normal function of APP.
This work was supported, in whole or in part, by National Institutes of Health Grants AG019070 and AG021495 (to G. T.) and AG12376 and AG032179 (to E. H. K.) from NIA. This work was also supported by the American Health Assistance Foundation Alzheimer's Disease Research Program and the Alzheimer's Association (to G. T.) and by the Glenn Foundation for Medical Research (to E. H. K.). This is the first article of eleven in the Thematic Minireview Series on the Molecular Basis of Alzheimer Disease. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: APP, amyloid precursor protein; AD, Alzheimer disease; Aβ, amyloid β-protein; AICD, APP intracellular domain; ER, endoplasmic reticulum; TGN, trans-Golgi network; APPs, secreted APP; LRP, lipoprotein receptor-related protein; EGF, epidermal growth factor; APLP, amyloid precursor-like protein.
References
- 1.Allinson, T. M., Parkin, E. T., Turner, A. J., and Hooper, N. M. (2003) J. Neurosci. Res. 74 342–352 [DOI] [PubMed] [Google Scholar]
- 2.Vassar, R. (2004) J. Mol. Neurosci. 23 105–114 [DOI] [PubMed] [Google Scholar]
- 3.Iwatsubo, T. (2004) Curr. Opin. Neurobiol. 14 379–383 [DOI] [PubMed] [Google Scholar]
- 4.Selkoe, D. J., and Wolfe, M. S. (2007) Cell 131 215–221 [DOI] [PubMed] [Google Scholar]
- 5.Sisodia, S. S. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 6075–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koo, E. H., and Squazzo, S. L. (1994) J. Biol. Chem. 269 17386–17389 [PubMed] [Google Scholar]
- 7.Small, S. A., and Gandy, S. (2006) Neuron 52 15–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riddell, D. R., Christie, G., Hussain, I., and Dingwall, C. (2001) Curr. Biol. 11 1288–1293 [DOI] [PubMed] [Google Scholar]
- 9.Ehehalt, R., Keller, P., Haass, C., Thiele, C., and Simons, K. (2003) J. Cell Biol. 160 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vetrivel, K. S., Cheng, H., Lin, W., Sakurai, T., Li, T., Nukina, N., Wong, P. C., Xu, H., and Thinakaran, G. (2004) J. Biol. Chem. 279 44945–44954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vetrivel, K. S., Cheng, H., Kim, S. H., Chen, Y., Barnes, N. Y., Parent, A. T., Sisodia, S. S., and Thinakaran, G. (2005) J. Biol. Chem. 280 25892–25900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo, E. H., Sisodia, S. S., Archer, D. R., Martin, L. J., Weidemann, A., Beyreuther, K., Fischer, P., Masters, C. L., and Price, D. L. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buxbaum, J. D., Thinakaran, G., Koliatsos, V., O'Callahan, J., Slunt, H. H., Price, D. L., and Sisodia, S. S. (1998) J. Neurosci. 18 9629–9637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamal, A., Stokin, G. B., Yang, Z., Xia, C. H., and Goldstein, L. S. (2000) Neuron 28 449–459 [DOI] [PubMed] [Google Scholar]
- 15.Lazarov, O., Morfini, G. A., Lee, E. B., Farah, M. H., Szodorai, A., DeBoer, S. R., Koliatsos, V. E., Kins, S., Lee, V. M., Wong, P. C., Price, D. L., Brady, S. T., and Sisodia, S. S. (2005) J. Neurosci. 25 2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez, R. G., Soriano, S., Hayes, J. D., Ostaszewski, B., Xia, W., Selkoe, D. J., Chen, X., Stokin, G. B., and Koo, E. H. (1999) J. Biol. Chem. 274 18851–18856 [DOI] [PubMed] [Google Scholar]
- 17.Miller, C. C., McLoughlin, D. M., Lau, K. F., Tennant, M. E., and Rogelj, B. (2006) Trends Neurosci. 29 280–285 [DOI] [PubMed] [Google Scholar]
- 18.Pietrzik, C. U., Yoon, I. S., Jaeger, S., Busse, T., Weggen, S., and Koo, E. H. (2004) J. Neurosci. 24 4259–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ando, K., Iijima, K. I., Elliott, J. I., Kirino, Y., and Suzuki, T. (2001) J. Biol. Chem. 276 40353–40361 [DOI] [PubMed] [Google Scholar]
- 20.Lee, M. S., Kao, S. C., Lemere, C. A., Xia, W., Tseng, H. C., Zhou, Y., Neve, R., Ahlijanian, M. K., and Tsai, L. H. (2003) J. Cell Biol. 163 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, K., Li, Y., Bennett, M., McKay, M., Zhu, X., Shern, J., Torre, E., Lah, J. J., Levey, A. I., and Kahn, R. A. (2003) J. Biol. Chem. 278 36032–36040 [DOI] [PubMed] [Google Scholar]
- 22.Schmidt, V., Sporbert, A., Rohe, M., Reimer, T., Rehm, A., Andersen, O. M., and Willnow, T. E. (2007) J. Biol. Chem. 282 32956–32964 [DOI] [PubMed] [Google Scholar]
- 23.Rogaeva, E., Meng, Y., Lee, J. H., Gu, Y., Kawarai, T., Zou, F., Katayama, T., Baldwin, C. T., Cheng, R., Hasegawa, H., Chen, F., Shibata, N., Lunetta, K. L., Pardossi-Piquard, R., Bohm, C., Wakutani, Y., Cupples, L. A., Cuenco, K. T., Green, R. C., Pinessi, L., Rainero, I., Sorbi, S., Bruni, A., Duara, R., Friedland, R. P., Inzelberg, R., Hampe, W., Bujo, H., Song, Y. Q., Andersen, O. M., Willnow, T. E., Graff-Radford, N., Petersen, R. C., Dickson, D., Der, S. D., Fraser, P. E., Schmitt-Ulms, G., Younkin, S., Mayeux, R., Farrer, L. A., and St George-Hyslop, P. (2007) Nat. Genet. 39 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattson, M. P. (1997) Physiol. Rev. 77 1081–1132 [DOI] [PubMed] [Google Scholar]
- 25.Saitoh, T., Sundsmo, M., Roch, J. M., Kimura, N., Cole, G., Schubert, D., Oltersdorf, T., and Schenk, D. B. (1989) Cell 58 615–622 [DOI] [PubMed] [Google Scholar]
- 26.Ninomiya, H., Roch, J. M., Sundsmo, M. P., Otero, D. A., and Saitoh, T. (1993) J. Cell Biol. 121 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meziane, H., Dodart, J. C., Mathis, C., Little, S., Clemens, J., Paul, S. M., and Ungerer, A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12683–12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roch, J. M., Masliah, E., Roch-Levecq, A. C., Sundsmo, M. P., Otero, D. A., Veinbergs, I., and Saitoh, T. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 7450–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung, A. Y., Koo, E. H., Haass, C., and Selkoe, D. J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 9439–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossjohn, J., Cappai, R., Feil, S. C., Henry, A., McKinstry, W. J., Galatis, D., Hesse, L., Multhaup, G., Beyreuther, K., Masters, C. L., and Parker, M. W. (1999) Nat. Struct. Biol. 6 327–331 [DOI] [PubMed] [Google Scholar]
- 31.Kwak, Y. D., Brannen, C. L., Qu, T., Kim, H. M., Dong, X., Soba, P., Majumdar, A., Kaplan, A., Beyreuther, K., and Sugaya, K. (2006) Stem Cells Dev. 15 381–389 [DOI] [PubMed] [Google Scholar]
- 32.Caille, I., Allinquant, B., Dupont, E., Bouillot, C., Langer, A., Muller, U., and Prochiantz, A. (2004) Development (Camb.) 131 2173–2181 [DOI] [PubMed] [Google Scholar]
- 33.Ghiso, J., Rostagno, A., Gardella, J. E., Liem, L., Gorevic, P. D., and Frangione, B. (1992) Biochem. J. 288 1053–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storey, E., Spurck, T., Pickett-Heaps, J., Beyreuther, K., and Masters, C. L. (1996) Brain Res. 735 59–66 [DOI] [PubMed] [Google Scholar]
- 35.Yamazaki, T., Koo, E. H., and Selkoe, D. J. (1997) J. Neurosci. 17 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young-Pearse, T. L., Bai, J., Chang, R., Zheng, J. B., LoTurco, J. J., and Selkoe, D. J. (2007) J. Neurosci. 27 14459–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho, A., and Sudhof, T. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2548–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopan, R., and Ilagan, M. X. (2004) Nat. Rev. Mol. Cell Biol. 5 499–504 [DOI] [PubMed] [Google Scholar]
- 39.Cao, X., and Sudhof, T. C. (2001) Science 293 115–120 [DOI] [PubMed] [Google Scholar]
- 40.Cao, X., and Sudhof, T. C. (2004) J. Biol. Chem. 279 24601–24611 [DOI] [PubMed] [Google Scholar]
- 41.Baek, S. H., Ohgi, K. A., Rose, D. W., Koo, E. H., Glass, C. K., and Rosenfeld, M. G. (2002) Cell 110 55–67 [DOI] [PubMed] [Google Scholar]
- 42.Pardossi-Piquard, R., Petit, A., Kawarai, T., Sunyach, C., Alves da Costa, C., Vincent, B., Ring, S., D'Adamio, L., Shen, J., Muller, U., St George-Hyslop, P., and Checler, F. (2005) Neuron 46 541–554 [DOI] [PubMed] [Google Scholar]
- 43.Liu, Q., Zerbinatti, C. V., Zhang, J., Hoe, H. S., Wang, B., Cole, S. L., Herz, J., Muglia, L., and Bu, G. (2007) Neuron 56 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y. W., Wang, R., Liu, Q., Zhang, H., Liao, F. F., and Xu, H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10613–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma, Q. H., Futagawa, T., Yang, W. L., Jiang, X. D., Zeng, L., Takeda, Y., Xu, R. X., Bagnard, D., Schachner, M., Furley, A. J., Karagogeos, D., Watanabe, K., Dawe, G. S., and Xiao, Z. C. (2008) Nat. Cell Biol. 10 283–294 [DOI] [PubMed] [Google Scholar]
- 46.Zheng, H., Jiang, M., Trumbauer, M. E., Sirinathsinghji, D. J., Hopkins, R., Smith, D. W., Heavens, R. P., Dawson, G. R., Boyce, S., Conner, M. W., Stevens, K. A., Slunt, H. H., Sisoda, S. S., Chen, H. Y., and Van der Ploeg, L. H. (1995) Cell 81 525–531 [DOI] [PubMed] [Google Scholar]
- 47.Dawson, G. R., Seabrook, G. R., Zheng, H., Smith, D. W., Graham, S., O'Dowd, G., Bowery, B. J., Boyce, S., Trumbauer, M. E., Chen, H. Y., Van der Ploeg, L. H., and Sirinathsinghji, D. J. (1999) Neuroscience 90 1–13 [DOI] [PubMed] [Google Scholar]
- 48.Anliker, B., and Muller, U. (2006) Neurodegener. Dis. 3 239–246 [DOI] [PubMed] [Google Scholar]
- 49.Wang, P., Yang, G., Mosier, D. R., Chang, P., Zaidi, T., Gong, Y. D., Zhao, N. M., Dominguez, B., Lee, K. F., Gan, W. B., and Zheng, H. (2005) J. Neurosci. 25 1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, G., Gong, Y. D., Gong, K., Jiang, W. L., Kwon, E., Wang, P., Zheng, H., Zhang, X. F., Gan, W. B., and Zhao, N. M. (2005) Neurosci. Lett. 384 66–71 [DOI] [PubMed] [Google Scholar]
- 51.Wang, B., Yang, L., Wang, Z., and Zheng, H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14140–14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashley, J., Packard, M., Ataman, B., and Budnik, V. (2005) J. Neurosci. 25 5943–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hornsten, A., Lieberthal, J., Fadia, S., Malins, R., Ha, L., Xu, X., Daigle, I., Markowitz, M., O'Connor, G., Plasterk, R., and Li, C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herms, J., Anliker, B., Heber, S., Ring, S., Fuhrmann, M., Kretzschmar, H., Sisodia, S., and Muller, U. (2004) EMBO J. 23 4106–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]