Abstract
The GAL genes, which encode the enzymes required for normal galactose metabolism in yeast, are transcriptionally regulated by three proteins: Gal4p, an activator; Gal80p, an inhibitor; and Gal3p, a galactose sensor. These proteins control the switch between inert and active gene expression. The transcriptional activation function of Gal4p is rendered inactive in the presence of Gal80p. Here we present the three-dimensional structure of a complex between the acidic activation domain of Gal4p and Gal80p. The transactivation domain initiates with an extended region of polypeptide chain followed by two turns of an amphipathic α-helix. It fits into and across a deep cleft within the Gal80p dimer with the protein-protein interface defined primarily by hydrophobic interactions. A disordered loop in the apo-Gal80p structure (Asp-309 to Ser-316) becomes well-defined upon binding of the transactivation domain. This investigation provides a new molecular scaffold for understanding previous biochemical and genetic studies.
In yeast, the GAL genes encode the enzymes of the Leloir pathway, which are required for the conversion of galactose into a metabolically useful form, glucose 6-phosphate (1). The regulation of these genes in response to the organism being challenged with galactose has served as a research paradigm for eukaryotic transcriptional control for over 50 years (1-5). Three key protein components form the GAL regulatory switch: a transcriptional activator, Gal4p; a transcriptional inhibitor, Gal80p; and an inducer, Gal3p. When yeast cells are grown in the absence of galactose, the GAL genes are, for the most part, transcriptionally inert. Under these conditions, Gal4p is produced in the cell and is tethered upstream of the GAL genes (6), but its activity is inhibited by its interaction with Gal80p (7). When galactose is available as a carbon source, the GAL genes are transcribed, both rapidly and to a high level (8). Although the presence of galactose within the cell triggers the activation of Gal4p, neither Gal4p nor Gal80p function as the galactose sensor. Instead, Gal3p serves in this capacity by binding both galactose and ATP and adopting the conformation required for its interaction with Gal80p (1, 9). The net result of this interaction is that Gal4p becomes active, and transcription of the GAL genes proceeds.
Gal4p is a large protein of 881 amino acids with the first ∼100 residues functioning in DNA recognition and dimerization. The last C-terminal residues serve as an acidic transactivation domain (TAD)3 that is required, ultimately, for the recruitment of RNA polymerase II to initiate transcription (10). The amino acid residues comprising the TAD also provide the binding platform for Gal80p (11, 12). Both Gal3p and Gal80p are smaller proteins containing ∼520 and ∼450 amino acids, respectively.
Until very recently, the only three-dimensional structural information available for any of these key regulatory components of the GAL genetic switch was that of an N-terminal 65-residue fragment of Gal4p bound to DNA (13) and the dimerization domain of the same protein (residues 50-106) (14). Through the efforts of our laboratories, however, we now have an excellent homology model for Gal3p based on the structure of the highly similar galactokinase enzyme Gal1p (15). In addition, we have determined the three-dimensional structure of Gal80p from Kluyveromyces lactis to 2.1-Å resolution (16). Strikingly, the overall architecture of Gal80p is similar to that of glucose-fructose oxidoreductase, an enzyme in the sorbitol-gluconate pathway (17). From our combined biochemical and structural studies, we have shown that the K. lactis Gal80p is a dimer with an extensive subunit-subunit interface that buries a total surface area of ∼4400 Å2. Each subunit of the dimeric protein adopts a two-domain architecture with the N-terminal motif containing a classical Rossmann fold and the C-terminal domain dominated by a nine-stranded mixed β-sheet. A pronounced cleft separates the two domains, and on the basis of past mutational studies (18, 19), we postulated in our initial studies that this cleft represents the binding site for the Gal4p TAD. Additionally, in the initial structural analysis of the K. lactis Gal80p, two short loop regions (Asp-245 to Gly-248 and Asp-309 to Ser-316) and two larger regions (Gly-328 to Glu-362 and Leu-394 to Lys-413) were shown to be disordered (16). One of these loops, Asp-309 to Ser-316, was situated near the proposed Gal4p binding site.
Here, we present the crystal structure of K. lactis Gal80p bound to a peptide that mimics the Gal4p TAD. This study provides a molecular model for understanding the biochemical characteristics of the GAL genetic switch, which, to date, is the best understood system for eukaryotic transcriptional control.
EXPERIMENTAL PROCEDURES
X-ray Structural Analysis—The K. lactis Gal80p was cloned, overexpressed, and purified according to previously published procedures (16). The protein utilized for crystallization contained a His tag at the C terminus with the following sequence: LEHHHHHH. Crystals of Gal80p in complex with a 22-mer of the following sequence, TQQLFNTTTMDDVYNYIFDNDE, representing amino acids Thr-844 to Glu-865 of the K. lactis Gal4p TAD (See Fig. 1A), were grown from 20-25% (w/v) pentaerythritol propoxylate 5/4 and 100 mm MES (pH 6.5) via the hanging drop method of vapor diffusion. They belonged to the space group P21 with unit cell dimensions of a = 101.1 Å, b = 160.5 Å, c = 132.6, β = 94.7° and eight subunits (or four dimers) in the asymmetric unit. X-ray data from flashed-cooled crystals were collected at the Structural Biology Center Beamline 19-ID to 3.0-Å resolution (Advanced Photon Source, Argonne National Laboratory, Argonne, Illinois). These data were processed and scaled with HKL2000 (20). Relevant x-ray data collection statistics are presented in Table 1.
FIGURE 1.
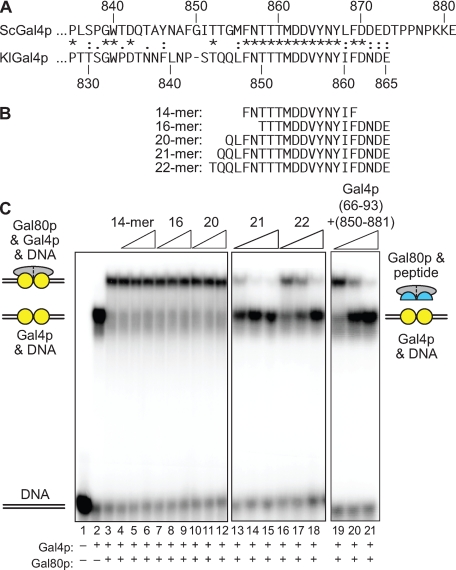
The interaction of Gal80p with peptides representing the TAD of Gal4p. A, amino acid sequence alignment of the activation and Gal80p interaction domains of Gal4p from S. cerevisiae (top) and K. lactis (bottom). B, sequence and location of some of the peptides used in this study. C, ability of each peptide to inhibit the formation of the Gal4p-Gal80p complex was assayed using an electrophoretic mobility shift assay. The locations of the free DNA, the Gal4p-DNA complex, and the Gal80p-Gal4p-DNA complex are indicated. Peptides corresponding to the activation domain of KlGal4p (B) were added as shown, and the loss of the Gal80p-Gal4p-DNA and reappearance of the Gal4p-DNA complex indicated that the peptide was capable of interacting with Gal80p and preventing its association with the larger Gal4p derivative.
TABLE 1.
X-ray data collection statistics
| X-ray data set | |
|---|---|
| Wavelength (Å) | 0.98000 |
| Resolution limits (Å) | 30.0-3.0 (3.11-3.0)a |
| No. independent reflections | 75005 (4730) |
| Completeness (%) | 88.5 (55.8) |
| Redundancy | 4.5 (2.1) |
| Avg I/Avg σ(I) | 7.2 (2.0) |
| Rsymb (%) | 9.8 (24.9) |
Statistics for the highest resolution bin.
Rsym = (∑|I - Ī|/∑I) × 100.
The structure of the Gal4p TAD-Gal80p complex was solved by molecular replacement with the software package Phaser (21) and using the apo-Gal80p model as the search probe. The electron densities for the eight monomers in the asymmetric unit were averaged with the software package DM (22). On the basis of this averaged map, a model for the complex was manually adjusted using Coot (23). At this stage, the peptide was not built into the model, although there was clear electron density for it. The model constructed on the basis of this “averaged” electron density map was placed back into the asymmetric unit in all eight positions and refined with the program TNT (24) using NCS symmetry restraints. After refinement, the electron density map calculated with 2Fo-Fc coefficients was once again averaged, and in this averaged map, the position of the peptide was immediately obvious as was the ordering of the Asp-309/Ser-316 loop. The peptide was then built into the “averaged” electron density. This model, now with bound peptide, was placed back into the eight equivalent positions in the asymmetric unit and manually adjusted with Coot. Subsequent NCS-restrained least-squares refinement of the Gal4p TAD-Gal80p model resulted in an overall R-factor of 23.1% using all x-ray data from 30 to 3.0 Å resolution (R-free of 28.9%). Relevant refinement statistics are presented in Table 2.
TABLE 2.
Least-squares refinement statistics
| Resolution limits (Å) | 30.0-3.0 |
| R-factora (overall) %no. reflections | 23.1/74,757 |
| R-factor (working) %no. reflections | 22.8/67,196 |
| R-factor (free) %no. reflections | 28.9/7561 |
| No. protein atoms | 25015 |
| No. peptide atoms | 952 |
| Weighted R.M.S. deviations from ideality | |
| Bond lengths (Å) | 0.009 |
| Bond angles (deg) | 2.2 |
| Trigonal planes (Å) | 0.005 |
| General planes (Å) | 0.011 |
| Torsional angles (deg)b | 22.0 |
R-factor = (∑|Fo - Fc|/∑|Fo|) × 100, where Fo is the observed structure-factor amplitude, and Fc is the calculated structure-factor amplitude.
The torsional angles were not restrained during the refinement.
Ramachandran statistics for all eight subunits in the asymmetric unit indicate that 81.3% of the residues adopt ϕ, ψ angles in the “most favored,” 18.5% in the “additionally allowed,” and 0.2% in the “generously allowed” regions. There was none in the disallowed region. The discussion of the TAD-binding pocket presented below refers to the model built on the basis of the averaged electron density map.
Mobility Shift Assay—Reaction mixes (10 μl) containing 20 mm HEPES pH 7.5, 150 mm NaCl, 10% w/v glycerol, 10 μm ZnSO4, 5 mm MgSO4, 100 μg/ml bovine serum albumin, 10 pm 32P-labeled stranded oligonucleotide probe (5′-GATCCCGGGAAGCGCTTCCCGAATT-3′; representing a single Gal4p-binding site), and the indicated proteins (Gal4p(1-93 + 768-881), 1.9 μm; Gal4p(66-93 + 850-881), 0.9-8 μm; Saccharomyces cerevisiae Gal80p, 1.3 μm; K. lactis Gal80p, 1.7 μm) were incubated for 30 min at room temperature and then subjected to electrophoresis through a pre-run 5% polyacrylamide gel containing 0.5× TBE, 1% v/v glycerol for 90 min at 150 V. Gels were analyzed by autoradiography. Peptides were synthesized by Eurogentec (Liège, Belgium). Each was dissolved at a final concentration of 14 mg/ml and used in the mobility shift assays at final concentrations of 40-400 μm.
RESULTS AND DISCUSSION
To define a minimal peptide from the sequence of the K. lactis Gal4p that is capable of interacting with Gal80p and that might be useful in a structural analysis, we performed a series of electrophoretic mobility shift assays using labeled DNA encompassing a single Gal4p-binding site (Fig. 1). The addition of Gal80p to the Gal4p-DNA complex resulted in the formation of a supershifted complex. The presence of this slower migrating complex could be inhibited by versions of Gal4p that were capable of interacting with Gal80p but did not possess a competent DNA binding domain, e.g. a fusion protein composed of amino acids 66-93 + 850-881 of the S. cerevisiae Gal4p sequence (Fig. 1C, lanes 19-21). The ability of peptides to out-compete the Gal4p-Gal80p complex, resulting in a complex composed of DNA and Gal4p alone, was measured. Peptides ending at the C terminus of the K. lactis Gal4p and composed of 21 or more amino acids were found to be capable of interacting with Gal80p in this assay. Shorter peptides, e.g. a 16-mer encompassing residues Thr-850 to Glu-865, were found to be incapable of competitively displacing Gal80p from Gal4p (Fig. 1C, lanes 7-9). We therefore attempted to crystallize Gal80p from K. lactis in the presence of either the 21-mer or the 22-mer peptide.
Suitably diffracting crystals were subsequently obtained with the 22-mer peptide. These crystals belonged to the space group P21 and contained eight subunits in the asymmetric unit. The complex structure was solved by molecular replacement and refined to a nominal resolution of 3.0 Å. Electron densities corresponding to the TAD peptides were visible in all eight subunits in the initial maps. To further improve the quality of the protein phases, the electron densities for the eight monomers were averaged with the software package DM (22). Unbiased electron density corresponding to the TAD peptide as observed in this “averaged” electron density map is presented in Fig. 2A. Although a 22-mer was utilized in the crystallization trials, only 14 residues of the Gal4p TAD were visible. Given the shapes of the side chain densities as well as their surrounding chemical environments, the observed peptide corresponds to Leu-847 through Ile-860 of the KlGal4p sequence (Fig. 1A).
FIGURE 2.
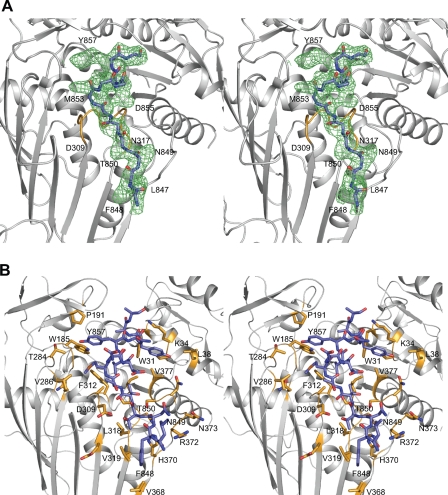
The interaction between the Gal4p TAD and Gal80p. A, stereo view of unbiased electron density corresponding to the bound Gal4p TAD before model building. The “averaged” electron density map was contoured at ∼1σ. A polyglycine chain, displayed with carbons, nitrogens, and oxygens in slate, blue, and red, respectively, was positioned into the map for the sake of clarity in viewing the electron density. The Asp-309/Ser-316 loop that becomes ordered upon Gal4p binding is highlighted in gold. B, close-up stereo view of the Gal80p TAD binding region. Those residues of Gal80p lying within ∼5 Å of the Gal4p TAD (slate) are displayed in gold. This figure and Figs. 3, 4, 5, 6 were prepared with the software package PyMOL (34).
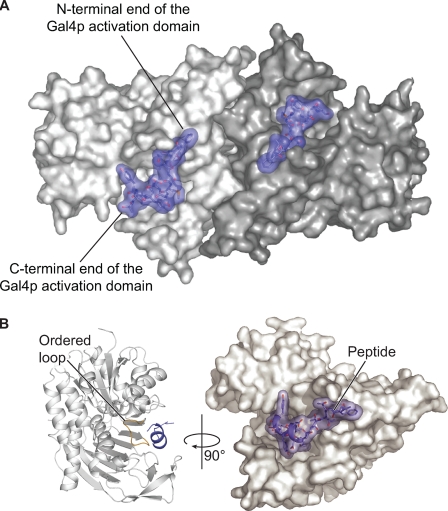
A surface representation of the Gal4p TAD-Gal80p complex is presented in Fig. 3A. Like that observed for the apo-Gal80p structure, the complex crystallizes as a tight dimer with overall dimensions of ∼55 × 75 × 110 Å. The large mixed β-sheets found in the C-terminal domains of the Gal80p subunits provide extensive subunit-subunit interactions. The two Gal4p TAD peptides bind on the same side of the dimer with their N-terminal ends separated by ∼24 Å and their C-terminal ends situated ∼55 Å apart.
FIGURE 3.
The location of the Gal4p TAD binding site in Gal80p. A, Gal80p dimer is highlighted with one subunit in white and the other in gray. The Gal4p peptides are displayed in slate. B, single Gal80p subunit with bound peptide is displayed in a ribbon representation (left) or in a surface representation (right and rotated ∼90°). The Gal80p loop that becomes ordered upon Gal4p TAD binding is highlighted in yellow (left).
As displayed in Fig. 3B, the Gal4p peptide adopts a partial helical conformation that binds into, and across, the pronounced cleft that separates the N- and C-terminal domains of Gal80p. A total of 750 Å2 of surface area for the peptide is buried upon binding to Gal80p, which represents ∼40% of its total. The loop within Gal80p, formed by Asp-309 to Ser-316, that was previously unresolved (16) becomes ordered upon peptide binding, and the backbone density for Asp-245 to Gly-248 becomes better defined. With the exception of these two regions, however, the apo- and peptide-bound forms of the K. lactis Gal80p are exceedingly similar such that their α-carbons superimpose with an R.M.S.D. of ∼1.2 Å.
A close-up stereo view of the Gal80p TAD binding site is presented in Fig. 2B. As can be seen, the side chain of Phe-848 resides in a hydrophobic patch formed by Val-319 and Val-368 and participates in a stacking interaction with the side chain of His-370. The backbone carbonyl group of Asn-849 appears to play a role in the ordering of the Asp-309/Ser-316 loop by interacting with the side chain of Asn-317. Additionally, the side chain of Thr-850 possibly interacts with the side chain of Asp-309, again helping to order the Asp-309/Ser-316 loop. There are hydrophobic interactions between Gal80p and the Gal4p TAD that most likely function in reducing the flexibility of the Asp-309/Ser-316 loop, namely those formed by the side chains of Val-856 and Tyr-857 with Trp-185 and Phe-312. The side chains of Asp-854, Asp-855, and Asn-858 project away from the Gal80p cleft, whereas the side chains of Met-853 and Tyr-857 participate in an intramolecular stacking interaction. Finally, the side chain of Tyr-859, covered on one side by Lys-34 and on the other by Val-377, forms a T-shaped stacking interaction with Trp-31. With the exception of Trp-31, the C-terminal domain contributes all of the Gal80p residues involved in TAD recognition.
Gal80p has a similar molecular structure to that described for glucose-fructose oxidoreductase from Z. mobilis (17). This enzyme contains a tightly bound NADP+, which is required for the reduction of fructose to sorbitol with the concomitant oxidation of glucose to gluconolactone. Given that the K. lactis Gal80p has been shown to associate with dinucleotides,4 we attempted to crystallize it in the presence of NADP+, NADPH, NAD+, or NADH, but have thus far been unsuccessful. Recently, however, the structure of Gal80p from S. cerevisiae was reported in complex with NAD+ and a 9-mer representing the TAD of S. cerevisiae Gal4p (25).
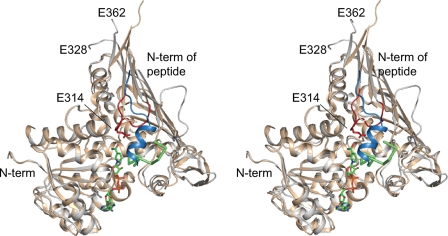
Both the K. lactis and S. cerevisiae versions of Gal80p are dimers, and they share a high degree of structural homology (an R.M.S.D. of ∼2.1 Å for 344 α-carbons per subunit) as would be expected from their amino acid sequence similarities (58% amino acid identity and 82% similarity over their entire length). Importantly, however, these two proteins appear to bind the Gal4p TADs in completely different orientations, which may be a function of NAD binding to the S. cerevisiae Gal80p (Fig. 4). In the S. cerevisiae Gal4p TAD-NAD-Gal80p complex, the peptide reportedly interacts with the nicotinamide portion of the dinucleotide (25), whereas in K. lactis Gal80p, the Gal4p peptide is situated away from the Rossmann fold. Additionally, in the K. lactis Gal80p, the Asp-309/Ser-316 loop moves toward where the cofactor is located in the S. cerevisiae Gal80p model with Glu-314 from the loop projecting near the nicotinamide ring (Fig. 4). Interestingly, the equivalent Asp-309/Ser-316 loop in the S. cerevisiae Gal80p is still disordered even in the presence of the 9-mer. In the structural analysis of the S. cerevisiae Gal4p TAD-NAD-Gal80p complex, the side chain densities for the 9-mer were not well-defined. As such, it is not possible to highlight in this report the detailed differences in protein-protein interactions observed between the S. cerevisiae and K. lactis Gal4p TAD-Gal80p complexes.
FIGURE 4.
Comparison of the Gal80p binding sites for the Gal4p TADs. A superposition of the Gal80p models from S. cerevisiae (wheat) and K. lactis (white) is shown in stereo. The bound nucleotide and peptide in the S. cerevisiae Gal80p are displayed in green, whereas the bound peptide and the Asp-309/Ser-316 loop in the K. lactis Gal80p are shown in slate and red, respectively. Glu-314 in the K. lactis Gal80p is depicted in a stick representation.
The Gal4p TAD is regarded as an archetypal example of an acidic transactivation domain. In addition, nine amino acids of the Gal4p TAD (corresponding to residues 862-870 of the S. cerevisiae Gal4p, DDVYNYLFD) have been identified as a common sequence motif defining the transactivation domain in a variety of proteins from yeast, animal, and viruses (26). This sequence represents the last seven amino acids of the peptide in our structure (Asp-854 to Ile-860 of the K. lactis Gal4p TAD). It can thus be suggested that archetypal TADs adopt helical conformations in which the acidic side chains are predominantly located on one surface and the hydrophobic residues on another.
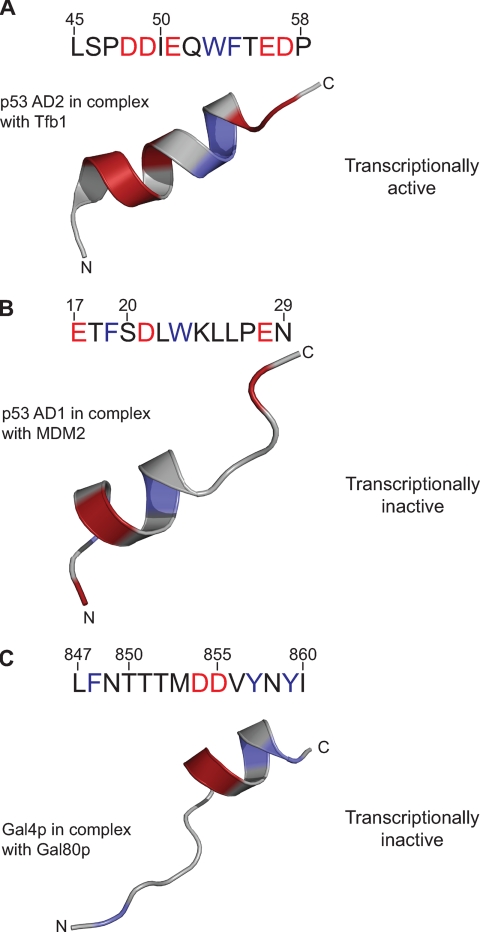
Acidic RNA pol II transactivation domains invariably contain a preponderance of both acidic and bulky hydrophobic amino acids (27). In the absence of binding partners, TADs generally do not possess well-defined secondary structures. However, they have been observed to adopt secondary structure on interaction with other proteins, either proteins that inhibit their function or proteins that they interact with as part of the activation process (28, 29). Previously, activation domain 1 (amino acids 15-29) of p53 was solved in the presence of the inhibitor MDM2 (28) and activation domain 2 of the same protein (amino acids 45-58) solved as a complex with a subunit of the general transcription factor TFIIH (30). A comparison of the structures of these activation domains with the Gal4p TAD is shown in Fig. 5. Additional structural information is known for the E1A TAD in complex with Rb (31) and for the CREB activation domain bound to the KIX domain of CBP (32). The predominant feature of each of these short TADs is an α-helix, which is approximately amphipathic. However, the length of the helix functioning as a TAD is different in each case, as is the extent of the amphipathic character.
FIGURE 5.
Comparison of the structures of the transactivation domains of p53 and Gal4p. A, sequence and NMR structure of the p53 TAD. The structure of the peptide was determined in the presence of the Tfb1 subunit of TFIIH (structural data taken from PDB accession code 2GS0) (30). Here, and in B and C, only the structure of the activation domain peptide is shown as a schematic model. In both the sequence and the structure, the acidic amino acids have been colored red and the large hydrophobic residues in blue. B, sequence and structure of activation domain 1 of p53 bound to the transcriptional inhibitor MDM2 (structural data taken from PDB accession code 1YCR) (28). C, sequence and structure, as determined in this study, of the TAD of Gal4p. As in A, in both the sequence and the structure the acidic amino acids have been colored red and the large hydrophobic residues in blue.
Prior to the recent structural analyses of the Gal4p TAD-Gal80p complexes from both K. lactis and S. cerevisiae, the C-terminal region of the S. cerevisiae Gal4p was extensively studied by mutagenic analysis (33). Each residue within the Gal4p region between Met-855 and Asp-870 (Fig. 1A) was mutated to a cysteine, and the effects of these changes were monitored both in terms of the ability of the protein to activate transcription and on the ability of the protein to interact with Gal80p. Most single point mutations within this region demonstrated little impact on the ability of Gal4p to activate transcription. However, specific point mutations within Gal4p (e.g. mutations at Phe-856, Thr-859, and Met-861) rendered the protein incapable of interacting with Gal80p (33). These amino acids correspond to Phe-848, Thr-851, and Met-853 in the K. lactis Gal4p TAD sequence. As indicated previously, Phe-848 participates in a series of hydrophobic interactions with Gal80p residues Val-319, Val-368, and His-370. Replacement with a cysteine residue would create a substantial hole in this region. The side chain hydroxyl of Thr-851 is located near the backbone nitrogen of Asn-317 within Gal80p. Its replacement with a cysteine would result in the loss of a potential hydrogen bond that might be critically important in reducing the flexibility of the Asp-309/Ser-316 loop. Finally, Met-853 is deeply buried within the Gal80p cleft and surrounded by the side chains of Trp-185 and Phe-312. Again, the replacement of this residue with a cysteine would create a destabilizing hole in the binding cleft between the peptide and Gal80p.
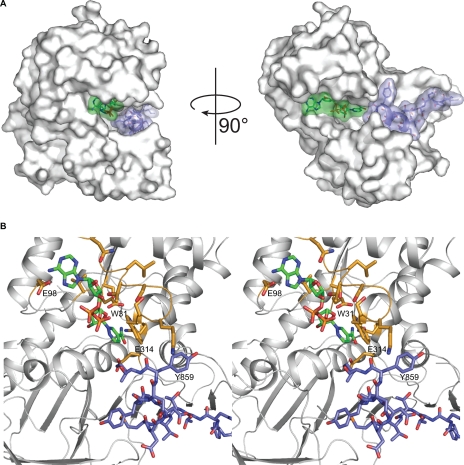
Given that the K. lactis Gal80p reportedly binds dinucleotides, we modeled NAD into our complex structure, based on its location described by Kumar et al. (25) to examine what types of interactions might occur between the ligand and the Gal4p TAD (Fig. 6). Notably, in the model the side chain of Trp-31 from the Rossmann fold domain forms a stacking interaction with both the peptide (Tyr-859) and the nicotinamide ring of the dinucleotide suggesting a possible route of communication between the two binding sites. Furthermore, it is possible that the unobserved residues at the C-terminal end of the peptide used for this study interact with the NAD in vivo or in the absence of NAD, fold into the dinucleotide binding cleft.
FIGURE 6.
The potential interaction between the K. lactis Gal80p, the K. lactis Gal4p activation peptide, and NAD. A, Gal80p monomer showing the location of the Gal4p peptide (slate) and the modeled NAD (green). B, close-up stereo view of the Gal80p region (white ribbon and gold bonds) surrounding the modeled NAD (green) and the observed Gal4p peptide (slate). Glu-98 in the K. lactis Gal80p is a serine residue in the S. cerevisiae Gal80p.
What is the purpose of dinucleotide binding to Gal80p? Kumar et al. (25) have clearly shown the presence of NAD within the structure of the S. cerevisiae Gal80p. They also demonstrate, however, that NAD has no effect on the formation of the Gal4p TAD-Gal80p complex using a pull-down assay. Instead, the presence of NADP has been suggested to be refractory to the formation of this complex. The K. lactis Gal80p has also been shown to associate with dinucleotides, but again it is uncertain as to the physiological role that the dinucleotide plays with respect to either the function of Gal80p or its complex with the Gal4p TAD. It is apparent, however, that Gal80p evolved from an oxidoreductase enzyme (16). Whether Gal80p has simply retained the ability to interact with dinucleotides as part of this evolutionary process or rather dinucleotides play critical roles in the regulation of GAL gene expression is unclear at the present time. What is undeniable in the case of the K. lactis Gal80p, however, is that dinucleotides are not required for Gal4p TAD binding as evidenced by our crystal structure.
Taken together, the results described here demonstrate that an archetypal acidic transactivation domain adopts a predominantly helical conformation that is amphipathic in nature. In addition to defining the conformation of the K. lactis Gal4p TAD, our model of the complex suggests that the binding of a dinucleotide to Gal80p, if indeed it is physiologically relevant, need not necessarily preclude the interaction of Gal80p with Gal4p.
Acknowledgments
We thank C. Chang and F. Rotella for assistance with the x-ray data collection. Results in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by the University of Chicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. The insightful comments of D. Timson, I. Rayment, and A. Steinberg are gratefully acknowledged.
The atomic coordinates and structure factors (code 3E1K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant DK47814 (to H. M. H.). This work was also supported by the Wellcome Trust and BBSRC (to R. J. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TAD, transactivation domain; MES, 4-morpholineethanesulfonic acid; PDB, Protein Data Bank; R.M.S.D., root mean square deviation.
Dr. Karin D. Breunig, personal communication.
References
- 1.Sellick, C. A., and Reece, R. J. (2005) Trends Biochem. Sci. 30 405-412 [DOI] [PubMed] [Google Scholar]
- 2.Traven, A., Jelicic, B., and Sopta, M. (2006) EMBO Rep. 7 496-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston, M. (1987) Microbiol. Rev. 51 458-476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubio-Texeira, M. (2005) FEMS Yeast Res. 5 1115-1128 [DOI] [PubMed] [Google Scholar]
- 5.Lohr, D., Venkov, P., and Zlatanova, J. (1995) Faseb J. 9 777-787 [DOI] [PubMed] [Google Scholar]
- 6.Giniger, E., Varnum, S. M., and Ptashne, M. (1985) Cell 40 767-774 [DOI] [PubMed] [Google Scholar]
- 7.Platt, A., and Reece, R. J. (1998) EMBO J. 17 4086-4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yarger, J. G., Halvorson, H. O., and Hopper, J. E. (1984) Mol. Cell Biochem. 61 173-182 [DOI] [PubMed] [Google Scholar]
- 9.Zenke, F. T., Engles, R., Vollenbroich, V., Meyer, J., Hollenberg, C. P., and Breunig, K. D. (1996) Science 272 1662-1665 [DOI] [PubMed] [Google Scholar]
- 10.Ptashne, M., and Gann, A. (2002) Genes and Signals, Cold Spring Harbor Laboratory Press, New York
- 11.Lue, N. F., Chasman, D. I., Buchman, A. R., and Kornberg, R. D. (1987) Mol. Cell. Biol. 7 3446-3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma, J., and Ptashne, M. (1987) Cell 50 137-142 [DOI] [PubMed] [Google Scholar]
- 13.Marmorstein, R., Carey, M., Ptashne, M., and Harrison, S. C. (1992) Nature 356 408-414 [DOI] [PubMed] [Google Scholar]
- 14.Hidalgo, P., Ansari, A. Z., Schmidt, P., Hare, B., Simkovich, N., Farrell, S., Shin, E. J., Ptashne, M., and Wagner, G. (2001) Genes Dev. 15 1007-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thoden, J. B., Sellick, C. A., Timson, D. J., Reece, R. J., and Holden, H. M. (2005) J. Biol. Chem. 280 36905-36911 [DOI] [PubMed] [Google Scholar]
- 16.Thoden, J. B., Sellick, C. A., Reece, R. J., and Holden, H. M. (2007) J. Biol. Chem. 282 1534-1538 [DOI] [PubMed] [Google Scholar]
- 17.Kingston, R. L., Scopes, R. K., and Baker, E. N. (1996) Structure 4 1413-1428 [DOI] [PubMed] [Google Scholar]
- 18.Melcher, K. (2005) Genetics 171 469-476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilauri, V., Bewley, M., Diep, C. Q., and Hopper, J. E. (2005) Genetics 169 1903-1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 21.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowtan, K., and Main, P. (1998) Acta Crystallogr. Sect. D 54 487-493 [DOI] [PubMed] [Google Scholar]
- 23.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 24.Tronrud, D. E., Ten Eyck, L. F., and Matthews, B. W. (1987) Acta Crystallogr. Sect. A 43 489-501 [Google Scholar]
- 25.Kumar, P. R., Yu, Y., Sternglanz, R., Johnston, S. A., and Joshua-Tor, L. (2008) Science 319 1090-1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piskacek, S., Gregor, M., Nemethova, M., Grabner, M., Kovarik, P., and Piskacek, M. (2007) Genomics 89 756-768 [DOI] [PubMed] [Google Scholar]
- 27.Mapp, A. K., Ansari, A. Z., Ptashne, M., and Dervan, P. B. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3930-3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kussie, P. H., Gorina, S., Marechal, V., Elenbaas, B., Moreau, J., Levine, A. J., and Pavletich, N. P. (1996) Science 274 948-953 [DOI] [PubMed] [Google Scholar]
- 29.Uesugi, M., Nyanguile, O., Lu, H., Levine, A. J., and Verdine, G. L. (1997) Science 277 1310-1313 [DOI] [PubMed] [Google Scholar]
- 30.Di Lello, P., Jenkins, L. M., Jones, T. N., Nguyen, B. D., Hara, T., Yamaguchi, H., Dikeakos, J. D., Appella, E., Legault, P., and Omichinski, J. G. (2006) Mol. Cell 22 731-740 [DOI] [PubMed] [Google Scholar]
- 31.Liu, X., and Marmorstein, R. (2007) Genes Dev. 21 2711-2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radhakrishnan, I., Perez-Alvarado, G. C., Parker, D., Dyson, H. J., Montminy, M. R., and Wright, P. E. (1997) Cell 91 741-752 [DOI] [PubMed] [Google Scholar]
- 33.Ansari, A. Z., Reece, R. J., and Ptashne, M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13543-13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA