Abstract
Membrane binding by prothrombin, mediated by its N-terminal fragment 1 (F1) domain, plays an essential role in its proteolytic activation by prothrombinase. Thrombin is produced in two cleavage reactions. One at Arg320 yields the proteinase meizothrombin that retains membrane binding properties. The second, at Arg271, yields thrombin and severs covalent linkage with the N-terminal fragment 1.2 (F12) region. Covalent linkage with the membrane binding domain is also lost when prethrombin 2 (P2) and F12 are produced following initial cleavage at Arg271. We show that at the physiological concentration of prothrombin, thrombin formation results in rapid release of the proteinase into solution. Product release from the surface can be explained by the weak interaction between the proteinase and F12 domains. In contrast, the zymogen intermediate P2, formed following cleavage at Arg271, accumulates on the surface because of a ∼20-fold higher affinity for F12. By kinetic studies, we show that this enhanced binding adequately explains the ability of unexpectedly low concentrations of F12 to greatly enhance the conversion of P2 to thrombin. Thus, the utilization of all three possible substrate species by prothrombinase is regulated by their ability to bind membranes regardless of whether covalent linkage to the F12 region is maintained. The product, thrombin, interacts with sufficiently poor affinity with F12 so that it is rapidly released from its site of production to participate in its numerous hemostatic functions.
Thrombin, the key effector serine proteinase of the blood coagulation cascade, is produced by specific and limited proteolysis of the zymogen, prothrombin. The physiologically relevant catalyst for this reaction is the prothrombinase complex consisting of the serine proteinase, factor Xa, and the cofactor, factor Va, assembled on membranes in the presence of Ca2+ (1). In addition to facilitating the assembly of the enzyme complex, membranes containing acidic or amino phospholipids play an important role in mediating the delivery of prothrombin to the membrane-bound enzyme (1, 2). This arises from the ability of prothrombin to bind to these membranes through the fragment 1 (F1)2 domain present at its N terminus (1, 3, 4).
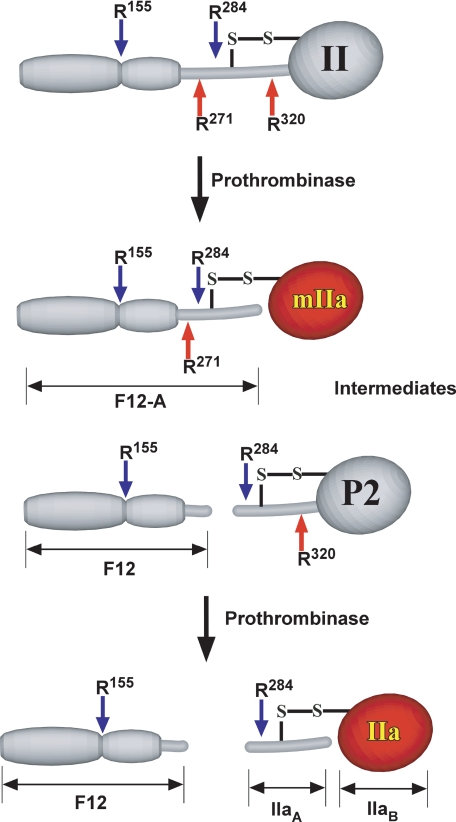
Thrombin, derived from the C-terminal half of prothrombin, is produced as a result of cleavages3 following Arg271 and Arg320 (1, 3, 5). Cleavage at Arg320 converts the zymogen to a proteinase, whereas cleavage at Arg271 severs covalent linkage with the N-terminal fragment 1.2 (F12) domain harboring the membrane binding site (Scheme 1). Covalent linkage of the C-terminal domain with F12 is also lost in the zymogen intermediate, prethrombin 2 (P2), produced following cleavage only at Arg271 (Scheme 1). In contrast, meizothrombin (mIIa), produced following cleavage only at Arg320 is covalently linked to the membrane binding domain through a disulfide bond (Scheme 1). Accordingly, both prothrombin and mIIa are established to bind to membranes, and this binding interaction impacts their utilization as substrates by prothrombinase (2, 5).
SCHEME 1.
Intermediates and products formed upon cleavage of human prothrombin by prothrombinase. The red arrows denote the Arg271 and Arg320 sites cleaved by prothrombinase. The blue arrows denote Arg155 and Arg284 sites that are susceptible to feedback cleavage by IIa and mIIa. The competent catalytic domain in mIIa and IIa is shaded red to distinguish it from the zymogen form of the domain in II and P2.
The ability of thrombin or P2 to bind reversibly to the fragment 2 (F2) domain has been established in a series of studies as well as by x-ray crystallography (6–10). Initial studies at low ionic strength, with prothrombin fragments of bovine origin, provided evidence for a subnanomolar affinity for either interaction (6). Such high affinity interactions have implied a fundamental role for F2-mediated binding of thrombin or P2 to F12 in bridging the C-terminal domain either in the zymogen or proteinase states to the membrane-binding domain and consequently the membrane surface. This idea is supported by the greatly enhanced cleavage of P2 by prothrombinase in the presence of an equimolar concentration of F12 (7, 11–13). Furthermore, light scattering studies have provided evidence indicating that the interaction between thrombin and F12 allows membrane binding by the product and its nearly quantitative retention on the membrane surface at the site of prothrombin activation (14). A key regulatory event in the form of feedback cleavage by thrombin at Arg155 between the F1 and F2 domains (Scheme 1) has been proposed to be necessary for the release of nascent thrombin from the membrane surface (14). Membrane-bound thrombin is likely to be sequestered from and to exhibit different preferences for the range of biological substrates acted on by thrombin in solution. Thrombin released by cleavage at Arg155 would also be expected to be essentially saturated with F2 bound to the anion binding exosite II region of the proteinase. This interaction is known to significantly impact active site function, inhibition of thrombin by anti-thrombin III and possibly platelet aggregation (6, 15–17). Therefore, the proposed tight interactions with F2 have unexpected and important implications for the numerous functions of thrombin in hemostasis that have largely been established with the purified proteinase in the absence of F2 or F12 (5, 18).
A series of more recent and careful studies with human prothrombin derivatives, conducted at physiological pH and ionic strength (or NaCl), now indicate that the interactions between F2 and thrombin or P2 are 1000-fold (or more) weaker than initially reported in the bovine system (8, 9). Based on the possible concentrations of the various species expected during the activation of prothrombin, these findings suggest that noncovalent interactions with the F2 domain either on its own or within F12 probably play a minor role in retaining thrombin on the membrane surface or in modulating the function of thrombin in solution. These ideas are consistent with the need for very high concentrations of F12 to observe effects on platelet aggregation catalyzed by human thrombin (17). However, despite the somewhat tighter affinity reported for the binding of F12 to P2 (19), the available data present a paradox because they do not provide an obvious explanation for the ability of a stoichiometric equivalent of F12 to greatly and saturably enhance the cleavage of 10-7 m P2 by human prothrombinase (12, 13, 20). Thus, it remains possible that interactions with F12 measured in solution are further substantially modulated when F12 is membrane-bound.
In this study, we focus on the role played by interactions with F12 in regulating function by affecting partitioning of substrate and product species to the membrane surface. We have employed a series of recombinant prothrombin derivatives to investigate the fate of membrane-bound intermediates and products during the activation of human prothrombin by prothrombinase, the role of cleavage at Arg155 in regulating their fates, and the kinetic mechanism underlying the ability of F12 to regulate P2 cleavage by prothrombinase.
EXPERIMENTAL PROCEDURES
Materials—Hen egg l-α-phosphatidylcholine (PC) and porcine brain l-α-phosphatidylserine (PS) were purchased from Avanti Polar Lipids (Alabaster, AL). Small unilamellar phospholipids vesicles composed of 75% (w/w) PC and 25% (w/w) PS (PCPS) were prepared and characterized as described (21). For some kinetic studies, large unilamellar vesicles of the same composition (PCPSLUV) were prepared by extrusion, as previously detailed (12). Proteinase inhibitors dansyl-l-arginine-N-(3-ethyl-1,5-pentanediyl)amide (DAPA) and d-phenylalanyl-l-prolyl-l-arginine chloromethyl ketone (FPRck) were from Hematologic Technologies (Essex Junction, VT) and Calbiochem, respectively. The peptidyl substrate, H-d-phenylalanyl-l-pipecolyl-l-arginine-p-nitroanilide was from Chromogenix (West Chester, OH). Human plasma used for the isolation of proteins was a generous gift of the Plasmapheresis Unit of the Hospital of the University of Pennsylvania. All kinetic measurements were conducted in 20 mm Hepes, 0.15 m NaCl, 5 mm Ca2+, 0.1% (w/v) polyethylene glycol 8000, pH 7.5 (Assay Buffer), at 25 °C.
Recombinant Proteins—Prothrombin variants were cloned into an appropriately adapted pcDNA 3.1(+) vector using the Gateway cloning system (Invitrogen) (12). The donor cassette in the pENTR entry vector contained a Kozak sequence, followed by sequences encoding a translational start site, signal peptide, propeptide, and the mature protein followed by 97 bases encoding the prothrombin 3′-untranslated region. This cassette was used with appropriate mutagenic primers and the QuikChange mutagenesis kit (Stratagene) to substitute codons for Gln in place of Arg155 and Arg284 (IIQ155/Q284) or to prepare the triple mutant (IITM) containing codons for Gln at Arg155, Arg271, and Arg284. A similar strategy was employed to substitute Ala for Ser195 (IIA195) or Gln for Arg320 (IIQ320) or in place of Arg271 and Arg320 (IIQQ). The integrity of each cassette was verified by sequencing both before and after λ-integrase-mediated transfer to the pcDNA destination vector. Previously described strategies were utilized for transfection of HEK293 cells with these constructs, selection of stable cell lines, and their expansion for large scale protein production in serum-free medium supplemented with 10 μg/ml vitamin K (Abbott) (12). Prothrombin variants were purified from 20 liters of conditioned medium, as previously detailed (12). N-terminal sequence analysis (performed at the Emory University microchemical facility) indicated that all variants possessed a correctly processed NH2 terminus. Quantitative analysis of 4-carboxyglutamic acid content following base hydrolysis (22, 23) yielded values for the variants that were indistinguishable from the values observed with prothrombin isolated from plasma.
Plasma Proteins—Human factors X, V, and II were purified as described (13). Factors Xa and Va were prepared and characterized as described before (20, 24). The derivatives F12, F2, P2, mIIa, and IIa were prepared by preparative cleavage of prothrombin purified from plasma (IIPL) and subsequent repurification using established procedures (25). An equivalent strategy using IIA195 as substrate was employed to generate IIaA195 and P2A195. Inactivated derivatives of IIa and mIIa (IIai and mIIai) were prepared by covalent modification with FPRck as described (12). Protein concentrations were determined using the following molecular weights and extinction coefficients (E280 mg-1·cm2): human Xa, 1.16 and 45,300 (26); human Va, 1.78 and 173,000 (27); F12, 1.12, 34,800; F2, 1.28 and 12,800 (25); P2, P2A195, IIa, IIai, and IIaA195, 1.94 and 37,500 (25, 28); mIIai, 1.42 and 72,000; IIPL and all recombinant prothrombin variants, 1.42 and 72,000 (25). Prothrombin variants were exchanged into assay buffer by centrifugal gel filtration before use.
Prolonged Digestion of Prothrombin Variants—Reaction mixtures (100 μl) in assay buffer contained the indicated prothrombin variant (3.0 μm), 50 μm PCPS, 30 nm Va, and either 0 or 3 nm Xa. Following incubation for 30 min at 25 °C, steady state light scattering intensity was measured on samples (20 μl) prepared by diluting with an equal volume of 50 μm PCPS in assay buffer using a Viscotek DLS instrument (Protein Solutions). Scattering intensity was normalized to that observed with 50 μm PCPS alone. In parallel, samples (15 μl) were mixed with an equal volume of 125 mm Tris, 20% (v/v) glycerol, 2% (w/v) SDS, 0.02% (w/v) bromphenol blue, 50 mm EDTA, 83 mm dithiothreitol, pH 6.8. After heating at 89 °C for 3 min, the samples were analyzed by SDS-PAGE using 4–12% NOVEX BisTris gels (Invitrogen) run with MES buffer. Protein bands were visualized by staining with Coomassie Brilliant Blue.
Kinetics of Cleavage of Prothrombin Variants—Changes in right angle light scattering during prothrombin activation were measured with a PTI QuantaMaster fluorescence spectrophotometer (Photon Technologies International) in 1 × 1-cm stirred quartz cells maintained at 25 °C. Samples in Assay Buffer (2.5 ml) containing 1.4 μm prothrombin variant, 50 μm PCPS, and 30 nm Va were initiated with 1 nm Xa. Light scattering was continuously monitored using λEX = λEM = 320 nm. Limits of the scattering signal were independently established using reaction mixtures containing 50 μm PCPS alone or 50 μm PCPS with 1.4 μm prothrombin, mIIai, F12, or F2. Scattering intensities were normalized using the signal observed with PCPS alone. Parallel reaction mixtures were subsampled and processed by SDS-PAGE as described above. Protein bands were visualized with Colloidal Blue stain (Invitrogen) following the instructions of the manufacturer.
Isothermal Titration Calorimetry—Measurements were performed using a MCS-ITC instrument (Microcal, Cambridge, MA) at 25 °C. Both interacting species were extensively dialyzed into 20 mm Hepes, 0.15 m NaCl, 5 mm Ca2+, pH 7.5, and the dialysate was used for subsequent dilutions and control experiments. All samples were degassed and thermally equilibrated before use. The cell (1.3298 ml) contained either 25.94 μm IIaA195, 20.8 μm IIaA195, or 12.97 μm P2A195. For experiments with IIaA195, the injection syringe contained 608 μm F12 or 1015 μm F2. For titrations with P2A195, the injection syringe contained 465 μm F12. Heat flow was measured with continuous stirring, following injections of F12 or F2 spaced at 210-s intervals. In a typical experiment, an initial small volume injection (2 μl) was used to purge trapped air, followed by 16 larger injections (10.1 μl) of titrant. Trivial heat flow due to titrant dilution and buffer mismatch was determined from an identical series of injections of F12 or F2 into dialysate.
Steady State Kinetic Constants for P2 Cleavage—Reaction mixtures containing increasing concentrations of P2 (14 samples; 0–25 μm), 50 μm PCPS, and 40 nm Va in Assay Buffer at 25 °C were initiated with 5 nm Xa. Six samples (10 μl each) were serially withdrawn over a 3-min period and quenched by mixing with 90 μl of Assay Buffer lacking Ca2+ but containing 50 mm EDTA. Following further dilution in the same buffer, concentrations of thrombin present in the quenched samples were inferred from initial velocity measurements of peptidyl substrate cleavage as described (29). Initial velocities were determined from the linear appearance of thrombin as a function of time. For product inhibition studies with IIai, initial velocities were determined with increasing P2 (14 concentrations; 0–20 μm) in the presence of 0, 6, 12, and 24 μm IIai.
Steady State Kinetic Studies of P2 Cleavage in the Presence of F12—Initial velocities for thrombin formation were determined using reaction mixtures containing 1.4 μm P2 and increasing concentrations of F12, 30 μm PCPS, 30 nm Va, and 10 pm Xa at 25 °C. Alternatively, initial velocities were determined using increasing concentrations of P2 and different fixed concentrations of F12 with prothrombinase assembled using 27 nm Va, 31 μm PCPSLUV, and 3.8 pm Xa.
Assessment of Xa-mediated Cleavage of II at Position 155—Reaction mixtures containing 1.4 μm IIQQ 50 μm PCPS, and varying concentrations of Va (0, 5, and 30 nm) in Assay Buffer at 25 °C were initiated with 5 nm Xa. A fourth reaction mixture with similar concentrations of IIQQ and PCPS but with 5 nm Va was initiated with 30 nm Xa. Two samples (30 μl each) were withdrawn at 0 and 20 min and quenched by mixing with an equal volume of 125 mm Tris, 20% (v/v) glycerol, 2% (w/v) SDS, 0.02% (w/v) bromphenol blue, and 50 mm EDTA. Half of the quenched samples were treated with 83 mm dithiothreitol, pH 6.8. After heating at 89 °C for 3 min, the samples were analyzed by SDS-PAGE as above and stained with Colloidal Blue stain (Invitrogen).
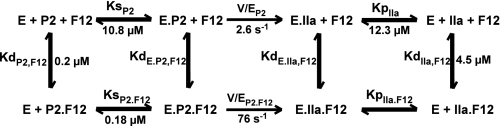
Data Analysis—For all kinetic studies, concentrations of PCPS and Va were chosen to be in excess of the stoichiometries and equilibrium dissociation constants for the interactions with Xa that lead to prothrombinase assembly (30). Hence, the concentration of prothrombinase was considered equal to the limiting concentration of Xa. At low concentrations of enzyme, kinetic complexity arising from partitioning of substrate to vesicles lacking enzyme was minimized using PCPSLUV (12). Nonlinear least squares analysis using the Levenberg-Marquardt algorithm (31) was employed to fit parameters to the indicated explicit equations, which are reported ±95% confidence limits. Steady state kinetic constants were determined by analysis according to the Henri-Michaelis-Menten equation or by analysis of product inhibition profiles using the rate expression for classical competitive inhibition (32). Heat flow traces were analyzed using Origin scripts provided by Microcal (Northampton, MA) to yield ΔH, Kd, and stoichiometry for the binding of titrant to independent and noninteracting sites. ΔG and ΔS were given by the relationships ΔG = RTlnKd and ΔG =ΔH - TΔS. Global analysis according to Scheme 2 was performed by combining non-least squares approaches with the numerical solution of ordinary differential equations using Dynafit version 3.28.050, a generous gift of Petr Kuzmic (BioKin, Pullman, WA) (33), to derive the indicated fitted parameters and the corresponding linear approximations of their 95% confidence limits. The data presented are representative of at least two similar experiments performed at an equivalent level of detail and frequently with different protein preparations.
SCHEME 2.
Pathways for P2 cleavage in the presence and absence of F12. The kinetic scheme is annotated with experimentally determined constants from initial velocity, product inhibition, and equilibrium binding measurements more fully described in Table 2.
RESULTS
Changes in Vesicle Size Associated with Prothrombin Cleavage—A series of prothrombin variants were employed to establish relationships between prothrombin cleavage by prothrombinase and steady state light scattering intensity (Fig. 1). At the concentrations employed, the intensity of scattered light is predominantly affected by vesicle size, dictated by the equilibrium distribution of prothrombin and its derived fragments between solution and the membrane surface as well as the molecular weights of the membrane-bound species (34, 35). Measurements done in the absence of added prothrombin or in complete reaction mixtures containing EDTA established minor contributions of membrane-bound prothrombinase or solution phase proteins to the scattering signal.
FIGURE 1.
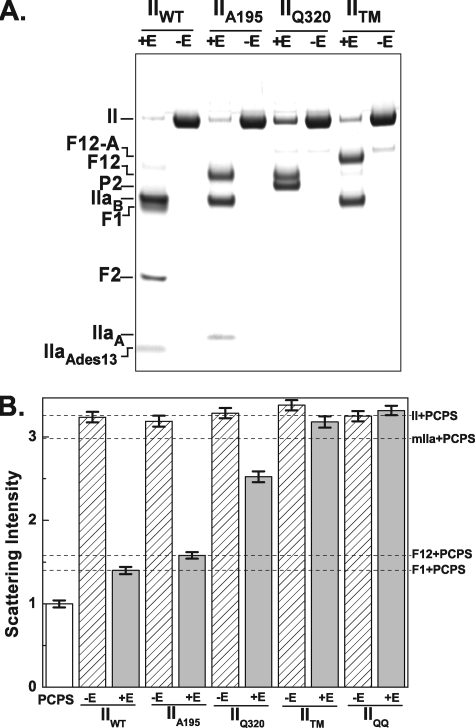
Prolonged action of prothrombinase on prothrombin variants. The indicated prothrombin variants (3 μm) were incubated at 25 °C for 30 min with 30 nm Va, 50 μm PCPS (-E), or 3 nm Xa, 30 nm Va, 50 μm PCPS (+E). A, prothrombin cleavage was assessed by SDS-PAGE following disulfide bond reduction with bands visualized following staining with Coomassie Brilliant Blue. The various fragments are identified in the left margin. B, right angle light scattering intensities measured for the same samples diluted as described under “Experimental Procedures.” Scattering intensities were normalized by setting the signal observed with 50 μm PCPS to 1. Error bars (±2 S.D.), the variation in signal over a 3-min measurement period. Because light scattering intensity is highly dependent on experimental conditions, the error bars provide appropriate confidence limits for assessing changes within an experiment but are probably inaccurate in describing variation between experiments done on different days. Intensities measured with the indicated purified proteins (1.4 μm) plus 50 μm PCPS are indicated by the dashed lines and in the right margin.
Prolonged cleavage of IIWT by prothrombinase in the absence of thrombin inhibitors yielded bands corresponding to those expected from further processing of the terminal products, thrombin and F12, by thrombin produced in the reaction (Fig. 1A). Bands observed following disulfide bond reduction corresponded to F1 and F2 (resulting from cleavage by thrombin at Arg155) and to thrombin B chain and thrombin A chain lacking 13 residues from its NH2 terminus (resulting from cleavage by thrombin at Arg284). In this case, light scattering intensity decreased to the level observed with the same concentrations of purified F1 and PCPS (Fig. 1B). Prolonged cleavage of IIA195 yielded F12 and the two chains derived from thrombin (Fig. 1A). Because IIaA195 produced in this reaction is devoid of catalytic activity, further processing at Arg155 and Arg284 was not evident. Light scattering intensity decreased to the level observed with purified F12 and PCPS (Fig. 1B). As expected, cleavage of IIQ320 with prothrombinase resulted in the formation of F12 and P2 (Fig. 1A). However, this reaction yielded a smaller decrease in scattering intensity in comparison with that observed with the cleavage of IIA195 (Fig. 1B). Additionally, cleavage of IITM to yield an mIIa derivative resistant to further proteolytic processing or the addition of prothrombinase to cleavage resistant IIQQ was accompanied by relatively minor changes in light scattering intensity (Fig. 1B), consistent with the equivalent size of prothrombin and mIIa as well as the established ability of these species to bind PCPS with similar affinity (36, 37). Our findings indicate that thrombin formation, irrespective of further processing at Arg155, leads to a decrease in inferred size of the PCPS vesicle. Limiting scattering intensities in line with those expected for vesicle-bound F1 or F12 suggest that the bulk of thrombin produced readily dissociates from the membrane surface even when F12 is produced as a stable product. Thus, cleavage at Arg155 seemingly plays an undetectable role in regulating the release of nascent thrombin from the site of its production. In contrast, the smaller decrease in light scattering evident upon formation of P2 plus F12 implies that a significant fraction of P2 may remain membrane-bound through interactions mediated by F12.
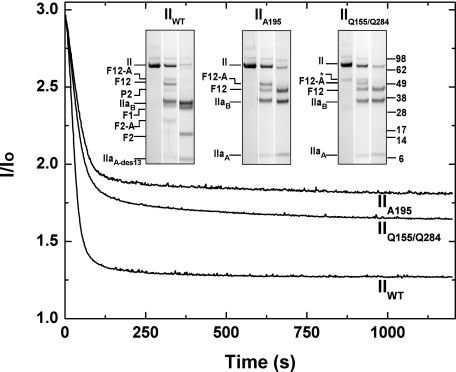
Kinetics of Cleavage and Release of Prothrombin-derived Species from the Membrane Surface—More detailed approaches correlated changes in light scattering intensity with the kinetics of prothrombin cleavage at its physiological concentration. The addition of prothrombinase to IIWT in the absence of thrombin inhibitors led to a rapid decrease in normalized light scattering intensity (Fig. 2). Selected lanes from an SDS-PAGE analysis of the full time course of cleavage are presented to illustrate that the decrease in light scattering was contemporaneous with prothrombin cleavage, and a limiting light scattering signal was achieved upon quantitative cleavage at Arg155 to produce F1. Accordingly, the limiting intensity of scattered light was similar to the reference signal obtained with a purified mixture of F1 and PCPS. Similar experiments were performed with IIA195 that yields inactive thrombin and IIQ155/Q284 that yields active proteinase but cannot be processed at residues 155 and 284. The addition of prothrombinase to either substrate also yielded a rapid decrease in light scattering but instead to a limiting value, in approximate agreement with the signal observed with a purified mixture of F12 and PCPS (Fig. 2). For either substrate, the selected lanes from an SDS-PAGE analysis of the full time course (Fig. 2) illustrate that the scattering changes occurred on the same time scale as substrate cleavage and yielded thrombin and F12 as the final products. Variability in the limiting scattering signal seen with the two species probably reflects some differences in the extent of substrate cleavage (Fig. 2). These findings are generally consistent with the interpretation that thrombin is readily and rapidly released from the membrane surface upon its formation regardless of the proteolysis of F12 to F1 and F2 through cleavages mediated by thrombin. In agreement with recent observations (8, 9), these findings also imply that potential interactions between thrombin and the F2 domain within F12 are relatively weak and play a minor role in retaining thrombin on the membrane surface at the site of its formation.
FIGURE 2.
Prothrombin activation monitored by light scattering in the absence or presence of feedback cleavages by thrombin. Light scattering intensity was continuously monitored during proteolytic cleavage of the indicated prothrombin variants (1.4 μm) by 1 nm prothrombinase (1 nm Xa, 30 nm Va, 50 μm PCPS). Traces were normalized by setting the scattering intensity observed in the absence of added prothrombin to 1. Insets, selected lanes (reaction times of 0, 0.5, and 20 min following initiation) assembled from a full time course analyzed by SDS-PAGE following disulfide bond reduction for each indicated variant. The various polypeptide species are indicated in the left margin of each inset, and the migration of molecular weight markers (Mr × 10-3) is indicated on the right. The asterisk denotes a band of unestablished identity present in the preparation of IIQ155/Q284.
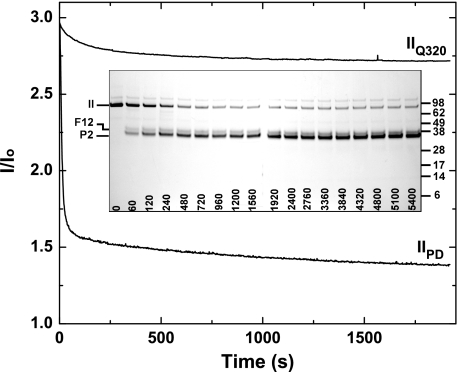
Similar studies with IIQ320 required higher concentrations of prothrombinase for effective cleavage on this time scale (Fig. 3). This is consistent with the established ability of prothrombinase to cleave at the Arg271 site in intact prothrombin with a significantly reduced Vmax (12). In comparison with prothrombin isolated from plasma used as a reference under the same conditions, cleavage of IIQ320 to yield P2 and F12 produced a small change in light scattering that correlated with the formation of products (Fig. 3). The limiting light scattering signal was substantially greater than that observed with a purified mixture of F12 and PCPS. These findings imply that in contrast to thrombin, the interaction between P2 and F12 is sufficiently tight to allow a sizable fraction of P2 to remain membrane-bound.
FIGURE 3.
Light scattering changes associated with the formation of P2 plus F12. Light scattering progress curves for the cleavage of either 1.4 μm IIQ320 or of 1.4 μm prothrombin isolated from plasma (IIPD) following the addition of 5 nm prothrombinase (5 nm Xa, 30 nm Va, 50 μm PCPS). Scattering intensity was normalized to the signal observed in the absence of added prothrombin. Inset, SDS-PAGE analysis of the cleavage of IIQ320 yielding P2 and F12. The migration of molecular weight markers (Mr × 10-3) is indicated in the right margin.
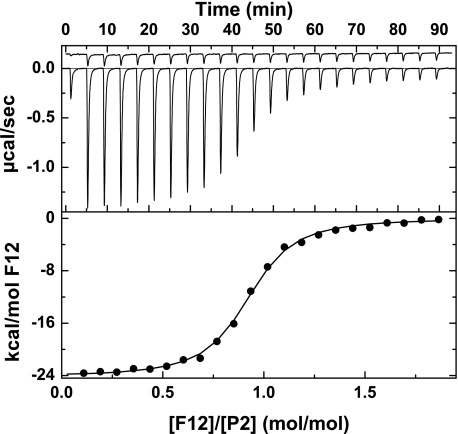
Binding of Thrombin and P2 to the F2 Domain within F12—Implied differences in the equilibrium dissociation constants for the binding of thrombin and P2 to F12 were pursued by isothermal titration calorimetry. We employed P2A195 and IIaA195 to permit appropriate comparisons without the need to covalently inactivate thrombin. Further, both P2A195 and IIaA195 possessed an N-terminal sequence consistent with cleavage at Arg271 with no evidence for proteolysis at Arg284 (data not shown). Incremental additions of F12 to P2A195 yielded large changes in heat flow that decreased as the concentration of F12 exceeded that of P2A195 (Fig. 4). Small contributions from heats of dilution were corrected for by parallel injections of F12 into buffer (Fig. 4). The data could be adequately described by the interaction of titrant with independent, noninteracting sites with the indicated thermodynamic parameters (Table 1). F12 binds P2A195 with an equilibrium dissociation constant of ∼10-7 m and a stoichiometry of ∼1 in an enthalpically driven reaction. Similar experiments with IIaA195 revealed a ∼20-fold lower affinity of the proteinase for F12 that could completely be accounted for by a weaker interaction between IIaA195 and F2 (Table 1). Consistent with recent findings (8, 9, 19), the interaction between P2A195 and F2 was also weak and comparable with the affinities observed with IIaA195 (Table 1). The thermodynamic constants are consistent with a large ordering effect observed in the interaction of P2 with F12 that is substantially reduced with IIaA195 or the binding of F2 to either species. Our findings suggest an indirect effect of the F1 domain in modulating the binding of the F2 domain to the zymogen but not the proteinase. The measured equilibrium constants also provide a quantitative explanation for the results of the light scattering studies, indicating the ability of F12 to retain significant concentrations of P2 but not thrombin on the membrane surface.
FIGURE 4.
Thermodynamic measurement of the binding of P2 to F12. Top, heat flow traces measured by isothermal titration calorimetry following sequential injections (10.1 μl) of 465 μm F12 into a cell containing 12.97 μm P2A195 (lower trace) or buffer (upper trace). The first injection was 2 μl. All components were in assay buffer lacking polyethylene glycol. Bottom, binding isotherm constructed by integrating the heat flow traces shown in the top. The line is drawn following analysis of the binding of F12 to independent and noninteracting sites using the fitted parameters listed in Table 1.
TABLE 1.
Calorimetric measurements of the binding of fragment 1.2 or fragment 2 to thrombin or prethrombin 2
| Liganda | Titrant | Kd ± S.D. | n ± S.D.b | ΔH ± S.D. | ΔG ± S.D. | ΔS ± S.D. |
|---|---|---|---|---|---|---|
| μm | mol/mol | kcal·mol–1 | kcal·mol–1 | cal·K–1·mol–1 | ||
| P2S195A | F12 | 0.20 ± 0.01 | 0.90 ± 0.01 | –24.2 ± 0.06 | –9.14 ± 0.02 | –51.0 ± 0.2 |
| IIaS195A | F12 | 4.54 ± 0.3 | 0.93 ± 0.02 | –13.6 ± 0.4 | –7.28 ± 0.04 | –21.2 ± 1.2 |
| P2S195A | F2 | 1.59 ± 0.13 | 1.02 ± 0.02 | –11.0 ± 0.25 | –7.90 ± 0.07 | –10.4 ± 1.11 |
| IIaS195A | F2 | 3.14 ± 0.12 | 1.20 ± 0.01 | –11.4 ± 0.1 | –7.50 ± 0.02 | –13.0 ± 0.4 |
The species in the cell is denoted as ligand, and the species in the syringe is denoted as titrant
Mol of titrant bound/mol of ligand at saturation
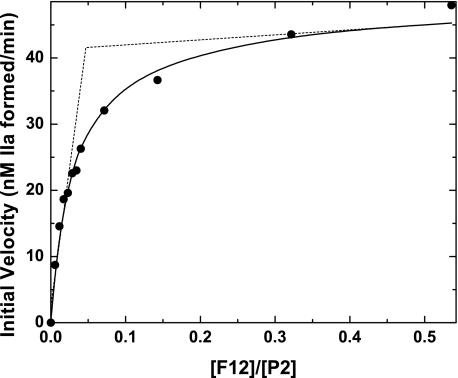
Functional Effects F12 on P2 Cleavage—The relatively tight interaction between P2 and F12 and membrane binding by the resultant complex provides a rational explanation for the established ability of F12 to enhance P2 cleavage by prothrombinase (12, 13). However, initial velocity studies of P2 cleavage by prothrombinase in the presence of increasing concentrations of F12 revealed features that appeared inconsistent with the physical measurements (Fig. 5). The initial rate for thrombin formation was increased greatly and saturably with increasing concentrations of F12. Surprisingly, the rate saturated at 0.1–0.2 equivalents of F12, well below the 1:1 stoichiometry established by calorimetry (Fig. 5). The equivalents of F12 required for saturating rate effects varied with the fixed concentration of P2 (not shown). On the surface, these peculiar findings suggest fundamental problems in reconciling physical measurements with the functional contributions of F12 to prothrombinase function. It is possible that a large fraction of P2 is dysfunctional as a substrate despite its ability to bind F12. This possibility is not consistent with the nearly quantitative conversion of P2 to thrombin both in the presence and absence of F12. Alternatively, the observations could reflect kinetic limitations on the system despite agreement of the component steps with thermodynamic principles and independently established equilibrium constants and stoichiometries.
FIGURE 5.
Enhanced conversion of P2 to thrombin in the presence of F12. Initial rates of thrombin formation were measured in reaction mixtures containing 1.4 μm P2, increasing concentrations of F12, and 10 pm prothrombinase (10 pm Xa, 30 nm Va, 30 μm PCPS). The solid line is drawn following analysis according to Scheme 2 using the fixed and fitted parameters listed in Table 2. The dashed lines are drawn following a linear regression of the limits of the data to illustrate that saturation is achieved at a substoichiometric equivalent of F12.
Kinetic Analysis of P2 Cleavage by Prothrombinase—The simplest possible kinetic model accounting for the action of prothrombinase on P2 either in the absence or presence of F12 is illustrated in Scheme 2. It is assumed that one equivalent of F12 binds per mole of P2, converting it to a substrate that is cleaved with higher catalytic efficiency (Scheme 2). An equivalent assumption applies for the binding of F12 to thrombin, except that the interactions related to thrombin are not informative for initial velocity measurements. Furthermore, although extensive evidence in the literature indicates a more complex binding pathway for either substrate to prothrombinase through both exosite and active site interactions, the overall process can be adequately described employing the rapid equilibrium assumption and the simplified pathways as illustrated (13). The caveat here is that the inferred rate constant for the catalytic step also contains an additional binding term and is therefore referred to as V/E rather than kcat (38).
Initial velocity and product inhibition studies were used to establish the kinetic constants for the cleavage of P2 and inhibition by the product (Table 2). Kinetic constants for the cleavage of P2 bound to F12 under equivalent conditions were taken from Orcutt and Krishnaswamy (12). These values along with measured thermodynamic constants were used as the quantitative basis to test for the ability of Scheme 2 to describe the function of F12 in regulating P2 cleavage by prothrombinase.
TABLE 2.
Prethrombin 2 cleavage in the absence and presence of fragment 1.2
|
Constanta
|
Units
|
Measured
valueb
|
Sourcec
|
Fitted
termsd
|
|
|---|---|---|---|---|---|
| Fig. 5 | Fig. 6 | ||||
| Kd(P2·F12) | nm | 200 ± 10 | a | F | 165 ± 19 |
| n (mol F12/mol P2) | 0.90 ± 0.01 | a | F (1) | F (1) | |
| Kd(IIa·F12) | μm | 4.54 ± 0.3 | a | F | F |
| n (mol F12/mol IIa) | 0.93 ± 0.02 | a | F (1) | F (1) | |
| Ks(P2) | μm | 10.8 ± 0.6 | b | F | F |
| V/EP2 | s–1 | 2.62 ± 0.08 | b | F | F |
| Kp(IIa) | μm | 12.3 ± 0.3 | c | F | F |
| Ks(P2·F12) | nM | 176 ± 11 | d | 45.8 ± 6.3 | 158 ± 7 |
| V/EP2·F12 | s–1 | 76 ± 1 | d | 98 ± 4 | 114 ± 2 |
| Kd(E·P2,F12) | nm | 2.5 ± 0.4 | e | 0.7 ± 0.2 | 2.4 ± 0.4 |
| Kp(IIa·F12) | NDe | NF | NF | ||
| Kd(E·IIa,F12) | ND | NF | NF | ||
Symbolic constants correspond to those illustrated in Scheme 2
Measured values are presented ±95% confidence limits
Parameters are derived from isothermal titration calorimetry (a), derived from initial velocity studies (b), derived from product inhibition studies (c), taken from Orcutt and Krishnaswamy (12) (d), and calculated from Kd(P2·F12), Ks(P2), and Ks(P2·F12) (e)
Fitted terms are listed ±linear approximations of the 95% confidence limits. F denotes terms that were fixed using the measured values or fixed at the value indicated in parentheses. NF, not fitted
ND, not determined
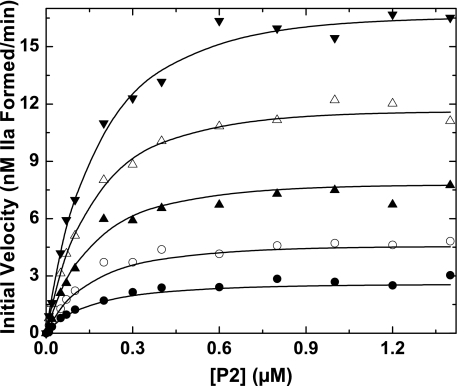
This scheme could provide a reasonable description of the data in Fig. 5 despite the assumption of a unit stoichiometry for the interaction of F12 with P2. The informative fitted parameters from this analysis were in approximate agreement with the independently measured terms. Under these conditions, Ks(P2,F12) is probably compromised because of transport limitations arising from the use of low concentrations of enzyme and small unilamellar PCPS vesicles (12). This potential limitation was circumvented by analysis of a more comprehensive data set obtained using PCPSLUV and varying concentrations of P2 at different fixed concentrations of F12 (Fig. 6). These data could also be adequately described following global analysis according to Scheme 2 (Fig. 6). The informative parameters that could be reliably fitted were in good agreement with the independently measured terms (Table 2). Thus, Scheme 2, grounded in independently determined equilibrium constants and stoichiometries for the relatively weak binding of F12 to thrombin or its ∼20-fold tighter interaction with P2, can adequately describe the major enhancing effects of F12 on P2 cleavage by prothrombinase under a wide range of conditions. The explanation for substoichiometric concentrations of F12 to saturably enhance P2 cleavage lies in the fact that the rate approaches a limiting value when the concentration of the P2·F12 complex significantly exceeds Ks(P2·F12), regardless of the total concentrations of P2 and F12 present. Major rate enhancements arising from the ability of F12 to bind P2 and facilitate its interaction with membranes paradoxically occur at substantially lower concentrations of F12 than those necessary to saturate P2. These findings also highlight the foregone dangers of inferring details of protein-protein interactions solely from functional studies with incomplete consideration of the kinetic behavior of the system.
FIGURE 6.
Kinetics of P2 cleavage in the presence of F12. Initial velocities of thrombin formation were measured in reaction mixtures containing increasing concentrations of P2, 3.8 pm prothrombinase (3.8 pm Xa, 27 nm Va, 31 μm PCPSLUV) using concentrations of F12 fixed at 24 nm (•), 48 nm (○), 100 nm (▴), 200 nm (▵), and 500 nm (▾). The lines are drawn following analysis according to Scheme 2 using the fixed and fitted parameters listed in Table 2.
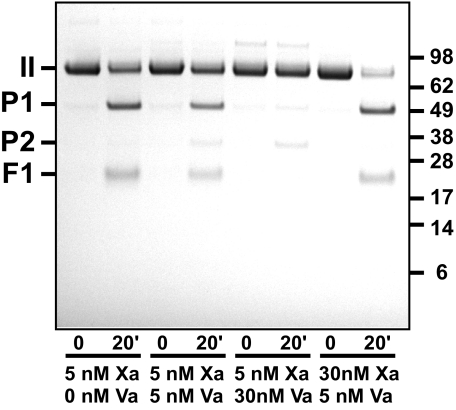
Cleavage at Arg155—Both thrombin and mIIa are well known to catalyze cleavage at Arg155 in prothrombin and its derivatives (25, 39, 40). Thus, high concentrations of proteinase produced during prothrombin activation necessitate the use of thrombin inhibitors such as DAPA to selectively inhibit this feedback reaction (41). This becomes necessary because loss of the F1 domain associated with cleavage at Arg155 impacts product formation from prothrombin, mIIa, and P2 plus F12 catalyzed by prothrombinase (21, 42). We noted that cleavage at Arg155 could not be fully explained by the activities of thrombin or mIIa. This observation was most directly documented with IIQQ, which cannot be cleaved at Arg320 or Arg271 at an appreciable rate by Xa or by prothrombinase (12). Prolonged incubation of IIQQ with Xa and PCPS revealed the formation of bands arising from the cleavage of prothrombin at Arg155 and confirmed by N-terminal sequence analysis (Fig. 7). These products continued to be seen even in the presence of high concentrations of DAPA or with IIA195 but were not produced when PCPS was omitted or when IIQ155/Q284 was used as a substrate (not shown). Furthermore, cleavage at Arg155 was reduced and eventually eliminated with increasing concentrations of Va (Fig. 7). These findings indicate that Arg155 can be effectively cleaved by Xa in the presence of membranes, but this activity of Xa is lost upon its interaction with Va on the membrane surface. This conclusion is best illustrated by comparison of the findings made with 5 nm Xa plus 30 nm Va versus 30 nm Xa plus 5 nm Va (Fig. 7). Despite the fact that both conditions are expected to yield 5 nm prothrombinase, bands arising from cleavage at Arg155 were only seen under conditions in which excess free Xa was present. Thus, free Xa itself can catalyze cleavage at Arg155 and regulate prothrombinase function by altering the partitioning of substrate species to the membrane surface.
FIGURE 7.
Cleavage of prothrombin at Arg155 by Xa but not prothrombinase. SDS-PAGE analysis following incubation of 1.4 μm IIQQ with the indicated concentrations of Xa and Va and 50 μm PCPS. Bands denoted as P1 and F1 were identified by N-terminal sequence analysis. Traces of P2 evident following prolonged incubation with prothrombinase arise from slow but detectable cleavage at position 271 despite substitution of Arg with Gln. Migration positions of the molecular weight markers (Mr × 10-3) are indicated in the right margin.
DISCUSSION
We show that prothrombin and both possible reaction intermediates (Scheme 1) bind comparably to membranes during the action of prothrombinase on prothrombin. In the case of prothrombin and mIIa, membrane binding arises from the well known contributions of the F1 domain covalently linked to the rest of the molecule. Membrane binding by P2 arises from a reversible but relatively tight interaction with F12, which can bridge the singly cleaved intermediate to the membrane surface. The large rate-enhancing effects of F12 on P2 cleavage by prothrombinase can be quantitatively accounted for by the interaction of F12 with P2. Since membrane binding plays an important role in the utilization of each of these substrates by prothrombinase, loss of the F1 domain arising from thrombin-mediated cleavage at Arg155 is expected to be important for the regulation of their cleavage. On the other hand, thrombin binds the F2 domain within F12 with ∼20-fold lower affinity and is therefore readily released from the membrane surface upon its formation. Nearly quantitative product release is independent of prior cleavage at Arg155 previously proposed to represent a key regulatory reaction of thrombin necessary to facilitate release of thrombin from the membrane surface (14). Our findings reconcile physical parameters for the reversible interaction between thrombin or P2 and the F2 domain within F12 with the ability of the activation peptide to regulate the fates of membrane-bound substrates and products in the action of prothrombinase on prothrombin.
The equilibrium dissociation constants for interactions of thrombin or P2 with F2 or F12 are in agreement with those determined in careful studies with human proteins (8, 9, 19). We speculate that the somewhat higher affinity we report for the binding of F12 to P2 could reflect the fact that P2 prepared by preparative proteolysis of IIA195 or generated in situ with IIQ320 is not proteolyzed at Arg284 and therefore possesses an authentic NH2 terminus. The thermodynamic measurements also point to a major ordering effect associated with the formation of the F12·P2 complex, seen to a far lesser extent upon binding of F12 to thrombin. This idea is consistent with the previous suggestion that one mechanism underlying enhanced substrate function upon binding of F12 to P2 is probably related to changes in substrate structure (7). F2 and by extension, F12, bind to residues within the anion binding exosite II region in thrombin and P2 (10). The reduction in affinity for F12 that accompanies the conversion of the zymogen to the proteinase mirrors the increase in affinity for hirugen, a prototypic ligand for anion binding exosite I (9, 19). Thus, the conversion of zymogen to proteinase is accompanied by reciprocal changes of approximately equal magnitude in liganding at these two exosites.
The ∼20-fold tighter interaction of F12 with P2 in comparison with thrombin lies at the heart of the ability of F12 to tether substantial concentrations of P2 but not thrombin to the membrane surface. However, our findings are at variance with those in a previous report dealing with the activation of bovine prothrombin (14). Those studies documented that conversion of prothrombin to thrombin plus F12 by prothrombinase measured in the presence of DAPA was not accompanied by changes in light scattering indicating the replacement of membrane-bound prothrombin by an equivalent concentration of thrombin bound to F12 (14). A rapid decrease in light scattering observed in the absence of DAPA was interpreted in terms of an essential role for feedback cleavage at Arg155 in mediating thrombin release from the membrane surface (14). These differences with our findings could clearly reflect the intrinsic properties of fragments derived from bovine versus human prothrombin. This possibility is highlighted by the ∼10-fold higher affinity along with obvious differences in thermodynamic parameters previously measured for the binding of bovine F2 to bovine P2 by calorimetric approaches (7). Support for such possible species-specific differences is provided by x-ray structures indicating differences in contacts between human thrombin complexed with bovine F2 in comparison with its complex with human F2 (10). There is also a difference in one of the key residues of anion binding exosite II in the proteinase domain of the two thrombin species found at the interface with F2. Alternatively, the previous light scattering studies could also have been impacted by the optical activity of DAPA at the wavelength used for the measurements and its change in fluorescence upon binding to newly formed thrombin. In recognition of this possible complexity, our conclusions are based on approaches conducted with mutant derivatives of prothrombin that obviate the need for DAPA or other inhibitors to block the feedback reactions of thrombin.
Conclusions derived from reaction progress assessed by light scattering could have been strengthened by a more detailed quantitative analysis relating concentrations of various substrate and product species with scattering intensity. We have deliberately chosen to document agreement between substrate cleavage and the scattering change in a rough way, focusing instead on the limiting scattering signal when the reaction was nearly complete with a relatively stable population of reactants and products. This approach was necessitated by the complex relationship between scattering intensity, vesicle size, concentrations of various membrane-bound species, and the changing distributions between these species during reaction progress (34, 35). However, the conclusions derived from these measurements can be fully rationalized by the measured equilibrium constants for the binding of F12 to either thrombin or P2, which predict that >70% of the P2 versus ∼17% of thrombin would be expected to be bound to F12 at the highest concentrations of these species (1.4 μm) that can be produced in blood. The activation of most thrombin substrates in clotting blood is expected to occur during the initiation phase, when less than 2 nm thrombin is produced (43). Because F12 is not necessarily expected to accumulate in vast excess of thrombin (Scheme 1), only very minor concentrations of thrombin will probably be complexed with F12 when the reactions of thrombin in blood coagulation are most relevant. Concentration estimates for P2 produced as an intermediate under physiological conditions are not available. However, the higher affinity for the 1:1 interaction between P2 and F12 provides an adequate quantitative explanation for the enhancing effects of F12 on P2 cleavage by prothrombinase, at least under defined conditions. Kinetic measurements under a wide range of conditions can be adequately accounted for by Scheme 2 and the associated independently determined kinetic constants despite the fact that the dependence of reaction rate on the concentrations of P2 and F12 superficially appears inconsistent with this model. An alternative possibility is that F12 functions by directly binding E and thus facilitating P2 cleavage with increased efficiency. This possibility requires formal consideration because of reports implicating the F1 domain in binding interactions between the substrate and Va and the ability of added F1 to modulate cleavage of prothrombin species lacking this domain (44–46). Fitting according to this type of model failed to provide an adequate description of the experimental data (not shown).
Cleavage at Arg155 and the associated release of the F1 domain is expected to play an important regulatory role in modulating the membrane-dependent delivery of prothrombin, mIIa, and P2·F12 to prothrombinase. In addition to thrombin, which has long been recognized to be the proteinase responsible for cleavage at this site (47, 48), our studies reveal that cleavage at this site can also be accomplished by Xa bound to membranes. It is presently unclear whether this reaction is specific for human Xa acting on human prothrombin or also has bearing on the initial report that cleavage of bovine prothrombin at the equivalent site can be accomplished by Xa (49). Because this function of Xa is substantially reduced upon its assembly into prothrombinase, the rate of cleavage at Arg155 is dependent on the free concentration of Xa and not the concentration of prothrombinase. Consequently, processing of substrate species at Arg155 and the associated loss in their membrane binding is expected to predominate under conditions when Xa is in excess and Va is limiting but not when Va is in excess to saturably form prothrombinase. Because loss of membrane binding by prothrombin resulting from cleavage at Arg155 is associated with a change in the relative contributions of the two pathways for bond cleavage by prothrombinase (42), different cleavage intermediates are expected to be observed at the same concentration of prothrombinase assembled with either Xa in excess or Va in excess. These ideas probably have bearing on the fragments produced as well as the inferred pathway for prothrombin cleavage, depending on whether the concentration of Va is limiting or when it is present at saturating concentrations.
In summary, our findings are consistent with the interpretation that all substrate species, prothrombin and both possible intermediates, can bind membranes through interactions mediated by the fragment 1 domain. In the case of P2, membrane binding is mediated by a reversible association of higher affinity with F12, which adequately explains the ability of F12 to yield large enhancements in reaction rate at physiologically relevant concentrations. In contrast, upon formation of thrombin, there is an accompanying ∼20-fold reduction in the affinity of F12 facilitating the dissociation of the bulk of thrombin from F12 even at the highest possible concentrations of products that can be achieved in blood. This reduced affinity for F12 allows for the release of thrombin from the membrane surface and the site of its production.
This work was supported, in whole or in part, by National Institutes of Health (NHLBI) Grants HL-74124 and HL-47465 (to S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: F1, fragment 1; DAPA, dansyl-l-arginine-N-(3-ethyl-1,5-pentanediyl)amide; FPRck, d-phenylalanyl-l-prolyl-l-arginine chloromethyl ketone; F12, fragment 1.2; F2, fragment 2; II, prothrombin; IIA195, recombinant II containing Ala in place of the catalytic Ser; IIPD, II isolated from plasma; IIQ155/Q284, recombinant II containing Gln substitutions at Arg155 and Arg284;IIQ320, recombinant II containing Gln in place of Arg320; IIQQ, recombinant II containing Gln in place of Arg320 and Arg271; IITM, a triple mutant of II containing Gln in place of Arg at positions 155, 284, and 271; IIWT, recombinant wild type II; IIai, thrombin inactivated with FPRck; mIIa, meizothrombin; mIIai, mIIa inactivated with FPRck; P2, prethrombin 2; IIaA195, thrombin produced from IIA195; P2A195, P2 produced from IIA195; PC, l-α-phosphatidylcholine; PS, l-α-phosphatidylserine; PCPS, small unilamellar vesicles containing 75% (w/w) PC and 25% (w/w) PS; PCPSLUV, large unilamellar PCPS vesicles; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MES, 4-morpholineethanesulfonic acid.
Cleavage sites in prothrombin are denoted based on consecutive numbering of the residues in mature prothrombin. The catalytic serine is numbered according to homology between the catalytic domain and chymotrypsinogen (50).
References
- 1.Mann, K. G., Jenny, R. J., and Krishnaswamy, S. (1988) Annu. Rev. Biochem. 57 915-956 [DOI] [PubMed] [Google Scholar]
- 2.Mann, K. G., Nesheim, M. E., Church, W. R., Haley, P., and Krishnaswamy, S. (1990) Blood 76 1-16 [PubMed] [Google Scholar]
- 3.Jackson, C. M., and Nemerson, Y. (1980) Annu. Rev. Biochem. 49 765-811 [DOI] [PubMed] [Google Scholar]
- 4.Gitel, S. N., Owen, W. G., Esmon, C. T., and Jackson, C. M. (1973) Proc. Natl. Acad. Sci. U. S. A. 70 1344-1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenny, N. S., Lundblad, R. L., and Mann, K. G. (2006) Thrombin. In Colman, R. W., Marder, V. J., Clowes, A. J., George, J. N., and Goldhaber, S. Z. (eds) Hemostasis and Thrombosis: Basic Principles and Clinical Practice, pp. 193-213 Lippincott Williams & Wilkins, Philadelphia
- 6.Myrmel, K. H., Lundblad, R. L., and Mann, K. G. (1976) Biochemistry 15 1767-1773 [DOI] [PubMed] [Google Scholar]
- 7.Krishnaswamy, S., and Walker, R. K. (1997) Biochemistry 36 3319-3330 [DOI] [PubMed] [Google Scholar]
- 8.Bock, P. E. (1992) J. Biol. Chem. 267 14974-14981 [PubMed] [Google Scholar]
- 9.Anderson, P. J., Nesset, A., and Bock, P. E. (2003) J. Biol. Chem. 278 44482-44488 [DOI] [PubMed] [Google Scholar]
- 10.Arni, R. K., Padmanabhan, K., Padmanabhan, K. P., Wu, T. P., and Tulinsky, A. (1993) Biochemistry 32 4727-4737 [DOI] [PubMed] [Google Scholar]
- 11.Brufatto, N., and Nesheim, M. E. (2003) J. Biol. Chem. 278 6755-6764 [DOI] [PubMed] [Google Scholar]
- 12.Orcutt, S. J., and Krishnaswamy, S. (2004) J. Biol. Chem. 279 54927-54936 [DOI] [PubMed] [Google Scholar]
- 13.Orcutt, S. J., Pietropaolo, C., and Krishnaswamy, S. (2002) J. Biol. Chem. 277 46191-46196 [DOI] [PubMed] [Google Scholar]
- 14.Nesheim, M. E., Abbott, T., Jenny, R., and Mann, K. G. (1988) J. Biol. Chem. 263 1037-1044 [PubMed] [Google Scholar]
- 15.Walker, F. J., and Esmon, C. T. (1979) J. Biol. Chem. 254 5618-5622 [PubMed] [Google Scholar]
- 16.Fredenburgh, J. C., Stafford, A. R., and Weitz, J. I. (1997) J. Biol. Chem. 272 25493-25499 [DOI] [PubMed] [Google Scholar]
- 17.Dasgupta, S. K., and Thiagarajan, P. (2007) J. Thromb. Thrombolysis 24 157-162 [DOI] [PubMed] [Google Scholar]
- 18.Stubbs, M. T., and Bode, W. (1995) Trends Biochem. Sci. 20 23-28 [DOI] [PubMed] [Google Scholar]
- 19.Anderson, P. J., and Bock, P. E. (2003) J. Biol. Chem. 278 44489-44495 [DOI] [PubMed] [Google Scholar]
- 20.Krishnaswamy, S., Church, W. R., Nesheim, M. E., and Mann, K. G. (1987) J. Biol. Chem. 262 3291-3299 [PubMed] [Google Scholar]
- 21.Walker, R. K., and Krishnaswamy, S. (1994) J. Biol. Chem. 269 27441-27450 [PubMed] [Google Scholar]
- 22.Price, P. A. (2002) Methods Enzymol. 91 13-17 [DOI] [PubMed] [Google Scholar]
- 23.Camire, R. M., Larson, P. J., Stafford, D. W., and High, K. A. (2000) Biochemistry 39 14322-14329 [DOI] [PubMed] [Google Scholar]
- 24.Buddai, S. K., Toulokhonova, L., Bergum, P. W., Vlasuk, G. P., and Krishnaswamy, S. (2002) J. Biol. Chem. 277 26689-26698 [DOI] [PubMed] [Google Scholar]
- 25.Mann, K. G., Elion, J., Butkowski, R. J., Downing, M., and Nesheim, M. E. (1981) Methods Enzymol. 80 286-302 [DOI] [PubMed] [Google Scholar]
- 26.Di Scipio, R. G., Hermodson, M. A., and Davie, E. W. (1977) Biochemistry 16 5253-5260 [DOI] [PubMed] [Google Scholar]
- 27.Toso, R., and Camire, R. M. (2004) J. Biol. Chem. 279 21643-21650 [DOI] [PubMed] [Google Scholar]
- 28.Lundblad, R. L., Kingdon, H. S., and Mann, K. G. (1976) Methods Enzymol. 45 156-176 [DOI] [PubMed] [Google Scholar]
- 29.Krishnaswamy, S., and Betz, A. (1997) Biochemistry 36 12080-12086 [DOI] [PubMed] [Google Scholar]
- 30.Krishnaswamy, S. (1990) J. Biol. Chem. 265 3708-3718 [PubMed] [Google Scholar]
- 31.Bevington, P. R., and Robinson, K. D. (1992) Data Reduction and Error Analysis for the Physical Sciences, 2nd Ed., pp. 141-164 McGraw-Hill, New York
- 32.Segel, I. H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems, 1st Ed., pp. 100-124 John Wiley & Sons, Inc., New York
- 33.Kuzmic, P. (1996) Anal. Biochem. 237 260-273 [DOI] [PubMed] [Google Scholar]
- 34.Wei, G. J., Bloomfield, V. A., Resnick, R. M., and Nelsestuen, G. L. (1982) Biochemistry 21 1949-1959 [DOI] [PubMed] [Google Scholar]
- 35.Lim, T. K., Bloomfield, V. A., and Nelsestuen, G. L. (1977) Biochemistry 16 4177-4181 [DOI] [PubMed] [Google Scholar]
- 36.Armstrong, S. A., Husten, E. J., Esmon, C. T., and Johnson, A. E. (1990) J. Biol. Chem. 265 6210-6218 [PubMed] [Google Scholar]
- 37.Chen, Q., and Lentz, B. R. (1997) Biochemistry 36 4701-4711 [DOI] [PubMed] [Google Scholar]
- 38.Krishnaswamy, S. (2005) J. Thromb. Haemostasis 3 54-67 [DOI] [PubMed] [Google Scholar]
- 39.Doyle, M. F., and Haley, P. E. (1993) Methods Enzymol. 222 299-312 [DOI] [PubMed] [Google Scholar]
- 40.Doyle, M. F., and Mann, K. G. (1990) J. Biol. Chem. 265 10693-10701 [PubMed] [Google Scholar]
- 41.Nesheim, M. E., Katzmann, J. A., Tracy, P. B., and Mann, K. G. (1981) Methods Enzymol. 80 249-274 [DOI] [PubMed] [Google Scholar]
- 42.Malhotra, O. P., Nesheim, M. E., and Mann, K. G. (1985) J. Biol. Chem. 260 279-287 [PubMed] [Google Scholar]
- 43.Brummel, K. E., Paradis, S. G., Butenas, S., and Mann, K. G. (2002) Blood 100 148-152 [DOI] [PubMed] [Google Scholar]
- 44.Blostein, M. D., Rigby, A. C., Jacobs, M., Furie, B., and Furie, B. C. (2000) J. Biol. Chem. 275 38120-38126 [DOI] [PubMed] [Google Scholar]
- 45.Yegneswaran, S., Mesters, R. M., and Griffin, J. H. (2003) J. Biol. Chem. 278 33312-33318 [DOI] [PubMed] [Google Scholar]
- 46.Bukys, M. A., Orban, T., Kim, P. Y., Nesheim, M. E., and Kalafatis, M. (2008) Thromb. Haemost. 99 511-522 [PubMed] [Google Scholar]
- 47.Esmon, C. T., Owen, W. G., and Jackson, C. M. (1974) J. Biol. Chem. 249 606-611 [PubMed] [Google Scholar]
- 48.Downing, M. R., Butkowski, R. J., Clark, M. M., and Mann, K. G. (1975) J. Biol. Chem. 250 8897-8906 [PubMed] [Google Scholar]
- 49.Heldebrant, C. M., Butkowski, R. J., Bajaj, S. P., and Mann, K. G. (1973) J. Biol. Chem. 248 7149-7163 [PubMed] [Google Scholar]
- 50.Bode, W., Mayr, I., Baumann, U., Huber, R., Stone, S. R., and Hofsteenge, J. (1989) EMBO J. 8 3467-3475 [DOI] [PMC free article] [PubMed] [Google Scholar]