Abstract
The xanthine oxidoreductase gene (XOR) encodes an important source of reactive oxygen species and uric acid, and its expression is associated with various human diseases including several forms of cancer. We previously reported that basal human XOR (hXOR) expression is restricted or repressed by E-box and TATA-like elements and a cluster of transcriptional proteins, including AREB6-like proteins and DNA-dependent protein kinase (DNA-PK). We now demonstrate that the cluster contains the tumor suppressors SAFB1, BRG1, and SAF-A. We further demonstrate that SAFB1 silencing increases hXOR expression and that SAFB1 directly binds to the E-box. Multiple studies in vitro and in vivo including pulldown, immunoprecipitation and chromatin immunoprecipitation analyses indicate that SAFB1, Ku86, and BRG1 associate with each other. The results suggest that the SAFB1 complex binds to the hXOR promoter in a chromatin environment and plays a critical role in restricting hXOR expression via its direct interaction with the E-box, DNA-PK, and tumor suppressors. Moreover, we demonstrate that the cytokine, oncostatin M (OSM), induces the phosphorylation of SAFB1 and that the OSM-induced hXOR mRNA expression is significantly inhibited by silencing the DNA-PK catalytic subunit or SAFB1 expression. The present studies for the first time demonstrate that hXOR is a tumor suppressor-targeted gene and that the phosphorylation of SAFB1 is regulated by OSM, providing a molecular basis for understanding the role of SAFB1-regulated hXOR transcription in cytokine stimulation and tumorigenesis.
Xanthine oxidoreductase (XOR3; EC 1.1.3.22), a member of molybdoflavoprotein hydroxylases, is a rate-limiting enzyme in the catabolism of purines and a well defined source of reactive oxygen species (ROS) (1–3). Additionally, XOR has inorganic nitrate and nitrite reductase activities and generates nitric oxide (4). The enzyme is composed of two identical 150-kDa subunits, each of which contains four redox active centers; that is, two iron sulfur, one FAD, and one molybdopterin (1–3). It exists in two forms, an NAD-dependent dehydrogenase and an oxygen-dependent oxidase (XO). In mammals, the two forms are reversibly converted between each other by sulfhydryl oxidation, but the conversion of NAD-dependent dehydrogenase to XO by proteolytic modification is irreversible.
Beyond its role as a key enzyme in controlling purine metabolism, XOR plays important physiologic and pathologic roles by producing ROS and uric acid (UA). In physiologic conditions XOR-derived ROS function as mediators in signal transduction and gene transcription (5–8). XOR has been implicated in milk fat droplet enveloping and secretion, and regulation of cyclooxygenease-2, peroxisome proliferator-activated receptor γ, adipogenesis, and neuronal nitric-oxide synthase (NOS1)-controlled cardiac excitation-contraction coupling (9–12).
Importantly, a central role for XOR also has been demonstrated in a variety of clinical disorders and relevant animal models associated with oxidative stress or injury, including inflammation, infection, ischemic tissue injury, and various cancers (13–22). In acute renal failure, endotoxin-induced mucosal injury, viral pneumonia, ischemia-reperfusion injury, and cutaneous photosensitivity to hematoporphyrins, excess XOR-derived ROS have been demonstrated to cause tissue injury (15–22). In addition, XOR activity and gene expression are induced by LPS, cytokines, and hypoxia and during ischemia/reperfusion (23–27). Studies also indicate that dysregulation of XOR may be associated with heart failure, hypertension, and abnormal fat metabolism (11, 28).
ROS are known to attack DNA bases or deoxyribose residues to cause DNA damage or genetic mutations and in so doing play important roles in the development of cancer (29, 30). Previous studies demonstrate that XOR-derived ROS contribute to the increased cancer risk associated with oxidative stress (31, 32). As an example, XOR is involved in metabolism of alcohol and has been linked to the increased breast cancer risk in alcoholics. XOR-derived ROS may also contribute to the breast cancer caused by the prolonged exposure to environmental carcinogens (31–35). Recent studies show increased XOR activity in subjects with non-small cell lung cancer (36, 37).
In contrast, UA as a scavenger of ROS and a potent iron chelator (38–41) is a major antioxidant in plasma (38, 39, 42) and epithelial secretions (43). The high concentrations, however, predispose to hyperuricemia and crystalline deposition, the histopathologic hallmark of gout (44). Recent studies demonstrate that UA released from dying cells acts as a novel endogenous danger signal to alert the immune system by stimulating the maturation of dendritic cells and responses of T-cells to foreign antigens and by promoting tumor rejection (45, 46).
In view of the pathophysiologic importance of XOR-derived ROS and UA, it is not surprising that XOR activity is restricted in humans compared with rodents, at a level supporting longevity and health, as indicated by the fact that basal XOR activity is 100 times lower in humans than in nonprimate species (47–49). This suggests that XOR activity in humans must be controlled by unique mechanisms to preserve its activity for purine metabolism while minimizing the deleterious effects of XOR-derived ROS. To date, both post-translational and transcriptional mechanisms have been shown to play important roles in controlling human XOR (hXOR) activity. For post-translational mechanisms, loss of molybdenum, phosphorylation, and association with nitric-oxide synthase may partially account for the low activity (12, 50, 51). Our laboratory has focused on transcriptional mechanisms controlling hXOR by characterizing chromosomal location, genomic organization, and basal transcription (52–56). The studies demonstrate that the cis elements located in the hXOR promoter, including an E-box and TATA-like element, play critical roles in repressing basal promoter activity. A nuclear protein complex containing components of DNA-dependent protein kinase (DNA-PK, including Ku86, Ku70, and DNA-PK catalytic subunit (DNA-PKcs)) that participates in repair of damaged DNA and phosphorylation of transcription factors and AREB6-like proteins (proteins immuno-reactive to AREB6 antibody) interacts with the E-box of the hXOR promoter (55–59). Importantly, AREB6-like proteins play a key role in repressing XOR transcription by binding to the E-box (56). In the present study we identify and characterize the role of the “AREB6-like” proteins. The results demonstrate for the first time that hXOR is targeted by tumor suppressors including SAFB1, SAFA, and BRG1 in addition to DNA-PK. We also demonstrate that SAFB1 directly binds to the E-box and suppresses hXOR expression. Oncostatin M (OSM) increases the phosphorylation of SAFB1, which relieves basal suppression of hXOR expression, resulting in increased hXOR mRNA expression.
MATERIALS AND METHODS
Cells Culture and OSM Stimulation—PFSK-1, MDA-MB-231, and BEAS-2B (B2B) cells were used for the studies. They were obtained from ATCC and cultured according to ATCC recommendations with a supplement of 10% fetal bovine serum and penicillin/streptomycin. B2B cells were stimulated for 18 h by adding OSM (dissolved in DMSO) to a final concentration of 100 nm in the culture media with the addition of the same volume of DMSO as a control.
Purification of Nuclear Proteins Binding to the Probe EG—Nuclear extracts were prepared from cultured PFSK-1 cells as described previously (55, 60). Protein concentrations of the extracts were determined spectroscopically using Bio-Rad protein reagents. For DNA affinity purification the biotin-labeled DNA probe 3× EG (three repeats of probe EG, 47 bp for each repeat that contains the E-box, conjunct GTTTC, and Ku86 site) was generated by PCR amplification (56). The labeled probe was chemically conjugated to DYNAL streptavidin magnetic beads following the recommended protocol (DYNAL). The nuclear extracts were first desalted through a PD-10 column (Amersham Biosciences) and then purified by anion exchange using a Hi-Trap-Sepharose Q column following the recommended protocol (Amersham Biosciences). Nuclear proteins were incubated with the 3× EG-conjugated DYNAL beads. After three washes of the magnetic beads with buffer containing scrambled probes to remove the non-specifically bound proteins, specifically bound proteins were eluted in 100 μl of ice-cold buffer containing 20 mm Tris, pH 8.0, 1 mm EDTA, 10% glycerol, 1 mm dithiothreitol, 0.01% (v/v) Triton X-100, and 0.6 m NaCl. The eluted proteins and the molecular weight markers were mixed with 10 μl of 2 × Laemmli buffer, then boiled for 3 min, and while warm directly loaded onto SDS-polyacrylamide gels for electrophoresis. The proteins recovered from the gels were subject to NMR-mass spectrum analysis.
Expression of Recombinant Proteins—Recombinant GST-Ku86 and GST-Ku70 proteins were expressed in Escherichia coli using pGEX-2TK-Ku86 and pGEX-2TK-Ku70 vectors (gifts from Dr. Stephen Jackson, Cambridge, UK). Recombinant GST-SAFB1 C-terminal (SAFB1C, amino acid residues 400–916) was expressed in E. coli using a pGEX-2TK-SAFB1C vector modified from pGEX-2TK-SAFB1 (a gift from Dr. Oesterreich, Houston, TX). GST-fused AREB6 C-terminal (AREB6-C) and H-fragment (AREB6-H) were expressed using vectors modified from pGEX-3X-AREB6 (a gift from Dr. Kawakami, Kawachi, Japan) as described previously (56, 57). The recombinant proteins expressed in E. coli were purified using a GST purification kit (Amersham Biosciences). The purified proteins were further characterized and compared with their predicted molecular weights by SDS-polyacrylamide gel electrophoresis.
Electrophoretic Mobility Shift Assays (EMSA)—EMSA were employed to study the binding of recombinant SAFB1C, Ku86, and Ku70 proteins to DNA probes. For EMSA, the DNA probe EG (wild type) was used as a binding probe, whereas EG, EmG (mutant E-box) and Kum1 (mutant of the putative Ku86 binding site) were used as competitors, as described previously (55, 56). For gel-shift assays, 15–30 fmol of labeled binding probes were incubated for 30 min at room temperature with 200 ng of recombinant proteins in a 50-μl reaction mixture containing 4% glycerol, 10 mm Tris, pH 7.5, 10 mm NaCl, 0.5 mm dithiothreitol, 1 mm MgCl2, 0.5 mm EDTA, 0.05% Nonidet P-40, and 25 μg of bovine serum albumin plus 3 μg of poly[d(I-C)] or poly [d(A-T)] to reduce nonspecific binding. For competition studies, the samples were preincubated with unlabeled competitors for 10 min at room temperature before the addition of the labeled binding probes. For supershift analysis, incubation of EMSA reaction mixtures for 15 min at room temperature was followed by the addition of specific or irrelevant antibodies and incubation for an additional 30 min at room temperature. The reaction samples were electrophoresed and then transferred to nylon membranes by electroblotting. The oligonucleotides were fixed onto the membranes by baking at 100 °C for 30 min. The chemiluminescent signals were detected after using the DIG Gel Shift kit (Roche Applied Science) protocol.
Silencing the SAFB1 and DNA-PKcs Expression—To study the effect of silencing SAFB1 on hXOR expression in basal conditions, a pDS vector containing a cytomegalovirus promoter to express green fluorescent protein for fluorescent-activated cell sorting analysis and a U6 promoter for expression of siRNA was constructed by modifying vectors pcDNA3.1 (Invitrogen) and pU6 (Ambion). For preparation of DNA constructs (pDS-SAFB1) for SAFB1 silencing, synthesized oligonucleotides for RNA interference (siRNA) targeting SAFB1 were cloned into a pDS vector. The transfection of pDS-SAFB1 plasmids into MDA-MB-231 cells was performed using a Lipofectin Plus kit (Invitrogen) following the supplier's protocol. At 72 h after transfection, the cells were sorted using fluorescent-activated cell sorting. The silencing of SAFB1 expression in the sorted cells was confirmed using reverse transcription-PCR analysis. Cells transfected with the pDS plasmid (2 μg/well) containing insertion of nonspecific siRNA were used as a control.
To study the effects of silencing SAFB1 and DNA-PKcs on OSM-induced hXOR expression, double-stranded, chemically synthesized oligonucleotide mixtures (1 μm/well) for silencing SAFB1 or DNA-PKcs, purchased from Dharmacon, were used with relevant nonspecific oligonucleotide mixtures as controls (microarray-confirmed by the supplier). The transfections were performed in B2B cells using DharmaFECT® siRNA transfection reagents following the supplier's protocol.
Measurement of hXOR, SAFB1, and DNA-PKcs Transcripts—To determine the transcript levels, total RNA was isolated from cells using RNAgents Total RNA Isolation System (Promega) following the supplier's protocol and used for reverse transcription. For semiquantitative PCR analysis, the primers for amplification of XOR transcripts were described previously (55). The primer pairs 5′-TGACGGGGTCACCCACACTGTGCCCATCTA (forward) and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG (reverse) were used for amplification of β-actin transcripts (691 bp) and 5′-AATGGCGGAGACTCTGTCAG (forward) and 5′-CATCCCTCTTAAGGTTTGTAGA (reverse) for SAFB1 transcripts (1550 bp). The PCR conditions for amplification of XOR, SAFB1, and β-actin transcripts were the same as those for XOR (55) except that 30 cycles were used for amplification of β-actin. The PCR products were confirmed by sequencing analysis and electrophoresed on agarose gel. After ethidium bromide staining, the gel was imaged and subject to band density analysis. The β-actin density was used to standardize the levels of XOR and SAFB1 transcripts.
In addition, the real-time PCR was performed to quantify the expression of XOR, SAFB1, DNA-PKcs, and β-actin using Taq-Man Gene Expression assay kits (Applied Biosystems) following the supplier's protocol. The assay kits were Hs99999903_m1 for β-actin, Hs0016–1495_m1 for SAFB1, Hs01071747_m1 for XOR, and Hs00179161 for DNA-PKcs.
Western Blot Analysis and Protein Dephosphorylation—Western blot analysis of nuclear extracts was performed using an ECL kit (Amersham Biosciences) following the supplier's protocol. Except for SAFB1 (Upstate Biotechnology), all antibodies (Ku86, Ku70, DNA-PKcs, and BRG1) were purchased from Santa Cruz Biotechnology and used in a dilution range from 1:1000 to 1:3000. For determination of phosphorylated and nonphosphorylated SAFB1, the samples were treated with calf intestinal alkaline phosphatase (1 unit/sample) before the analysis.
GST Pulldown Assays and Immunoprecipitation—For GST pulldown, GST recombinant proteins were conjugated to glutathione-Sepharose beads (GE Healthcare). The conjugated beads were incubated with nuclear extracts at 4 °C for 2 h in a total volume of 400 μl of binding buffer (25 mm HEPES, pH 7.5, 10% glycerol, 150 mm NaCl, and 0.2 mm EDTA) containing 0.05% Triton X-100. After extensive washing with binding buffer, the beads were suspended in sample buffer for SDS-PAGE. Immunoprecipitation was performed using Ku86, SAFB1, or irrelevant (vimentin, Santa Cruz Biotechnology) antibodies following the protocol described in Bonifacino et al. (61). The precipitates were analyzed by immunoblotting as described above.
Chromatin Immunoprecipitation (ChIP) Assay—Approximately 2 × 108 PFSK-1 cells were collected for ChIP assays. Native protein-DNA complexes were cross-linked by formaldehyde treatment (1%) for 15 min. For ChIP assays, isolated chromatin was sonicated with 4–8 pulses for 15–20 s each using a Cole Parmer Ultrasonic Homogenizer 4710 with an output control set at 2.5. Equal aliquots of sonicated chromatin were used for immunoprecipitation with rabbit anti-DNA PKcs (Santa Cruz), Ku70 (Santa Cruz), Ku86 (Santa Cruz), or SAFB1 (Beth Lab) antibodies or rabbit IgG (negative control, Santa Cruz). The DNA fragments associated with immunoprecipitates were isolated and subject to PCR amplification with primers specific to the hXOR promoter sequences flanking the E-box/TATA-like element (forward, 5′-ACAGTCGCCTAGTGCCAAGTC; reverse, 5′-CGAACTCCAGGTACCTCACTC). 5 μg of each antibody were used for immunoprecipitation.
Statistical Studies—All experiments were repeated at least three times independently. Data are presented as the mean ± S.E.; statistical significance was determined by Student's t test. p values <0.05 were considered significant.
RESULTS
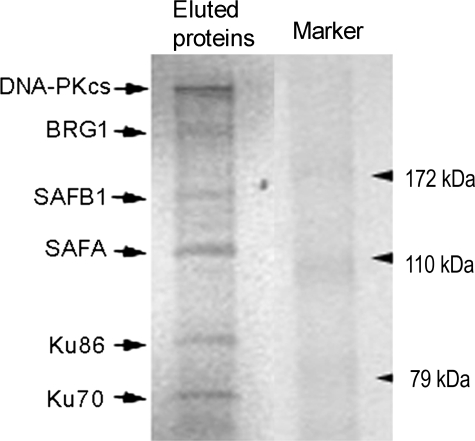
DNA Affinity Purification and Sequence Analysis of Eluted Nuclear Proteins Binding to Probe EG—Our previous studies demonstrated that components (Ku86, Ku70, and DNA-PKcs) of the DNA-PK complex and proteins immunoreactive to anti-AREB6 antibodies (AREB6-like proteins) bind to the hXOR promoter region containing the E-box and putative Ku86 sites (probe EG) (55). In the present investigation we further characterized the AREB6-like proteins from PFSK-1 nuclear extracts. Nuclear proteins binding to DNA probes containing three repeats of probe EG (probe 3× EG) conjugated to the DYNAL magnetic beads were eluted and separated by electrophoresis on a 10% polyacrylamide gel. Coomassie staining demonstrated that in addition to three known proteins, DNA-PKcs (350 kDa), Ku86 (86 kDa), and Ku70 (70 kDa), there were three unknown proteins with masses around 200, 150, and 120 kDa (Fig. 1). These unknowns were eluted and digested with trypsin to generate peptide fragments for sequencing. NMR-mass spectrum analysis of the peptides identified the unknown proteins as SAFA, SAFB1, and BRG1, as indicated in Fig. 1. The 120-kDa protein, SAFA, is consistent with its reported size (62). The 150-kDa protein is SAFB1, which has an aberrant migration on SDS-PAGE (apparent sizes 130∼150 kDa), although its calculated size is around 100 kDa (63). The 200 -Da protein is BRG1 (∼205 kDa) (64).
FIGURE 1.
Magnetic DNA affinity purification of nuclear proteins binding to probe 3× EG. The PFSK-1 nuclear proteins binding to the probe were eluted after purification with probe 3× EG-conjugated DYNAL magnetic beads and analyzed by electrophoresis. Six bands with sizes of around 350, 200, 150, 120, 86, and 70 kDa were observed.
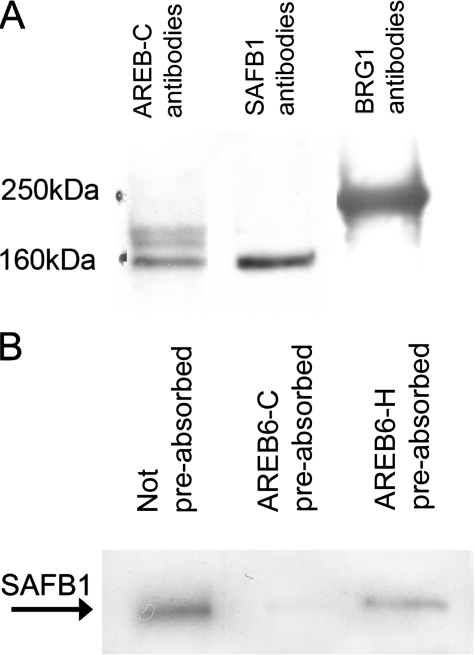
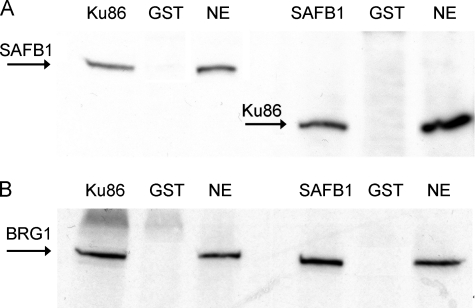
Proteins Immunoreactive to SAFB1 and BRG1 Antibodies Are Present in the Eluted Proteins and SAFB1 Is the AREB6-like Protein—To further confirm the NMR results, we used Western blotting to examine for the presence of the intact proteins in eluted nuclear extracts. These experiments demonstrated that proteins immunoreactive to SAFB1 and BRG1 antibodies were indeed present in the PFSK-1-eluted proteins (Fig. 2A) and in nuclear extracts from other cell types including B2B and MDA-MB-231 cells (data not shown). The presence of SAFA in the eluted nuclear proteins was not examined because a specific antibody is not currently available.
FIGURE 2.
A, detection of the eluted proteins from probe 3× EG-conjugated DYNAL magnetic beads using AREB6-C, SAFB1, and BRG1 antibodies. B, detection of SAFB1 was blocked by preabsorption of SAFB1 antibodies with AREB6-C extracts but not AREB6-H.
We previously reported the presence of AREB6-like proteins in the complex binding to the E-box/Ku86 region. However, NMR-mass spectrum analysis demonstrated no AREB6 in the eluted extracts, suggesting that SAFB1 and/or BRG1 are the AREB6-like proteins identified due to the cross-immunoreactivity between the proteins. Sequence analysis indicated that AREB6-C contains three zinc finger domains with the highest homology (60%) to the central region of SAFB1 that has an RNA recognition motif, possibly causing the cross-immunoreactivity of SAFB1 with AREB6-C antibody. As shown in Fig. 2A, AREB6-C antiserum detected a protein with a size similar to SAFB1 in the eluted proteins, suggesting that SAFB1 reacts with AREB6-C antibodies. This finding was further supported by results (Fig. 2B) showing that the detection of SAFB1 was blocked by preabsorption of SAFB1 antibodies with AREB6-C but not with AREB6-H peptides.
The Loss of SAFB1 Function Increases hXOR mRNA Expression—To study the functional role of SAFB1 in repressing basal hXOR expression, we next determined the effect of silencing SAFB1 on hXOR mRNA expression. In these experiments transfected MDA-MB-231 cells were sorted using fluorescent-activated cell sorting. In Fig. 3A we demonstrate that SAFB1 transcript expression was significantly lower in cells transfected with pTDS-SAFB1 siRNA compared with that in control cells transfected with pTDS-nonspecific siRNA, indicating that the SAFB1-targeted siRNA silenced SAFB1 expression. The silencing effect on SAFB1 protein levels was shown in Fig. 8B. The effect of silencing SAFB1 on hXOR expression was determined using reverse transcription-PCR. Fig. 3B shows that hXOR mRNA levels were substantially higher in cells with the silenced SAFB1 expression compared with those in the control cells.
FIGURE 3.
A, a representative image showing that SAFB1 expression is inhibited by SAFB1-targeted siRNA (SAFB1 siRNA) but not by the nonspecific siRNA (control). In addition, the expression of hXOR transcripts was greater in the cells with the silenced SAFB1 expression than that in the control cells. B, statistical results of three independent experiments (n = 3 for each experiment). *, indicates p < 0.05.
FIGURE 8.
Western blot images demonstrating that the phosphorylation of SAFB1 is up-regulated by OSM. A, the representative images showing that OSM (100 nm) increased the phosphorylated SAFB1 (P) that was dephosphorylated by calf intestinal alkaline phosphatase (CIAP) treatment but not the non-phosphorylated CIAP (non-P). B, the representative images showing that phosphorylated and nonphosphorylated SAFB1 were reduced by SAFB1-silencing (siRNA) but not by the nonspecific silencing (control). β-Actin was detected as a loading control.
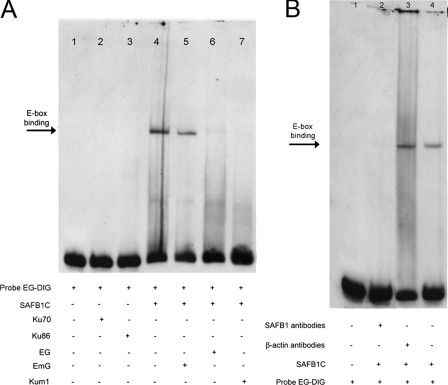
SAFB1 Binds to the hXOR Promoter—Previous studies indicated binding of AREB-6-like proteins to the E-box (56). Therefore, we surmised that SAFB1 could bind to the E-box. To assess this possibility, we determined the ability of recombinant SAFB1C to bind to the hXOR promoter. The binding specificity to the E-box was analyzed using DNA competitors. As shown in Fig. 4A, SAFB1C bound to the hXOR promoter (lane 4). The binding was competed by either wild-type (lane 6) or mutant Ku86 binding site competitors (lane 7). The competitor with a mutant E-box was unable to compete for the binding (lane 5). Moreover, supershift analysis (Fig. 4B) showed that the binding was blocked by SAFB1 antibodies but not by β-actin antibodies. Collectively, the results indicate that SAFB1 binds to the E-box.
FIGURE 4.
SAFB1C binds to the E-box. A, EMSA were performed using recombinant SAFB1C proteins. Probe EG contains both E-box and putative Ku86 sites. Binding to the E-box is indicated by an arrow and was specifically competed by the unlabeled probes EG (lane 6) or Kum1 (probe with mutant Ku86 site, lane 7), whereas the mutations of the E-box (probe EmG, lane 5) lost the ability to specifically compete. In comparison, Ku86 (lane 3) and Ku70 (lane 2) did not bind to the probe EG. B, lane 4 shows binding of SAFB1C to probe EG, as indicated by an arrow. The binding was blocked by SAFB1 antibodies (lanes 2) but not by nonspecific antibodies (lane 3).
Ku86 Alone Does Not Bind to the hXOR Promoter Containing the E-box/Ku86 Site—Our previous studies demonstrated a repressive effect of Ku86 on hXOR expression and its presence in the nuclear complex binding to the E-box/Ku86 region (55, 56). However, it still is not known whether the function of Ku86 is related to its direct binding to the putative Ku86 site. To address this issue, we assessed the ability of Ku86 to bind to the putative Ku86 site. As shown in Fig. 4A, neither Ku86 nor Ku70 binds to the putative Ku86 site (lanes 2 and 3). The results indicate that the direct binding of Ku86 to the promoter by itself likely does not occur.
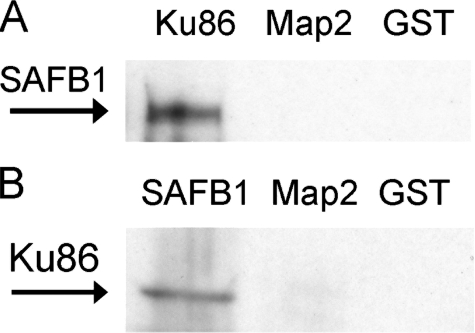
Ku86 Interacts with SAFB1 in Vitro—Our previous studies demonstrated protein-protein interactions between Ku86 and AREB6-like proteins (56). Therefore, if SAFB1 represents the AREB6-like protein, interactions between Ku86 and SAFB1 should be observed. To test this possibility we used recombinant Ku86 and SAFB1C for in vitro pulldown analyses. As shown in Fig. 5, SAFB1 in the nuclear extracts was pulled down by GST-Ku86 proteins. In addition, Ku86 was pulled down by GST-SAFB1. These results confirm interactions between Ku86 and SAF-B1.
FIGURE 5.
Representative immunoblots showing pulldown analysis of protein-protein interaction between SAFB1 and Ku86 proteins. A, nuclear proteins immunoreactive to SAFB1 antibodies, as indicated by an arrow, were pulled down by GST-Ku86 proteins. B, nuclear proteins immunoreactive to Ku86 antibodies, as indicated by an arrow, were pulled down by GST-SAFB1C proteins. Map2 and GST were used as negative controls.
Nuclear SAFB1, Ku86, and BRG1 Are Coimmunoprecipitated—To examine whether protein-protein interactions are present in vivo, we performed coimmunoprecipitation analyses using Ku86 and SAFB1 antibodies. The GST antibodies were used as nonspecific antibody controls. Fig. 6A shows the presence of SAFB1 and BRG1 in the coimmunoprecipitates with Ku86 antibodies, indicating interactions between Ku86, SAFB1, and BRG1 in vivo. Moreover, Ku86 and BRG1 coimmunoprecipitated with SAFB1 (Fig. 6B). The results establish the interaction between SAFB1, Ku86, and BGR1.
FIGURE 6.
Coimmunoprecipitation analysis of SAFB1, Ku86, and BRG1. A, nuclear proteins immunoreactive to SAFB1 and BRG1 antibodies, as indicated by the arrows, were present in the coimmunoprecipitates with Ku86 antibodies. B, nuclear proteins immunoreactive to Ku86 and BRG1 antibodies, as indicated by the arrows, were present in the coimmunoprecipitates with SAFB1 antibodies. GST was used as a negative control. SAFB1, Ku86, and BRG1 in the nuclear extracts (NE) were used as the positive controls.
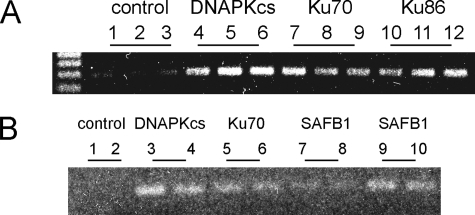
SAFB1, DNA-PKcs, Ku86, and Ku70 Are Associated with the hXOR Promoter in Chromatin—To determine whether the protein complex binds to the hXOR promoter in a chromatin environment, ChIP assays were performed in PFSK-1 cells. Cross-linked chromatin was immunoprecipitated with various rabbit antibodies or with rabbit IgG as a negative control, and PCR was used to amplify a 286-bp fragment of the hXOR promoter (–306 to –20). The data show that DNA-PKcs, Ku70, Ku86, and SAFB1 bind to the promoter (Fig. 7). Binding of BRG1 to the promoter was not observed (data not shown). These results demonstrate that the protein complex is associated with the hXOR promoter.
FIGURE 7.
Two independent representative ChIP assays of DNA-PKcs, Ku70, Ku86, and SAFB1 interactions with hXOR promoter in PFSK-1 cells. Bands are the PCR products (a 286-bp fragment of the hXOR promoter). A and B are images showing two independent results. In A each antibody essay was performed in triplicate (1–3, 4–6, 7–9, 10–12), whereas in B each was performed in duplicate (1 and 2, 3 and 4, 5 and 6, 7 and 8, 9 and 10).
SAFB1 Mediates the OSM-induced XOR mRNA Expression—Previous studies have shown that XOR mRNA expression is stimulated by tumor necrosis factor, interferon-γ, interleukin-6 and -1 in bovine epithelial cells (24). In the present investigation we sought to determine whether cytokine stimulation of hXOR expression was due to a loss of SAFB1-controlled basal repression. To test this possibility, we first examined the effects of OSM, a member of interleukin-6 family, on hXOR expression in human lung epithelial cells (B2B). The results using real-time quantitative PCR analysis show that OSM induced hXOR expression 3-fold (3.05 ± 0.44, n = 3) compared with the control. We then determined whether SAFB1 mediates OSM-induced hXOR mRNA expression by examining the effect of OSM on hXOR mRNA expression in B2B cells with SAFB1 expression silenced by chemically synthesized siRNAs. The results demonstrate that the siRNAs reduced the SAFB1 mRNA expression by 85 ± 1% (n = 3) in comparison with the control mixture. In B2B cells with silenced SAFB1 expression, OSM only moderately increased the hXOR mRNA expression (1.52 ± 0.47-fold, n = 3). This indicates that silencing the SAFB1 expression attenuates the OSM-induced hXOR expression, suggesting that SAFB1 mediates the OSM-induced hXOR mRNA expression.
OSM Stimulates Phosphorylation of SAFB1—Many tumor suppressors are regulated by phosphorylation. Therefore, it is possible that phosphorylation of SAFB1 mediates the OSM-induced hXOR expression. We examined this possibility by determining the effects of OSM on SAFB1 phosphorylation. As shown in Fig. 8, SAFB1 has both phosphorylated (P) and non-phosphorylated (non-P) forms, which was confirmed by calf intestinal alkaline phosphatase treatment and SAFB1 silencing. NMR analysis showed that the phosphorylation occurs at the serine residue of SAFB1 at amino acid residue 604 (data not shown). Fig. 8A demonstrates that OSM increased the phosphorylation of SAFB1. In addition, we found that the SAFB1 expression was not induced by OSM (data now shown). The results support the possibility that OSM induces hXOR expression by stimulating phosphorylation of SAFB1, resulting in a release of its basal repression of hXOR transcription.
Role of DNA-PKcs in OSM-induced hXOR mRNA Expression—To investigate whether DNA-PKcs is involved in OSM-induced hXOR mRNA expression, we studied the effect of silencing DNA-PKcs on hXOR in the B2B cells with or without OSM stimulation. First, we determined the silencing efficacy of the siRNA targeting to the DNA-PKcs with a nonspecific siRNA mixture as a control. The quantitative real-time PCR analysis showed that the siRNA inhibits DNA-PKcs mRNA expression ∼5-fold (4.92 ± 1.04, n = 5) compared with the control (p < 0.01). Moreover, this silencing reduced basal hXOR mRNA expression by ∼17% (0.87 ± 0.23-fold, n = 4) compared with the control (1.04 ± 0.17-fold, n = 4, p < 0.01). Importantly, under the silencing conditions, OSM was unable to induce hXOR mRNA (1.03 ± 0.08-fold, n = 4) compared with the control (0.96 ± 0.10-fold, n = 4). These results indicate that DNA-PKcs contributes to the regulation of hXOR expression particularly after OSM stimulation.
DISCUSSION
hXOR Is a Newly Identified Gene Targeted by Tumor Suppressors—Our previous studies demonstrate that hXOR is transcriptionally suppressed in basal conditions. A promoter region containing an E-box and a putative Ku86 site (E-box/Ku86) plays a key role in the basal repression. A protein cluster binding to the region includes three components (Ku86, Ku70, and DNA-PKcs) of the DNA-PK and AREB6-like proteins that share immunogenicity with the repressive transcriptional factor, AREB6 (55, 56). In the present study we demonstrate that a complex of six proteins (350, 200, 150, 120, 86, and 70 kDa) binds to the E-box/Ku86 region. NMR-mass spectrum analysis showed that in addition to the previously identified DNA-PKcs, Ku86 and Ku70, the tumor suppressors SAFB1, BRG1, and SAFA bind to the E-box/Ku86 region. Western blot analysis further demonstrated the presence of SAFB1 and BRG1 in nuclear extracts from various cell types (data not shown) in addition to their presence in the eluted nuclear proteins. The absence of AREB6 in the eluted proteins suggested that SAFB1, SAFA, or BRG1 might interact with the AREB6-C antibodies that were used in our previous study to characterize the components in the protein cluster. This possibility was confirmed by the results demonstrating that SAFB1 and AREB6-C antibodies both recognize proteins with a similar size and that the detection of SAFB1 was blocked by preabsorption of SAFB1 antibodies with AREB6-C extracts. More importantly, ChIP analysis further supported the presence of the tumor suppressor SAFB1 in the nuclear protein complex associated with hXOR promoter in the chromatin environment.
Tumor suppressors play crucial roles in determining susceptibility to cancers by regulating the expression of their target genes. Understanding the function and mechanisms of tumor suppressor-controlled gene expression is important for improving strategies for cancer prevention, diagnosis, and treatment. SAFB1 and BRG1 are recently identified scaffold proteins acting as components of chromatin. Mutation of SAFB1 or BRG1 occurs in various tumors, including breast and lung cancers (65, 66). However, the downstream genes targeted by these proteins are unknown. Our results for the first time indicate that hXOR is a gene targeted by the tumor suppressors, SAFB1 and BRG1.
Tumor suppressors generally function as repressive transcriptional factors limiting the expression of targeted or downstream genes including various oncogenes (such as Ras, C-MYC, and RET genes) that encode mediators of signaling pathways important for controlling cell growth, proliferation, and apoptosis (67–70). A mutation of a tumor suppressor leads to the activation of oncogenes, thereby causing neoplasia. A series of genetic alterations that can be either hereditary or caused by various environmental factors result in the inactivation of tumor suppressor genes and susceptibility to cancer (71–75). The present investigation provides a new paradigm supporting the possibility that genetic alterations of tumor suppressors increases the susceptibility to cancer by releasing the repression on ROS producers such as XOR. This is consistent with our unpublished observation showing that in a subpopulation of liver and breast cancers, XOR expression is markedly increased when compared with normal liver or breast tissues.
Repressive Effect of SAFB1 on hXOR Expression and Its Molecular Basis—Previous studies have demonstrated that mutation of the E-box significantly increases hXOR promoter activity and that AREB6-like proteins bind to the E-box, suggesting that these proteins may repress hXOR expression (55). Therefore, we expected that SAFB1, as an AREB6-like protein, might repressively control hXOR expression. This prediction was confirmed in the present investigation by the results demonstrating that silencing SAFB1 expression significantly increases hXOR expression. We next demonstrated that SAFB1C specifically binds to the E-box. The findings lead us to conclude that SAFB1 represses hXOR expression by directly binding to the E-box of the gene.
In addition to the results demonstrating that the E-box is a critical cis element in controlling hXOR, our previous study demonstrated that a functional loss of Ku86 enhanced hXOR promoter activity and mRNA expression, suggesting that Ku86 contributes to the repressive regulation of hXOR. Moreover, we observed that mutation of the Ku86 binding site does not significantly alter hXOR promoter activity (55, 56). These results indicate that the repressive function of Ku86 in controlling hXOR expression may be accounted for by its interactions with other components of the protein cluster binding to the E-box. Alternatively, its interaction with the putative Ku86 site on the promoter may require the presence of other protein components (e.g. SAFB1). This possibility is supported by two findings. 1) Ku86 did not directly bind to the putative Ku86 site by itself. However, our preliminary results indicate that in the presence of SAFB1, Ku86 binds to the promoter (data not shown). 2) SAFB1, Ku86, and BRG1 associate as a protein complex in vitro and in vivo.
Combining the results of our previous and current studies, we have identified the proteins binding to the E-box/Ku86 region of hXOR, including all three components of DNA-PK and three tumor suppressors, SAFA, SAFB1, and BRG1. Based on the findings, a modified working model is presented to illustrate the function of these proteins and related cis elements in the regulation of hXOR expression by the E-box region of the promoter (Fig. 9). In the model the transcription factors include DNA-PK, identified tumor suppressors, co-repressors, TFIID, and other unknown proteins. Among the proteins, SAFB1 directly binds to the E-box and may function as a core protein in repressing hXOR, as supported by the result showing that the silencing of SAFB1 expression elicits a significant increase of hXOR mRNA levels. Other components within the protein cluster such as Ku86 and BRG1 control hXOR expression by acting as partners via interaction with the key component, SAFB1. This model for the protein complex binding to the hXOR promoter in chromatin environment is supported by the ChIP assay results.
FIGURE 9.
A model illustrating the molecular basis for regulation of the hXOR promoter. In this model SAFB1 binds to the consensus E-box (ACAGGTG) and restricts hXOR promoter activity and interacts with other co-repressors, such as Ku86, SAFA, and BRG1. Ku86 is associated with Ku70 and DNA-PKcs. Based on EMSA, other unknown transcription factors that may be involved in the regulation of hXOR are labeled with capital letters (A–C).
SAFB1 Mediates the Stimulation of XOR Expression by OSM—The induction of XOR mRNA expression by cytokines has been demonstrated previously (24). However, the molecular basis for this induction is not known. Based on the model, in the present investigation we determined the effect of OSM on hXOR mRNA expression and SAFB1. The results show that OSM increases phosphorylated SAFB1 but does not affect the overall SAFB1 expression level. In addition, the effect of OSM on XOR expression was weaker than that of SAFB1 silencing. These results suggest that SAFB1 silencing and OSM affect XOR expression through different mechanisms. The effect of SAFB1 knockdown observed under basal conditions reflects the repressive effect of SAFB1 on XOR expression, whereas the effect of OSM is likely attributed to its induction of SAFB1 phosphorylation and not the repression of SAFB1 expression. The results also indicate that phosphorylation of SAFB1 plays an important role in mediating the induction of XOR mRNA expression in response to OSM stimulation. This is supported by the results showing that silencing the kinase DNA-PKcs significantly reduces the SAFB1-mediated increase of hXOR expression in response to OSM induction, which suggests that DNA-PKcs has a central role in OSM-induced phosphorylation of SAFB1. Our scenario is consistent with previous results showing that DNA-PKcs acts as a regulator of gene transcription by phosphorylation of transcription factors in addition to its well known roles in DNA repair and that DNA-PK associating with tumor suppressors in a cluster may regulate the activity of the suppressors via DNA-PKcs-modulated phosphorylation (76–81). Therefore, the most attractive possibility is that DNA-PKcs, present with SAFB1 in the nuclear complex, regulates the phosphorylation of SAFB1. This in turn affects the interaction of SAFB1 with protein partners and/or binding to the DNA.
In summary, the present study for the first time demonstrates that hXOR is a gene targeted by tumor suppressors. The concept that a gene encoding an enzyme that catalyzes purine metabolism is a target of tumor suppressors is novel and suggests a new pathway by which tumor suppressors control cell growth. Both ROS and UA have been implicated in tumorigenesis (31–37, 46). Equally important, mutations of SAFB1 and BRG1 have been reported in various tumors, including breast and lung cancers. Our results provide a molecular basis for further studying the role of hXOR in SAFB1 and BRG1 mutation-associated cancers and the importance of phosphorylation of tumor suppressors in hXOR-associated diseases including carcinogenesis.
Acknowledgments
We thank Drs. Jackson, Oesterreich, and Kawakami for plasmids pGEX-2TK-Ku70, pGEX-2TK-SAFB1 and pGEX-3X-AREB6.
This work was supported, in whole or in part, by National Institutes of Health Grant 5RO1HL40665. This work was also supported by the Veterans Administration Research Service. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: XOR, xanthine oxidoreductase; hXOR, human XOR; XO, oxygen-dependent oxidase; UA, uric acid; B2B, BEAS-2B; EMSA, electrophoretic mobility shift assays; DNA-PK, DNA-dependent protein kinase; DNA-PKcs, DNA-PK catalytic subunit; ROS, reactive oxygen species; OSM, oncostatin M; GST, glutathione S-transferase; siRNA, small interfering RNA; EG, 47 bp probe containing E-box, conjunct GTTTC and Ku 86 site.
References
- 1.Massey, V., Brumby, P. E., and Komai, H. (1969) J. Biol. Chem. 244 1682–1691 [PubMed] [Google Scholar]
- 2.Waud, W. R., and Rajagopalan, K. V. (1976) Arch. Biochem. Biophys. 172 365–379 [DOI] [PubMed] [Google Scholar]
- 3.Amaya, Y., Yamazaki, K., Sato, M., Noda, K., Nishino, T., and Nishino, T. (1990) J. Biol. Chem. 265 14170–14175 [PubMed] [Google Scholar]
- 4.Harrison, R. (2002) Free Radic. Biol. Med. 33 774–779 [DOI] [PubMed] [Google Scholar]
- 5.Knaus, U. G., Heyworth, P. G., Evans, T., Curnutte, J. T., and Bokoch, G. M. (1991) Science 254 1512–1515 [DOI] [PubMed] [Google Scholar]
- 6.Sulciner, D. J., Irani, K., Yu, Z.-X., Ferrans, V. J., Goldschmidt-Clermont, P. J., and Finkel, T. (1996) Biochem. J. 318 379–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyamoto, Y., Akaike, T., Yoshida, M., Goto, S., Horie, H., and Maeda, H. (1996) Proc. Soc. Exp. Med. 211 366–373 [DOI] [PubMed] [Google Scholar]
- 8.Sen, C. K., and Packer, L. (1996) FASEB J. 10 709–720 [DOI] [PubMed] [Google Scholar]
- 9.Vorbach, C., Scriven, A., and Capecchi, M. R. (2002) Genes Dev. 16 3223–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtsubo, T., Rovira, I. I., Starost, M. F., Liu, C., and Finkel, T. (2004) Circ. Res. 95 1118–1124 [DOI] [PubMed] [Google Scholar]
- 11.Cheung, K. J., Tzameli, I., Pissios, P., Rovira, I., Gavrilova, O., Ohtsubo, T., Chen, Z., Finkel, T., Flier, J. S., and Friedman, J. M. (2007) Cell Metab. 5 115–128 [DOI] [PubMed] [Google Scholar]
- 12.Khan, S. A., Lee, K., Minhas, K. M., Gonzalez, D. R., Raju, S. V., Tejani, A. D., Li, D., Berkowitz, D. E., and Hare, J. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15944–15948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambrosone, C. B. (2000) Antioxid. Redox Signal. 2 903–917 [DOI] [PubMed] [Google Scholar]
- 14.Kang, D. H. (2002) AACN Clin. Issues 13 540–549 [DOI] [PubMed] [Google Scholar]
- 15.Diamond, J. R., Bonventre, J. V., and Karnovsky, M. J. (1986) Kidney Int. 29 478–483 [DOI] [PubMed] [Google Scholar]
- 16.Kawamura, T., Yoshika, T., Bills, T., Fogo, A., and Ichikawa, I. (1991) Kidney Int. 40 291–301 [DOI] [PubMed] [Google Scholar]
- 17.Linas, S. L., Wittenburg, D., and Repine, J. E. (1990) Am. J. Physiol. 258 F711–F716 [DOI] [PubMed] [Google Scholar]
- 18.Paller, M. S., and Neumann, T. V. (1991) Kidney Int. 40 1041–1049 [DOI] [PubMed] [Google Scholar]
- 19.Rodell, T. C., Cheronis, J. C., Ohnemus, C. L., Piermattie, D. J., and Repine, J. E. (1987) J. Appl. Physiol. 63 2159–2163 [DOI] [PubMed] [Google Scholar]
- 20.Akaike, T., Ando, M., Oda, T., Doi, T., Ijiri, S., Araki, S., and Maeda, H. (1990) J. Clin. Investig. 85 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granger, D. N. (1988) Am. J. Physiol. 255 H1269–H1275 [DOI] [PubMed] [Google Scholar]
- 22.Athar, M., Elmet, C. A., Bickers, D. R., and Mukhtar, H. (1989) J. Clin. Investig. 83 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont, G. P., Huecksteadt, T. P., Marshall, B. C., Ryan, U. S., Michael, J. R., and Hoidal, J. R. (1992) J. Clin. Investig. 89 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeffer, K. D., Huecksteadt, T. P., and Hoidal, J. R. (1994) J. Immunol. 153 1789–1797 [PubMed] [Google Scholar]
- 25.Falciani, F., Ghezzi, P., Terao, M., Cazzaniga, G., and Garattini, E. (1992) Biochem. J. 285 1001–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassoun, P. M., Yu, F. S., Shedd, A. L., Zulueta, J. J., Thannickal, V. J., Lanzillo, J. J., and Fanburg, B. L. (1994) Am. J. Physiol. 266 L163–L171 [DOI] [PubMed] [Google Scholar]
- 27.Sanders, K. A., Huecksteadt, T., Xu, P., Sturrock, A. B., and Hoidal, J. R. (1999) Chest. 116 (suppl. 1) 56–61 [DOI] [PubMed] [Google Scholar]
- 28.Berry, C. E., and Hare, J. M. (2004) J. Physiol. (Lond.) 555 589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dizdaroglu, M., Jaruga, P., Birincioglu, M., and Rodriguez, H. (2002) Free Radic. Biol. Med. 32 1102–1115 [DOI] [PubMed] [Google Scholar]
- 30.Klaunig, J. E., and Kamendulis, L. M. (2004) Annu. Rev. Pharmacol. Toxicol. 44 239–267 [DOI] [PubMed] [Google Scholar]
- 31.Wright, R. M., McManaman, J. L., and Repine, J. E. (1999) Free Radic. Biol. Med. 26 348–354 [DOI] [PubMed] [Google Scholar]
- 32.Chae, Y. H., Ji, B. Y., Lin, J. M., Fu, P. P., Cho, B. P., and El-Bayoumy, K. (1999) Toxicology 12 180–186 [DOI] [PubMed] [Google Scholar]
- 33.Castro, G. D., Delgado de Layno, A. M., Costantini, M. H., and Castro, J. A. (2001) Toxicology 160 11–18 [DOI] [PubMed] [Google Scholar]
- 34.Ritter, C. L., Decker, R. W., and Malejka-Giganti, D. (2000) Toxicology 13 793–800 [DOI] [PubMed] [Google Scholar]
- 35.Ritter, C. L., Culp, S. J., Freeman, J. P., Marques, M. M., Beland, F. A., and Malejka-Giganti, D. (2002) Toxicology 15 536–544 [DOI] [PubMed] [Google Scholar]
- 36.Kaynar, H., Meral, M., Turhan, H., Keles, M., Celik, G., and Akcay, F. (2005) Cancer Lett. 227 133–139 [DOI] [PubMed] [Google Scholar]
- 37.Tsao, S. M., Yin, M. C., and Liu, W. H. (2007) Nutr. Cancer 59 8–13 [DOI] [PubMed] [Google Scholar]
- 38.Ames, B. N., Cathcart, R., Schwiers, E., and Hochstein, P. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 6858–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howell, R. R., and Wyngaarden, J. B. (1960) J. Biol. Chem. 235 3544–3551 [PubMed] [Google Scholar]
- 40.Kellogg, E. W., III, and Fridovich, I. (1977) J. Biol. Chem. 252 6721–6729 [PubMed] [Google Scholar]
- 41.Davis, K. J., Sevanian, A., Muakkassah-Kelly, S. F., and Hoschstein, P. (1986) Biochem. J. 235 747–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becker, B. F., Reinhoklz, N., Ozcelik, T., Leipert, B., and Gerlach, E. (1989) Pfluegers Arch. 415 127–138 [DOI] [PubMed] [Google Scholar]
- 43.Peden, D. B., Hohman, R., Brown, M. E., Mason, R. T., Berkebile, C., Fales, H. M., and Kaliner, M. A. (1990) Proc. Natl. Acad. Sci. U. S. A. 97 7638–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slot, O. (1994) Ugeskr Laeger. 156 2396–2401 [PubMed] [Google Scholar]
- 45.Shi, Y., Evans, J. E., and Rock, K. L. (2003) Nature 425 516–521 [DOI] [PubMed] [Google Scholar]
- 46.Hu, D. E., Moore, A. M., Thomsen, L. L., and Brindle, K. M. (2004) Cancer Res. 64 5059–5062 [DOI] [PubMed] [Google Scholar]
- 47.Wajner, M., and Harkness, R. A. (1989) Biochim. Biophys. Acta 991 79–84 [DOI] [PubMed] [Google Scholar]
- 48.Muxfeldt, M., and Schaper, W. (1987) Basic Res. Cardiol. 82 486–492 [DOI] [PubMed] [Google Scholar]
- 49.Abadeh, S., Killacky, J., Benboubetra, M., and Harrison, R. (1992) Biochim. Biophys. Acta 1117 25–32 [DOI] [PubMed] [Google Scholar]
- 50.Harrison, R. (1997) Biochem. Soc. Trans. 25 786–791 [DOI] [PubMed] [Google Scholar]
- 51.Sanders, S., Eisenthal, R. S., and Harrison, R. (1997) Eur. J. Biochem. 245 541–548 [DOI] [PubMed] [Google Scholar]
- 52.Xu, P., Huechsteadt, T. P., Harrison, R., and Hoidal, J. R. (1994) Biochem. Biophys. Res. Commun. 199 998–1004 [DOI] [PubMed] [Google Scholar]
- 53.Xu, P., Zhu, X. L., Huecksteadt, T. P., Brothman, A., and Hoidal, J. R. (1994) Genomics 23 289–291 [DOI] [PubMed] [Google Scholar]
- 54.Xu, P., Huecksteadt, T. P., and Hoidal, J. R. (1994) Genomics 34 173–180 [Google Scholar]
- 55.Xu, P., Lavallee, P., and Hoidal, J. R. (2000) J. Biol. Chem. 275 5918–5926 [DOI] [PubMed] [Google Scholar]
- 56.Xu, P., LaVallee, P. A., Lin, J. J., and Hoidal, J. R. (2004) J. Biol. Chem. 279 16057–16063 [DOI] [PubMed] [Google Scholar]
- 57.Ikeda, K., and Kawakami, K. (1995) Eur. J. Biochem. 233 73–82 [DOI] [PubMed] [Google Scholar]
- 58.Smith, G. C. M., and Jackson, S. P. (1999) Genes Dev. 13 916–934 [DOI] [PubMed] [Google Scholar]
- 59.Schild-Poulter, C., Pope, L., Giffin, W., Kochan, J. C., Ngsee, J. K., Traykova-Andonova, M., and Hache, R. J. G. (2001) J. Biol. Chem. 276 16848–16856 [DOI] [PubMed] [Google Scholar]
- 60.Andrews, N. C., and Faller, D. V. (1991) Nucleic Acids Res. 19 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonifacino, J. S., Dell'Agelica, E. C., and Springer, T. A. (2001) Curr. Protoc. Mol. Biol., Chapter 10, Section VI, Unit 10.16, online [DOI] [PubMed]
- 62.Romig, H., Fackelmayer, F. O., Renz, A., Ramsperger, U., and Richter, A. (1992) EMBO J. 11 3431–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Renz, A., and Fackelmayer, F. O. (1996) Nucleic Acids Res. 24 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hendricks, K. B., Shanahan, F., and Lees, E. (2004) Mol. Cell. Biol. 24 362–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oesterreich, S., Allredi, D. C., Mohsin, S. K., Zhang, Q., Lee, A. V., Osborne, C. K., and O'Connell, P. (2001) Br. J. Cancer. 84 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reisman, D. N., Sciarrotta, J., Wang, W., Funkhouser, W. K., and Weissman, B. E. (2003) Cancer Res. 63 560–566 [PubMed] [Google Scholar]
- 67.Kopnin, B. P. (2000) Biochemistry (Mosc) 65 2–27 [PubMed] [Google Scholar]
- 68.Elend, M., and Eilers, M. (1999) Curr. Biol. 9 936–938 [DOI] [PubMed] [Google Scholar]
- 69.Donovan, J., and Slingerland, J. (2000) Breast Cancer Res. 2 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sears, R. C., and Nevins, J. R. (2002) J. Biol. Chem. 277 11617–11620 [DOI] [PubMed] [Google Scholar]
- 71.Gasco, M., Shami, S., and Crook, T. (2002) Breast Cancer Res. 4 70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pharoah, P. D., Day, N. E., and Caldas, C. (1999) Br. J. Cancer. 80 1968–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosen, E. M., Fan, S., Pestell, R. G., and Goldberg, I. D. (2003) J. Cell. Physiol. 196 19–41 [DOI] [PubMed] [Google Scholar]
- 74.Ross, J. S., Linette, G. P., Stec, J., Clark, E., Ayers, M., Leschly, N., Symmans, W. F., Hortobagyi, G. N., and Pusztai, L. (2003) Expert Rev. Mol. Diagn. 3 573–585 [DOI] [PubMed] [Google Scholar]
- 75.Fu, Y. P., Yu, J. C., Cheng, T. C., Lou, M. A., Hsu, G. C., Wu, C. Y., Chen, S. T., Wu, H. S., Wu, P. E., and Shen, C. Y. (2003) Cancer Res. 63 2440–2446 [PubMed] [Google Scholar]
- 76.Giffin, W., Torrance, H., Saffran, H., MacLeod, H. L., and Hache, R. J. (1994) J. Biol. Chem. 269 1449–1459 [PubMed] [Google Scholar]
- 77.Giffin, W., Gong, W., Schild-Poulter, C., and Hache, R. J. (1999) Mol. Cell. Biol. 19 4065–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finnie, N., Gottlieb, T., Hartley, K., and Jackson, S. P. (1993) Biochem. Soc. Trans. 21 930–935 [DOI] [PubMed] [Google Scholar]
- 79.Decker, T., and Kovarik, P. (2000) Oncogene 19 2628–2637 [DOI] [PubMed] [Google Scholar]
- 80.Cowell, I. G. (1994) Trends Biochem. Sci. 19 38–42 [DOI] [PubMed] [Google Scholar]
- 81.Hanna-Rose, W., and Hansen, U. (1996) Trends Genet. 12 229–234 [DOI] [PubMed] [Google Scholar]