Abstract
Nonclassical secondary cell wall polysaccharides constitute a major cell wall structure in the Bacillus cereus group of bacteria. The structure of the secondary cell wall polysaccharide from Bacillus cereus ATCC 10987, a strain that is closely related to Bacillus anthracis, was determined. This polysaccharide was released from the cell wall with aqueous hydrogen fluoride (HF) and purified by gel filtration chromatography. The purified polysaccharide, HF-PS, was characterized by glycosyl composition and linkage analyses, mass spectrometry, and one- and two-dimensional NMR analysis. The results showed that the B. cereus ATCC 10987 HF-PS has a repeating oligosaccharide consisting of a →6)-α-GalNAc-(1→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→ trisaccharide that is substituted with β-Gal at O3 of the α-GalNAc residue and nonstoichiometrically acetylated at O3 of the N-acetylmannosamine (ManNAc) residue. Comparison of this structure with that of the B. anthracis HF-PS and with structural data obtained for the HF-PS from B. cereus type strain ATCC 14579 revealed that each HF-PS had the same general structural theme consisting of three HexNAc and one Hex residues. A common structural feature in the HF-PSs from B. cereus ATCC 10987 and B. anthracis was the presence of a repeating unit consisting of a HexNAc3 trisaccharide backbone in which two of the three HexNAc residues are GlcNAc and ManNAc and the third can be either GlcNAc or GalNAc. The implications of these results with regard to the possible functions of the HF-PSs are discussed.
The Bacillus cereus is a group of Gram-positive bacteria that includes Bacillus anthracis, B. cereus, and Bacillus thuringiensis strains. Members of this group are very closely related. In fact, on the basis of detailed phylogenetic analysis, it has been suggested that they all may belong to a single species (1). Despite the very close relatedness of B. cereus group members, there is considerable pathogenic variability. Some members of this group are not pathogenic, whereas others are opportunistic pathogens causing a range of conditions on a variety of hosts. Bacillus thuringiensis is an insect pathogen, and B. cereus is normally a soil-dwelling bacterium, which in rare cases, causes, in humans, usually nonfatal cases of food poisoning, sepsis, endophthalmitis, and occasionally severe or fatal pneumonia. One member, B. anthracis, causes anthrax in animals and humans and is considered a high threat bioterrorism agent.
This variability in pathogenicity and host range is largely attributed to the plasmid content of the B. cereus group members, which can vary in size and number (2). For example, the crystal toxin genes of B. thuringiensis are carried on a plasmid (3), and plasmids pXO1 and pXO2 contain the genes required for the production of the B. anthracis toxin proteins and γ-polyglutamate capsule, respectively (4, 5). Further, recent B. cereus isolates from cases of severe and fatal pneumonia were found to have the pXO1 plasmid (6, 7), and another report showed that B. cereus or B. thuringiensis isolates from cases of “anthrax-like” disease in gorillas contain both pXO1 and pXO2 (8, 9).
For numerous bacterial pathogens, both Gram-positive and Gram-negative, cell wall polysaccharides are known virulence factors. However, little work has been done on the cell wall polysaccharides from members of the B. cereus group. We recently showed that the polysaccharides released from the cell walls from members of the B. cereus group using aqueous hydrogen fluoride (HF)3 have carbohydrate compositions that vary qualitatively in a manner that is correlated, at least in part, to phylogenetic relatedness as determined by multilocus sequence typing (10). That work also revealed quantitative glycosyl composition differences that indicated that structural variation can occur in the cell wall carbohydrates among B. cereus strains belonging to the same clade and lineage. Interestingly, it was shown that the B. cereus isolates which caused severe or fatal pneumonia and contained the pXO1 plasmid had HF-released polysaccharide (HF-PS) glycosyl compositions that closely resembled that of B. anthracis (10), indicating that HF-PS structural conservation plays a role in pathogenic function. Further work has shown that antiserum against B. anthracis cross-reacts to some extent with the HF-PS from these B. cereus isolates, indicating that they are structurally related to the B. anthracis HF-PS.4 Future publications will describe the structures of the HF-PSs from these B. cereus isolates.
Recent structural determination of B. anthracis HF-PS showed that it was composed of a trisaccharide backbone consisting of two GlcNAc residues and one N-acetylmannosamine (ManNAc) residue and that this backbone is variably substituted with terminal Gal residues (10, 12). This structure falls into the class of nonclassical secondary cell wall polymers (SCWP) of Gram-positive bacteria, as defined by Schäffer and Messner (13), that are covalently attached to the peptidoglycan through a phosphate or pyrophosphate bond. The S-layer proteins are anchored to the cell wall by binding to the SCWP polysaccharide via a carbohydrate-binding domain known as the S-layer homology (SLH) domain (14). Secondary cell wall polysaccharide structures have been determined for a number of Gram-positive bacteria, including some B. cereus strains (13). Older reports described that B. cereus AHU 1356 produced a neutral carbohydrate composed of GlcNAc, ManNAc, GalNAc, and Glc (15). In addition, an acidic carbohydrate composed of GlcNAc, Gal, rhamnose (Rha), glycerol, and phosphate was also identified in this strain (16). Because of the considerable importance of bacilli with regard to public health, a more complete picture of these carbohydrates is needed. Structural determination of the HF-PSs is necessary as the first step into structure-function studies of these SCWP polysaccharides as well as to answer questions about their suitability for developing new and/or improved vaccines and diagnostic agents.
In order to determine the relationship of these polysaccharides with pathogenicity, it is necessary to systematically characterize their structures. In this effort, we have initially selected the HF-PS from pathogenic B. anthracis and compared it with the HF-PSs from two normally nonpathogenic strains, the dairy isolate B. cereus ATCC 10987 and the B. cereus type strain ATCC 14579. We selected B. cereus strain ATCC 10987 and the B. cereus type strain ATCC 14579, since the genome of strain B. cereus ATCC 10987 is 93.7%, similar to that of B. anthracis and 90.9% similar to the genome of the B. cereus type strain, ATCC 14579, and since strain ATCC 10987 also contains a plasmid that is similar to the B. anthracis pXO1 virulence plasmid but lacks the pathogenicity island (17). We have already published, in this journal, the structure of the HF-PSs from B. anthracis Ames, Sterne, and Pasteur (12).
Here, we report the structure of the HF-PS from B. cereus strain ATCC 10987 as well as partial structural data for the HF-PS from the B. cereus type strain ATCC 14579 and compare those structures with that of the HF-PS from B. anthracis. We have included the partial structural data of the HF-PS from B. cereus type strain ATCC 14579, since these data, together with the complete structure of the B. cereus ATCC 10987 described here and that reported for B. anthracis (12), reveal that the HF-PSs from these strains contain unique structural features that are present on a possibly B. cereus-conserved structural scaffold. The implications of these results with regard to possible functions are discussed.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Cultural Conditions—B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579 were provided from the Centers for Disease Control and Prevention culture collection. Cultures were grown were grown as described in our previous report (12).
Preparation of Bacillus Cell Wall Polysaccharides—The bacterial cell walls were prepared using a modified procedure described by Brown (18) as we previously reported (10, 12). The polysaccharides were released from the isolated cell walls by treatment with aqueous HF and purified by gel permeation chromatography, as described in our previous report (12).
Composition and Glycosyl Linkage Analysis of the Cell Wall Polysaccharides—Glycosyl composition analysis was done by the preparation and gas chromatography-mass spectrometric analysis of trimethylsilyl methyl glycosides (19). The trimethylsilyl methyl glycosides were analyzed by combined gas chromatography-mass spectrometry, as previously reported (12). The glycosyl linkage analysis was performed according to a modification of the method of Ciucanu and Kerek (20), as described in our previous report (12).
De-O-acetylation of the Cell Wall Polysaccharides—The HF-PS was deacetylated by mild hydrazinolysis. Approximately, 4 mg of sample was treated with 200 μl of anhydrous hydrazine (Pierce) at 37 °C for 1 h. The sample was cooled on an ice bath, and 2 ml of cold acetone (–70 °C) was added dropwise with mild shaking. The mixture was kept overnight at –4 °C for complete precipitation of de-O-acetylated HF-PS. The de-O-acetylated HF-PS was collected by centrifugation at 10,000 × g for 10 min at 7 °C. The supernatant was discarded, and precipitate was washed with cold acetone (–70 °C), followed by centrifugation. (This step was repeated two more times.) Finally, the precipitate was dried under nitrogen flow, dissolved in water, and lyophilized.
NMR Analysis—The HF-PS sample (2–3 mg) was dissolved in 0.5 ml of regular grade deuterium oxide (D2O 98.5%) and lyophilized. This process was done twice to exchange the hydroxyl and amide protons with deuterium. The sample was finally dissolved in 0.5 ml of 100% D2O (Cambridge Isotopes) and transferred to a 5-mm NMR tube. All one- and two-dimensional NMR spectra were acquired with a 600-MHz Varian Inova instrument using the standard software supplied by Varian. 1H-1H homonuclear two-dimensional experiments were done after perfect 90° pulse calibration and 3.5 K spectral width in both dimensions; however, 1H-13C HSQC data were acquired taking 3.5 and 12 K in direct and indirect dimensions, respectively.
Mass Spectrometry—Mass spectral analysis of the isolated HF-PS was determined using a matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometer from Applied Biosystems. The HF-PS was dissolved in a 1:1 mixture of methanol/water and mixed in equal proportion (v/v) with 0.5 m 2,5-dihydroxybenzoic acid as the matrix. About 0.7 μl of the mixture was loaded on each spot on a stainless steel MALDI plate and air-dried. The spectra were acquired in either the linear or reflectron-positive modes using a 337-nm N2 laser with acceleration voltage of 20 kV.
RESULTS
Isolation and Initial Analysis; Glycosyl Composition and Linkage Analysis—The HF-PS from the investigated B. cereus strains eluted as a single major peak within the void volume of the Bio-Gel P2 column; the peak fractions were collected, lyophilized, and used for detailed structural analysis. The HF-PS composition from B. anthracis Ames consisted of Gal, GlcNAc, and ManNAc in an approximate 3:2:1 ratio, as previously described for four B. anthracis strains (Ames, Pasteur, Sterne 34F2, and UT60) (10, 12). The composition of the HF-PS from B. cereus ATCC 10987 consisted of Gal, ManNAc, GlcNAc, and GalNAc in a 1.6:1.5:1.0:1.4 ratio (Table 1). A small amount of Glc was present due to contamination of the HF-PS preparation with a Glc-rich polysaccharide, as previously reported (10). The composition of B. cereus ATCC 14579 HF-PS consisted of Glc, ManNAc, GlcNAc, and GalNAc in approximately a 1.8:1.5:3.1:1.0 ratio (Table 1). Thus, the differences in glycosyl compositions of these two B. cereus HF-PSs shows that their structures are not identical to each other and that they also differ from that of the B. anthracis HF-PS, as reported earlier (10, 12). Moreover, these two B. cereus HF-PSs contain GalNAc, which is not present in the B. anthracis HF-PS, and the B. cereus ATCC 14579 HF-PS contains Glc, which is not found in either the B. cereus ATCC 10987 or in the B. anthracis HF-PSs. These results are consistent with our earlier report (12), in which proton NMR analysis also showed that these HF-PSs have different structures.
TABLE 1.
The glycosyl compositions of the HF-PS preparations from B. cereus strains ATCC 10987 and ATCC 14579
The compositions are given as relative mass percentage of total carbohydrate. ND, not determined.
|
Strain
|
Sugar composition
|
|||||
|---|---|---|---|---|---|---|
| Man | Glc | Gal | ManNAc | GlcNAc | GalNAc | |
| % | % | % | % | % | % | |
| B. cereus ATCC 10987 | ND | 8.6 | 26.7 | 25.3 | 16.4 | 23.0 |
| B. cereus ATCC 14579 | ND | 25.3 | ND | 15.4 | 44.9 | 14.4 |
Methylation analysis showed that the B. cereus ATCC 10987 HF-PS consisted of terminally linked Gal, 3,6-linked GalNAc, 4-linked ManNAc, and 4-linked GlcNAc. Minor components included terminally linked GlcNAc, ManNAc, and GalNAc, indicating the possibility of molecular microheterogeneity within the HF-PS preparation. The presence of minor amounts of these terminal HexNAc residues is probably due to variously sized HF-PS molecules that differ in their nonreducing terminal glycosyl residues. They are present as minor components, since the HF-PS consists of variously sized molecules of up to 7759 daltons (as shown by MS analysis and described further below) and, therefore, the HexNAc residues at the nonreducing termini of these differently sized molecules found in the HF-PS preparation would be present in small amounts relative to the glycosyl residues in the multiple repeat units.
For the B. cereus ATCC 14579 HF-PS, methylation analysis showed terminally linked Glc, terminally linked GlcNAc, 4-linked GlcNAc, 4-linked ManNAc, 6-linked GalNAc, 3,4-linked ManNAc, and 3,4,6-linked GlcNAc. Exact quantification of the partially methylated alditol acetates (PMAAs) in these samples was not possible due to the resistance to acid hydrolysis of the aminoglycosyl glycoside bonds in the permethylated HF-PS. The complexity of the glycosyl linkages in B. cereus ATCC 14579 HF-PS compared with the HF-PSs from B. anthracis and B. cereus ATCC 10987 indicates that it is significantly more heterogeneous and structurally complex.
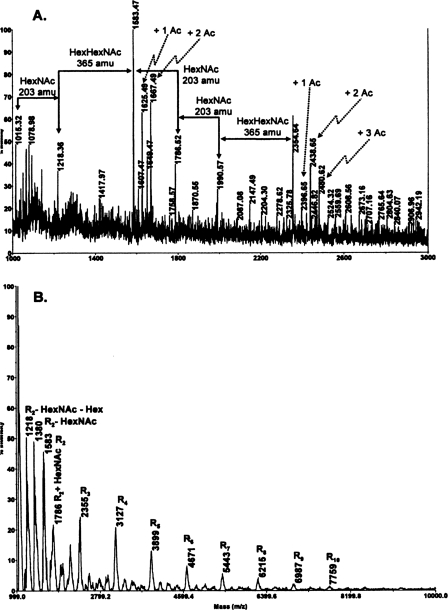
Structural Determination of the B. cereus ATCC 10987 HF-PS—The MALDI-TOF/TOF MS mass spectrum of the B. cereus ATCC 10987 HF-PS is shown in Fig. 1. The results show that this HF-PS preparation consists of a microheterogeneous mixture of molecules of varying sizes. The mass spectrum, obtained in the reflectron mode, shown in Fig. 1A reveals two ion clusters with the most intense ion in each cluster being the [M + Na]+ ions observed at m/z 1583.5 and at m/z 2354.6, respectively. The reflectron mode of analysis emphasizes lower molecular weight forms of the HF-PS; however, there are also higher molecular weight forms of the HF-PS, and these forms could be observed after de-O-acetylation of the HF-PS and MS analysis in the linear mode (shown in Fig. 1B and described further below). The difference between the m/z 2354.6 and 1583.5 ions, 772 mass units, exactly matches the mass of a (HexNAc)3(Hex)1 tetramer. These results suggest that the m/z = 1583.5 ion is due to an octasaccharide composed of two (HexNAc)3(Hex)1 tetrameric repeating units, and that the ion at m/z = 2354.6 corresponds to a molecule with three tetrameric repeat units. Based on glycosyl composition results described above, probable compositions for these masses are given in Table 2. In addition to these major ions for the two ion clusters, several other ions were observed. The ions at m/z 1625.5 and 1667.5 are 42 and 84 mass units greater, respectively, than the m/z 1583.5 ion, indicating that these ions are due to mono- and di-O-acetylation, respectively, of the ditetrasaccharide repeat structure. The O-acetyl groups, when present, are located on the ManNAc residues, as determined by NMR analysis, which is described below. The ions at m/z 1607.5 and m/z 1649.5 are due to minor amounts of dehydrated (–18 mass units) versions of these acetylated molecules. We also observe ions at m/z 1786.5 and 1990.6, which are consistent with one and two additional HexNAc residues, respectively, to the m/z 1583.5 ion. The difference between the m/z 1990.6 ion and the m/z 2354.6 ion is 365 mass units. This difference is consistent with the addition of a HexHexNAc disaccharide to the 1990.6 molecule.
FIGURE 1.
MALDI-TOF MS positive mode spectra of the HF-PS (A) and de-O-acetylated HF-PS (B) from B. cereus ATCC 10987. The proposed compositions for the various ions are given in Table 2. The spectrum shown in A was obtained in the reflectron mode, whereas the spectrum shown in B was obtained in the linear mode. R2, two (HexNAc)3(Hex)1 repeat units; R3, three repeat units, etc.
TABLE 2.
Proposed structures for the various molecular ions observed during MALDI-TOF MS analysis of the HF-PS from B. cereus ATCC 10987
The proposed structures are consistent with the glycosyl composition, linkage, and NMR data.
| Repeat units | Proposed structure | Observed [M + Na]+ ion |
|---|---|---|
| R1 + 1 | [ManNAc-GlcNAc(Gal)GalNAc]-ManNAc | 1015.3 |
| R1 + 2 | [ManNAc-GlcNAc-(GalGalNAc]-ManNAc-GlcNAc | 1218.4 |
| R2 + 0 | [ManNAc-GlNAc-(Gal)GalNc]-[ManNAc-GlcNc-(Gal)GalNAc] | 1583.5 |
| R2a + 0 + 1Ac | [(Ac)ManNAc-lcNAc-(Gal)GalAc]-[ManNAc-GlcAc-(Gal)GalNAc] | 1625.5 |
| R2a + 0 + 2Ac | [(Ac)ManNAc-GlNAc-(Gal)GalNc]-[(Ac)ManNAc-GcNAc-(Gal)GalNAc] | 1667.5 |
| R2 + 1 | [ManNAc-GlcNA-(Gal)GalNAc]-[anNAc-GlcNAc-(al)GalNAc]-ManNAc | 1786.5 |
| R2 + 2 | [ManNAc-GlcNAc-(Gl)GalNAc]-[ManAc-GlcNAc-(Gal)alNAc]-ManNAc-GlcNAc | 1990.6 |
| R3 + 0 | [ManNAc-GlcNAc-(Gal)GalNc]-[ManNAc-GlNAc-(Gal)GalNAc][ManNAc-GlcNAc-(Gal)GalNAc] | 2354.6 |
| R3 + 0 + 1Ac | [(Ac)ManNAc-GlcNAc-(Gal)GaNAc]-[ManNAc-GcNAc-(Gal)GalNc]-[ManNAc-GlcNAc-(Gal)GalNAc] | 2396.7 |
| R3 + 0 + 2Ac | [(Ac)ManNAc-GlcNAc-(Gal)GalNc]-[(Ac)ManNAcGlcNAc-(Gal)GalAc]-[ManNAc-GlcNAc-(Gal)GalNAc] | 2438.7 |
| R3 + 0 + 3Ac | [(Ac)ManNAc-GlcNAc-(Gal)GalNA]-[(Ac)ManNAc-GcNAc-(Gal)GalNc]-[(Ac)ManNAc-GlcNAc-(Gal)GalNAc] | 2480.6 |
The location of these acetyl (Ac) groups could be on any of one, two, or three of the various ManNAc residues. Rn, the number of repeating units. The number following the repeating units indicates the number of added HexNAc residues. Brackets enclose components of a single repeat unit.
Mass spectrometric analysis, in the linear mode, of the HF-PS after de-O-acetylation (Fig. 1B) shows that that there is considerable size heterogeneity in the HF-PS preparation. A series of [M + Na]+ ions are observed up to m/z 7759. Each of these ions differs from the previous ion by 772 mass units, which is the mass of the (HexNAc)3(Hex)1 repeat unit. These results show that the HF-PS polysaccharide consists of a mixture of molecules that vary from 2 to at least 10 repeating tetrasaccharide units. It is possible that this size heterogeneity is present in vivo or is produced during the release of the polysaccharide from the cell wall by aqueous HF. De-O-acetylation allows these ions to be observed by removing the considerable microheterogeneity caused by variation in acetylation, microheterogeneity that increases with increasing number of repeat units.
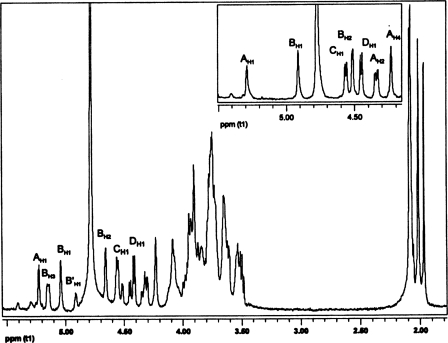
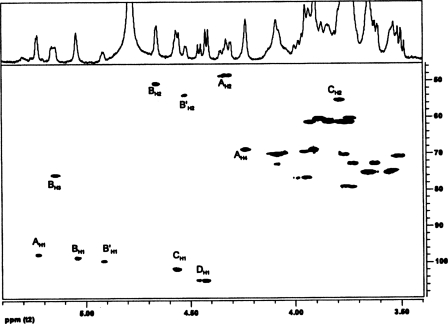
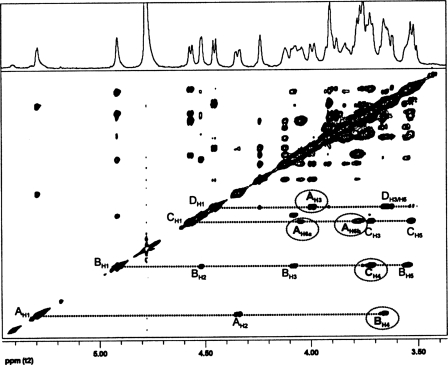
The anomeric configuration and the sequence of the glycosyl residues in the B. cereus ATCC 10987 were determined by both one- and two-dimensional NMR analysis of the HF-PS before and after removal of the O-acetyl groups. The complete proton and carbon assignments of this HF-PS were obtained by one-dimensional proton NMR (Fig. 2) and by two-dimensional gCOSY, TOCSY (Fig. 3), and gHSQC (Fig. 4) experiments. The proton spectrum shows four major anomeric H1 signals at δ 5.23, δ 5.04, δ 4.57, and δ 4.45 and a minor anomeric H1 signal at δ 4.92. Each of these anomeric protons is due to a unique glycosyl ring system in this HF-PS, and the gCOSY, TOCSY (Fig. 3), and gHSQC (Fig. 4) experiments allowed the proton and carbon assignments of each of these ring systems as explained below and given in Table 3.
FIGURE 2.
The proton NMR spectrum of the HF-PS from B. cereus ATCC 10987. The resonances A–D are the indicated respective resonances for the GalNAc, ManNAc, GlcNAc, and Gal residues of the HF-PS. The inset shows the anomeric region of the spectrum of the HF-PS after de-O-acetylation.
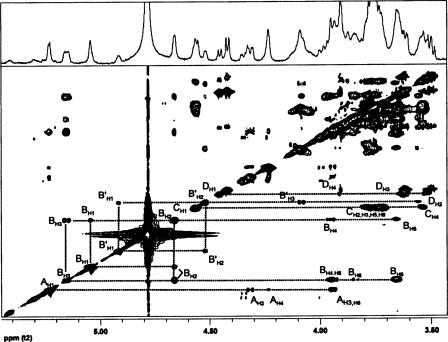
FIGURE 3.
The TOCSY spectrum of the HF-PS from B. cereus ATCC 10987. The protons for the various glycosyl ring systems are as labeled. The residues are labeled as defined in the legend to Fig. 2. The complete proton assignments were made from the TOCSY and COSY (spectrum not shown) data and are given in Table 3.
FIGURE 4.
The HSQC spectrum of the HF-PS from B. cereus ATCC 10987. The assignments are as indicated, and the labeling for the various glycosyl residues is as defined in the legend to Fig. 2. The carbon chemical shift assignments for the various glycosyl residues are given in Table 3.
TABLE 3.
1H and 13C chemical shifts for the B. cereus ATCC 10987 HF-PS
The values in parentheses are for the de-O-acetylated HF-PS.
| Residue | H1/C1 | H2/C2 | H3/C3 | H4/C4 | H5/C5 | H6a/C6 | H6b |
|---|---|---|---|---|---|---|---|
| α-d-GalNAc (A) | 5.23 | 4.32 | 4.23 | 4.23 | 3.93 | 4.06 | 3.78 |
| 97.6 | 48.2 | 68.7 | 68.7 | 70.0 | 69.5 | (3.78) | |
| (5.29) | (4.35) | (4.23) | (4.23) | (3.97) | (4.04) | ||
| (98.9) | (48.8) | (69.1) | 69.1 | (69.6) | (69.6) | ||
| β-d-ManNAc (B) | 5.04 | 4.66 | 5.15 | 3.94 | 3.65 | 3.92 | 3.83 |
| 98.6 | 50.8 | 75.8 | 75.5 | 75.6 | 61.4 | (3.83) | |
| (4.92) | (4.52) | (4.08) | (3.66) | (3.55) | (3.91) | ||
| (99.5) | (53.9) | (73.0) | (75.5) | (75.6) | (61.4) | ||
| β-d-GlcNAc (C) | 4.57 | 3.78 | 3.73 | 3.53 | 3.71 | 3.88 | 3.74 |
| 101.9 | 55.1 | 70.9 | 79.3 | 74.9 | 60.5 | (3.74) | |
| (4.57) | (3.77) | (3.72) | (3.51) | (3.70) | (3.88) | ||
| (101.6) | (55.4) | (70.9) | (79.3) | (74.9) | (60.5) | ||
| β-d-Gal (D) | 4.45 | 3.52 | 3.62 | 3.91 | 3.63 | 3.92 | 3.92 |
| 104.8 | 71.0 | 72.8 | 69.0 | 72.8 | 61.3 | (3.92) | |
| (4.46) | (3.52) | (3.62) | (3.91) | (3.60) | (3.92) | ||
| (105.0) | (71.0) | (72.8) | (69.0) | (72.8) | (61.3) |
The H1 signal at δ 5.23 (1H, s) is the most downfield signal in the spectrum and was assigned to the anomeric proton of an α-glycosyl residue (A). The H2 of residue A resonates at δ 4.32, as was confirmed from the gCOSY experiment (spectrum not shown). The chemical shift of the carbon to which this proton is attached was determined by gHSQC (Fig. 4) to be δ 48.2, indicating it to be nitrogen-bearing carbon consistent with a HexNAc residue. In addition, the downfield chemical shifts of C3 and C6 of this residue at δ 76.4 and 69.5, respectively, indicate that residue A is substituted at O3 and O6. The gCOSY and TOCSY (Fig. 3) spectra showed that H3 and H4 resonate at δ 3.94 and δ 4.23, respectively, and that the small overall J3,4 and J4,5 coupling constant of H4 (<9.6 Hz) indicates that residue A has a galacto-configuration. Therefore, residue A was assigned to be O3- and O6-substituted α-GalNAc, which is consistent with the glycosyl composition and linkage analysis described above showing the presence of a 3,6-linked GalNAc residue.
The next upfield anomeric proton signal at δ 5.04 (s, 1H) had a corresponding C1 chemical shift of δ 98.56. The characteristic downfield chemical shift of H2 at δ 4.66 with small J1,2 and J2,3 coupling constants indicated that this residue has a manno-configuration. The chemical shift of C2 is at δ 50.8 and shows that C2 is a nitrogen-bearing carbon. Therefore, residue B was assigned as ManNAc. Since both α/β-anomeric configurations of manno-glycosyl residues have low coupling constants, the β-anomeric configuration of residue B was confirmed by comparing the TOCSY with NOESY spectrum. The NOESY spectrum (not given) showed strong intraresidue NOE interactions of H1 at δ 5.04 to H2, H3, and H5 at δ 4.66, δ 5.15, and δ 3.65, respectively. These NOE interactions are consistent with the axial positions of H1, H3, and H5 of a β-anomeric configuration for residue B (the NOESY spectrum for the de-O-acetylated HF-PS is shown in Fig. 5 and also shows the H1/H3/H5 interactions). The downfield chemical shift of H3 at δ 5.15, which is attached to a carbon with a chemical shift of δ 75.8, is consistent with O-acetylation at O3 of this β-ManNAc residue. The presence of O-acetyl groups on this HF-PS is also consistent with the mass spectrometric data as described above.
FIGURE 5.
The NOESY spectrum of the de-O-acetylated HF-PS from B. cereus ATCC 10987. The assignments for the various NOE contacts are as indicated in the figure. The labeling of the various glycosyl residues is as defined in the legend to Fig. 2. The interresidue NOE contacts are circled and in boldface type.
The minor H1 anomeric signal at δ 4.92 was identified to be another β-ManNAc residue (B′) that does not bear an O-acetyl ester group on O3. The upfield chemical shifts of H2 at δ 4.53 (compared with δ 4.66 for residue B) and H3 at δ 4.08 (compared with δ 5.15 for residue B), respectively, are consistent with the lack of an O-acetyl group at O3 of this residue. The relative quantification of HF-PS bearing the O-acetyl group was done by comparing the H1 integral values of residues B and B′ from the proton NMR spectrum, and it was calculated that about 60% of the HF-PS was O-acetylated at O3 of the ManNAc residue. It is quite possible that the polysaccharide as found in the cell wall is completely O-acetylated and that the 60% value is due to partial removal of O-acetyl groups during its release by aqueous HF.
The next upfield signal at δ 4.57 is a doublet with a J1,2 coupling constant of 7.8 Hz, indicating it to have a β-anomeric configuration. The C1 chemical shift of δ 101.88 also supports the β-anomeric configuration. The H2 of this residue resonates at δ 3.65 and is attached to a nitrogen-bearing carbon at δ 55.1 (see Fig. 4), showing that this is a β-HexNAc residue. The TOCSY spectrum showed relatively large J2,3, J3,4, and J4,5 coupling constants, which is consistent with a gluco-configuration; therefore, this residue (C) was assigned as β-GlcNAc. The downfield chemical shift of C4 at δ 79.3 indicates that this residue is 4-substituted β-GlcNAc.
The remaining H1 anomeric signal has a proton chemical shift at δ 4.45 (1H, J1,2 = 7.8 Hz) and a C1 chemical shift at δ 104.75. These H1 and C1 chemical shifts and the large J1,2 coupling constant are consistent with this glycosyl residue (D) having a β-anomeric configuration. By comparing gCOSY and TOCSY (Fig. 3) spectra, it was found that the H4 signal resonates at δH 3.91 and has small J3,4 and J4,5 coupling constants (<9.6 Hz), showing that this residue has a galacto-configuration. Therefore, this residue (D) was assigned as Gal.
The NMR and mass spectral data just described indicate that this HF-PS preparation consists of a microheterogeneous mixture of molecules due to nonstoichiometric O-acetyl substitution at O3 of the ManNAc residue, to a varying number of tetrasaccharide repeating units, and to the addition of HexNAc and HexHexNAc saccharides. This microheterogeneity impacts the chemical shift values, which makes determining the glycosyl sequence by NMR analysis difficult. Therefore, in order to reduce the molecular heterogeneity, the sample was treated with anhydrous hydrazine, which removes the O-acetyl groups while leaving the rest of the structure intact. The removal of O-acetyl groups was confirmed in the NMR experiments, and the proton and carbon chemical shifts were determined by one-dimensional proton NMR analysis and two-dimensional gCOSY, TOCSY, and gHSQC NMR experiments (spectra not shown). These assignments are given in Table 3. The inset in Fig. 2 shows the proton spectrum of the anomeric region of the de-O-acetylated HF-PS. After hydrazine treatment, NMR analysis showed the presence of four glycosyl residues between δ 4.4 and δ 6.0 and three N-acetyl methyl protons near δ 2.0, indicating the presence of three N-acetylamino sugars. The H1 (δ 5.04) and the H3 (δ 5.15) resonances due to the presence of an acetylated β-ManNAc residue are both absent in the de-O-acetylated HF-PS.
Due to spectral simplicity of the de-O-acetylated HF-PS, the glycosyl sequence was determined from a NOESY experiment. The NOE spectrum (Fig. 5) showed a through space interresidue connectivity between H1 of residue A (δ 5.29) to H4 of residue B (δ 3.66) along with an intraresidue NOE with H2 at δ 4.35. This indicates that α-GalNAc (A) is linked to the 4-position of β-ManNAc residue (B)(i.e. α-GalNAc-(1→4)-β-ManNAc). This sequence is consistent with the methylation analysis, which shows that the β-ManNAc residue is 4-linked. The H1 of residue B (δ 4.92) showed an interresidue NOE with H4 of residue C (δ 3.72) along with a number of intraresidue NOE contacts. Hence, β-ManNAc (B) is connected to the 4-position of the β-GlcNAc residue (C)(i.e. α-GalNAc-(1→4)-β-ManNAc-(1→4)-β-GlcNAc). This result is consistent with the methylation data showing that the GlcNAc residue is 4-linked. The H1 of residue C had an interresidue NOE with H6a and H6b of residue A, indicating β-GlcNAc (C) to be linked to the O6 of α-GalNAc (residue A); i.e. →6)-α-GalNAc-(1→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→. The most upfield H1 signal of the β-Gal residue (D) at δ 4.46 has an interresidue NOE to H3 of residue A, indicating that β-Gal residue is connected to the O3 of the α-GalNAc residue; i.e. →6)-[β-Gal-(1→3)]α-GalNAc-(1→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→. The NOE contacts of the GlcNAc (C) and Gal (D) residues to O6 and O3, respectively, of the GalNAc residue (A) are consistent with the methylation data showing that the GalNAc residue is 3,6-linked. The NOESY experiment does not show any NOE contacts to the Gal residue from any of the H1 protons of the other glycosyl residues, indicating that Gal is a terminally linked glycosyl residue, a result that is also consistent with the methylation data. The combination of the composition, methylation, mass spectrometry, and NMR analyses indicates that the repeating unit of the B. cereus ATCC 10987 HF-PS has the structure of →6)-[β-Gal-(1→3)]α-GalNAc-(1→4)-[3-OAc]β-ManNAc-(1→4)-β-GlcNAc-(1→.
Comparison of the B. cereus ATCC 10987 HF-PS Structure with the Structural Features of the B. cereus ATCC 14579 HF-PS—Previously reported composition analysis (10) and proton NMR spectroscopy (12) as well as the composition and methylation analysis described above show that the HF-PS from the B. cereus type strain, ATCC 14579, has a different structure than the HF-PSs of B. cereus ATCC 10987 and B. anthracis. For example, it contains GalNAc, as is found in B. cereus ATCC 10987 HF-PS but not found in B. anthracis HF-PS, and it contains Glc rather than Gal, which is found in both B. cereus ATCC 10987 and in B. anthracis HF-PSs. The B. cereus ATCC 14579 HF-PS is similar to these latter HF-PSs in that it also contains both ManNAc and GlcNAc. However, despite the obvious structural differences in these HF-PSs, mass spectrometric analysis, using the reflectron mode, of the B. cereus ATCC 14579 HF-PS (Fig. 6) gives a spectrum with a pattern of ions very similar to that obtained for the B. cereus ATCC 10987 HF-PS (Fig. 1A). The ion with the greatest intensity is the [M + Na]+ ion at m/z 1583.4, as is the case for the B. cereus ATCC 10987 HF-PS. The ion at m/z 1786.4 is consistent with an added HexNAc residue (i.e. +203 mass units), and the ion at m/z 1989.6 is probably due to the addition of a second HexNAc residue. The ion at m/z 2354.6 is 365 mass units greater than the 1989.6 ion and is consistent with a HexHexNAc disaccharide added to the m/z 1989.6 structure. There are also ions consistent with the loss of a HexHexNAc from the 1583.5 ion (i.e. that at m/z 1218.4) and an ion at m/z 1015.3, which is consistent with the loss of a HexNAc from the 1218.4 ion. The proposed compositions for the pattern of ions are shown in Table 4. This pattern indicates that this HF-PS preparation consists of one, two, and three HexNAc3Hex trisaccharides, as observed for the B. cereus ATCC 10987 HF-PS (see Fig. 1A), with heterogeneity due to molecular species with added HexNAc and Hex residues. There is no evidence that the B. cereus ATCC 14579 HF-PS is acetylated, since ions with added increments of 42 mass units are not observed.
FIGURE 6.
MALDI-TOF MS spectrum, obtained in the reflectron mode, of the HF-PS from B. cereus ATCC 14579. The spectrum was acquired in the positive mode, and the proposed structures for the various ions are given in Table 4.
TABLE 4.
Proposed structures for the various molecular ions observed during MALDI-TOF MS analysis of the HF-PS from B. cereus ATCC 14579
| Repeat units | Proposed structure | Observed [M + Na]+ ion |
|---|---|---|
| R1a + 1 | [HexNAc3Glc]-HexNAc | 1015.3 |
| R1 + 2 | [HexNAc3Glc]-HexNAc2 | 1218.4 |
| R2 + 0 | [HexNAc3Glc]2 | 1583.5 |
| R2 + 1 | [HexNAc3Glc]-HexNAc | 1786.5 |
| R2 + 2 | [HexNAc3Glc]-HexNAc2 | 1989.4 |
| R3 + 0 | [HexNAc3Glc]3 | 2354.5 |
Rn, the number of repeating units. The number following the repeating units indicates the number of added HexNAc residues.
Fig. 7 compares the structures of the HF-PSs from B. anthracis (12) with the HF-PS structure of B. cereus ATCC 10987 and the proposed structural features for B. cereus ATCC 14579 HF-PS. From this comparison, a consensus structure is indicated for the HF-PSs from B. anthracis and B. cereus ATCC 10987 in which the repeating unit backbone consists of a [→6)-α-GlcNAc or →6)-α-GalNAc]-(1→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→ trisaccharide that is substituted by Gal residues and/or O-acetylated. Due to the increased structural heterogeneity of the B. cereus ATCC 14579 HF-PS, we do not yet know if it contains this same consensus structural feature; however, composition analysis together with the similarity of its mass spectrum with that of B. cereus ATCC 10987 HF-PS supports the possibility that these three HF-PSs share a general structural theme of a polysaccharide with an aminoglycosyl-rich backbone in which at least two of the aminoglycosyl residues are ManNAc and GlcNAc. The complete structure of the B. cereus ATCC 14579 HF-PS is under investigation and will be the subject of a subsequent report.
FIGURE 7.
A comparison of the B. cereus ATCC 10987 HF-PS structure with the published structure (12) for the HF-PSs from B. anthracis Ames, Sterne, and Pasteur and with a proposed structure for B. cereus ATCC 14579. It should be emphasized that the last structure has not been unambiguously determined and is currently under investigation. A consensus structure based on these structures is also given, in which X represents substitution of the aminoglycosyl backbone with other glycosyl residues, such as Gal or Glc, or with noncarbohydrate substituents, such as an O-acetyl group.
DISCUSSION
We have previously reported the structure of the HF-PS from B. anthracis, and comparison of NMR spectra showed that its structure varied from the HF-PS of B. cereus ATCC 10987 and ATCC 14579 (12). Here, we report the structure of a nonclassical SCWP polysaccharide isolated from B. cereus ATCC 10987 as well as structural data on this polysaccharide from the B. cereus type strain, ATCC 14579, and we have compared those structural data with the reported B. anthracis HF-PS structure (12). The results are summarized as follows: (i) the HF-PS from B. cereus ATCC 10987 is composed of a tetrasaccharide repeat unit consisting of a →6)-α-d-GalNAc-(1→4)-β-d-ManNAc-(1→4)-β-d-GlcNAc-(1→ trisaccharide in which the GalNAc residue is substituted at O3 with β-d-Gal and the ManNAc residue is 3-O-acetylated; (ii) there is heterogeneity in this HF-PS polysaccharide due to variation in the number of repeating units, the presence of O-acetyl groups, and the addition of ManNAc, GlcNAc, and GalGalNAc, respectively, to the direpeat and, possibly, mono- and trirepeat unit structures; (iii) in B. cereus ATCC 10987 and B. anthracis strains, there is a consensus HF-PS structural feature in that the repeating unit consists of a trisaccharide aminoglycosyl backbone in which two of the three aminoglycosyl residues are ManNAc and GlcNAc, whereas the third is either GlcNAc or GalNAc. Variability between the various HF-PS structures occurs in the substitution pattern of this trisaccharide with regard to both glycosyl and nonglycosyl substituents; (iv) the HF-PS from B. cereus ATCC 14579 is structurally more complex than those of B. cereus ATCC 10987 and B. anthracis. However, the similarities in the MS HF-PS ion patterns of these strains to that of the B. cereus ATCC 14579 HF-PS suggest that they share a general structural theme. Taken together, these data support the conclusion that the HF-PSs from B. anthracis strains, B. cereus ATCC 10987, and B. cereus ATCC 14579 each have unique structural features residing on an overall similar structural theme of a polysaccharide backbone that is rich in aminoglycosyl residues, at least two of which are ManNAc and GlcNAc.
The structures of nonclassical SCWP polysaccharides from bacilli have been described in a recent review by Schäffer and Messner (13). In that review, both common and variable structural features of these polysaccharides are discussed. The general structural feature for all of these polysaccharides is that they have repeating oligosaccharide units that are rich in aminoglycosyl residues and, further, that at least one of these residues has a manno-configuration and a second has a gluco-configuration. As we have shown here, this general structural theme is also present in the previously reported structure of B. anthracis HF-PS (12), the B. cereus ATCC 10987 HF-PS structure reported here, and composition, linkage, and MS analysis suggest that this theme is also likely for B. cereus ATCC 14579 HF-PS.
The fact that these HF-PSs each have unique structural features that are present on a common structural theme suggests that they are under functional constraints leading to structure conservation with modification that is possibly required for related or similar functions in various B. cereus and B. anthracis strains. It has been reported (14) that HF-PSs are involved in anchoring and/or targeting proteins to the cell surface, including the S-layer proteins via a carbohydrate-binding domain of these proteins (e.g. the SLH domain in the case of S-layer and other surface proteins). In the case of B. anthracis, it was reported that a mutant that is unable to add a pyruvate substituent to the HF-PS is defective in targeting the S-layer protein to the cell surface and is also adversely affected in cell division (14). Although it is not yet known if HF-PSs have essential functions for viability or virulence, some indirect evidence for their involvement in infection processes has already emerged. During B. cereus infection of insects, it has been noticed that the expression of an internalin-like protein is induced (21). This internalin-like protein is a candidate virulence protein that contains a SLH domain at its C-terminal end that presumably anchors the protein to the cell wall via binding to the secondary cell wall carbohydrates (21) that are covalently bound to the peptidoglycan (e.g. HF-PS). Recently, it was shown that another SLH-containing surface protein that is encoded on the pathogenicity island of pXO1, BslA, is involved in the attachment of B. anthracis to host cells (22). Thus, it is possible that the HF-PS is involved in localizing important virulence proteins to the bacterial cell surface. The variation on a common structural theme of the HF-PSs could be of high biological significance and account for the specificity of bacterium-host interactions; therefore, it will be an important future task to characterize the binding specificities of various SLH-containing proteins with HF-PSs from different B. cereus group members.
It is also known that phage endolysins have, in addition to a catalytic domain, a carbohydrate-binding domain that is thought to bind to a cell wall polysaccharide (23). The binding of the carbohydrate-binding domain of the endolysin to the polysaccharide is proposed to be required for the lytic activity and to determine the specificity of the endolysin for a particular bacterial host (23, 24). Therefore, structural specificity in the host bacterial cell wall polysaccharides would account for the specificity of the endolysins. This type of specificity was reported for the interaction of the γ-phage lysin, PlyG, with strains of B. anthracis (24). Since the HF-PS is the major cell wall polysaccharide of B. anthracis as well as of the B. cereus strains ATCC 10987 and ATCC 14579 and displays structural specificity, it seems likely that HF-PS is the carbohydrate ligand for the endolysins and accounts for their specificity. In fact, PlyG specifically lyses B. anthracis strains but only very poorly lyses B. cereus ATCC 10987 and does not lyse B. cereus strain ATCC 14579. Therefore, determining if the HF-PS is the endolysin ligand and characterizing the structural features required for binding of endolysins have important implications with regard to the functions of these polysaccharides and diagnostic and therapeutic applications of such bacteriophages (24, 25).
Experimentally addressing the functions of these HF-PSs depends on having different HF-PSs from various B. cereus group members so that their interaction with different proteins having SLH domains and with different lysins can be measured. The binding specificities of various HF-PSs with various proteins and endolysins are currently under investigation in our laboratory. The second major approach for characterizing HF-PS function is to create mutants in specific genes required for HF-PS synthesis. The creation of mutants affected in HF-PS synthesis is possible, since the available bacterial genome sequences allows the identification of putative genes that may be involved in HF-PS synthesis. Interestingly, a recent survey of gene differences in B. anthracis and B. cereus using suppression subtractive hybridization and bioinformatic analysis of Bacillus whole genome sequences found a glycosyltransferase group 1 family gene apparently specific to B. anthracis (26). Although the involvement of this gene in HF-PS biosynthesis is, at this point, not clear and currently under investigation in our laboratory, it is worth mentioning that from the few currently characterized bacterial galactosyltransferases some belong to the group family 1 glycosyltransferases (11, 27). Moreover, since all of the HF-PSs contain ManNAc, mutation of genes that encode for ManNAc biosynthetic proteins should prevent the synthesis of the HF-PS and allow evaluation of functional importance (i.e. whether or not the HF-PS has essential functions for bacterial growth, for anchoring/exporting S-layer and other surface proteins, for interacting with endolysins, or, in the case of a pathogen such as B. anthracis, for virulence). The preparation and analysis of such B. anthracis mutants is in progress.
This work was supported, in whole or in part, by National Institutes of Health, NIAID, Grant R21 AI059577 (to R. W. C.). This work was also supported in part by Department of Energy Grant DE-FG02-93ER20097 (to the CCRC). Patent pending (University of Georgia Research Foundation, Inc. and United States Centers for Disease Control and Prevention). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HF, hydrogen fluoride; HF-PS, HF-released polysaccharide; HexNAc, N-acetylhexose; Hex, hexose; MS, mass spectrometry; gCOSY, gradient 1H-1H correlated spectroscopy; TOCSY, total correlated spectroscopy; gHSQC, gradient heteronuclear single quantum coherence spectroscopy; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser effect spectroscopy; ManNAc, N-acetylmannosamine; SCWP, nonclassical secondary cell wall polymers; SLH, S-layer homology; Rha, rhamnose; MALDI, matrix-assisted laser desorption ionization; TOF, time of flight.
C. Leoff, E. Saile, J. Rauvolfova, C. P. Quinn, A. R. Hoffmaster, W. Zhong, A. S. Mehta, G. J. Boons, R. W. Carlson, and E. L. Kannenberg, submitted for publication.
References
- 1.Helgason, E., Okstad, O. A., Caugant, D. A., Johansen, H. A., Fouet, A., Mock, M., Hegna, I., and Kolsto, A. B. (2000) Appl. Environ. Microbiol. 66 2627–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasko, D. A., Altherr, M. R., Han, C. S., and Ravel, J. (2005) FEMS Microbiol. Rev. 29 303–329 [DOI] [PubMed] [Google Scholar]
- 3.Gonzales, J. M. J., Brown, B. J., and Carlton, B. C. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 6951–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okinaka, R. T., Cloud, K., Hampton, O., Hoffmaster, A. R., Hill, K. K., Keim, P., Koehler, T. M., Lamke, G., Kumano, S., Mahillon, J., Manter, D., Martinez, Y., Ricke, D., Svensson, R., and Jackson, P. J. (1999) J. Bacteriol. 181 6509–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mock, M., and Fouet, A. (2001) Annu. Rev. Microbiol. 55 647–671 [DOI] [PubMed] [Google Scholar]
- 6.Hoffmaster, A. R., Ravel, J., Rasko, D. A., Chapman, G. D., Chute, M. D., Marston, C. K., De, B. K., Sacchi, C. T., Fitzgerald, C., Mayer, L. W., Maiden, M. C. J., Priest, F. G., Barker, M., Jiang, L., Cer, R. Z., Rilstone, J., Peterson, S. N., Weyant, R. S., Galloway, D. R., Read, T. D., Popovic, T., and Fraser, C. M. (2004) Proc. Natl. Acad Sci. U. S. A. 101 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmaster, A. R., Gee, J. R., Marston, C. K., De, B. K., Popovic, T., Sue, D., Wilkins, P. P., Avashia, S. B., Drumgoole, R., Hill, K. K., Helma, C. H., Ticknor, L. O., Okinaka, R. T., and Jackson, P. J. (2006) J. Clin. Microbiol. 44 3352–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leendertz, F. H., Yumlu, S., Pauli, G., Boesch, C., Couacy-Hymann, E., Vigilant, L., Junglen, S., Schenk, S., and Ellerbrok, H. (2006) PLoS Pathog. 2 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klee, S. R., Özel, M., Appel, B., Boesch, C., Ellerbrok, H., Jacob, D., Holland, G., Leendertz, F. H., Pauli, G., Grunow, R., and Nattermann, H. (2006) J. Bacteriol. 188 5333–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leoff, C., Saile, E., Sue, D., Wilkins, P., Quinn, C. P., Carlson, R. W., and Kannenberg, E. L. (2008) J. Bacteriol. 190 112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue, T., Shingaki, R., Hirose, S., Waki, K., Mori, H., and Fukui, K. (2007) J. Bacteriol. 189 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhury, B., Leoff, C., Saile, E., Wilkins, P., Quinn, C. P., Kannenberg, E. L., and Carlson, R. W. (2006) J. Biol. Chem. 281 27932–27941 [DOI] [PubMed] [Google Scholar]
- 13.Schäffer, C., and Messner, P. (2005) Microbiology 151 643–651 [DOI] [PubMed] [Google Scholar]
- 14.Mesnage, S., Fontaine, T., Mignot, T., Delepierre, M., Mock, M., and Fouet, A. (2000) EMBO J. 19 4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amano, K., Hazama, S., Araki, Y., and Ito, E. (1977) Eur. J. Biochem. 75 513–522 [DOI] [PubMed] [Google Scholar]
- 16.Kojima, N., Araki, Y., and Ito, E. (1985) Eur. J. Biochem. 148 479–484 [DOI] [PubMed] [Google Scholar]
- 17.Rasko, D. A., Ravel, J., Okstad, O. A., Helgason, E., Cer, R. Z., Jiang, L., Shores, K. A., Fouts, D. E., Tourasse, N. J., Angiuoli, S. V., Kolonay, J., Nelson, W. C., Kolsto, A. B., Fraser, C. M., and Read, T. D. (2004) Nucleic Acids Res. 32 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, W. C. (1973) J. Bacteriol. 25 295–300 [Google Scholar]
- 19.York, W. S., Darvill, A. G., McNeil, M., Stevenson, T. T., and Albersheim, P. (1985) Methods Enzymol. 118 3–40 [Google Scholar]
- 20.Ciucanu, I., and Kerek, F. (1984) Carbohydr. Res. 131 209–217 [Google Scholar]
- 21.Fedhila, S., Daou, N., Lereclus, D., and Nielsen-LeRoux, C. (2006) Mol. Microbiol. 62 339–355 [DOI] [PubMed] [Google Scholar]
- 22.Kern, J. W., and Schneewind, O. (2008) Mol. Microbiol. 68 504–515 [DOI] [PubMed] [Google Scholar]
- 23.Low, L. Y., Yang, C., Perego, M., Osterman, A., and Liddington, R. C. (2005) J. Biol. Chem. 280 35433–35439 [DOI] [PubMed] [Google Scholar]
- 24.Schuch, R., Nelson, D., and Fischetti, V. A. (2002) Nature 418 884–889 [DOI] [PubMed] [Google Scholar]
- 25.Fischetti, V. A., Nelson, D., and Schuch, R. (2006) Nat. Biotech. 24 1508–1511 [DOI] [PubMed] [Google Scholar]
- 26.Kim, W., Kim, J.-Y., Cho, S.-L., Nam, S.-W., Shin, J.-W., Kim, Y.-S., and Shin, H.-S. (2008) J. Med. Microbiol. 57 279–286 [DOI] [PubMed] [Google Scholar]
- 27.Claesson, M. J., Li, Y., Leahy, S., Canchaya, C., van Pijkeren, J. P., Cerdeno-Tarraga, A. M., Parkhill, J., Flynn, S., O'Sullivan, G. C., Collins, J. K., Higgins, D., Shanahan, F., Fitzgerald, G. F., van Sinderen, D., and O'Toole, P. W. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6718–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]