Abstract
Amyloid oligomers are believed to play important causal roles in many types of amyloid-related degenerative diseases. Many different laboratories have reported amyloid oligomers that differ in size, morphology, toxicity, and method of preparation or purification, raising the question of the structural relationships among these oligomer preparations. The structural plasticity that has been reported to occur in amyloids formed from the same protein sequence indicates that it is quite possible that different oligomer preparations may represent distinct structural variants. In view of the difficulty in determining the precise structure of amyloids, conformation- and epitope-specific antibodies may provide a facile means of classifying amyloid oligomer structures. Conformation-dependent antibodies that recognize generic epitopes that are specifically associated with distinct aggregation states of many different amyloid-forming sequences indicate that there are at least two fundamentally distinct types of amyloid oligomers: fibrillar and prefibrillar oligomers. Classification of amyloid oligomers according to their underlying structures may be a more useful and rational approach than relying on differences in size and morphology.
A number of age-related degenerative diseases are characterized by the accumulation of misfolded proteins as amyloid deposits. Amyloid deposits are typically composed of 6–10-nm “cross-β”-fibrils, in which the polypeptide chain is arranged in β-sheets where the polypeptide is perpendicular to the fibril axis and hydrogen bonding is parallel (1). In AD,2 several types of amyloid deposits containing the Aβ peptide accumulate, including diffuse amyloid deposits, “cored,” “neuritic,” and “compact or burned out” senile plaques (2), and cerebrovascular amyloid deposits. The linkage of familial AD mutations to the increased production of more highly aggregation-prone Aβ42 supports a causal role of Aβ aggregation in disease (3), but the precise relationships between aggregation state and disease remain to be established. Many other age-related degenerative diseases are also characterized by the accumulation of amyloid deposits derived from a variety of other proteins. The hallmark lesions of Parkinson disease involve the accumulation of α-synuclein, whereas Huntington and other CAG triplet diseases are typified by the accumulation of polyglutamine-containing aggregates. This also includes prion diseases such as Creutzfeldt-Jakob disease with accumulation of misfolded prion protein, type II diabetes with accumulation of islet amyloid polypeptide, and amyotrophic lateral sclerosis with aggregated superoxide dismutase-1. Like AD, many of these diseases have both a sporadic and inherited form, and in many cases, the mutations associated with the familial forms are in the gene encoding the protein that accumulates or in genes directly related to its production, processing, or accumulation. Although these diseases are associated with different proteins of widely varying normal structure and function, they all involve the accumulation of abnormal aggregates containing β-sheet structure.
There is conflicting evidence for the role of macroscopic fibrillar amyloid deposits in pathogenesis. It has been reported that the extent of amyloid plaque accumulation does not correlate well with AD pathogenesis (4) and that a significant number of non-demented individuals have significant amounts of amyloid plaques. In some transgenic animal and cell culture models, pathological changes are frequently observed prior to the onset of amyloid plaque accumulation (5, 6). It has also been reported that soluble Aβ correlates better with dementia than insoluble fibrillar deposits (7, 8), suggesting that oligomeric forms of Aβ may represent the primary toxic species in AD. Indeed, soluble prefibrillar oligomers have been implicated as primary causative agents in many different degenerative diseases in which the accumulation of large fibrillar deposits may be either inert or protective (reviewed in Refs. 9 and 10). For Aβ, aggregates ranging from dimers up to particles of one million Da or greater have been reported in vitro (11–16). Electron microscopy and atomic force microscopy have identified spherical particles of ∼3–10 nm that appear at early times of incubation and disappear as mature fibrils appear (16–18). These spherical oligomers appear to represent intermediates in the pathway of fibril formation because they are transiently observed at intermediate times of incubation during fibril formation. Soluble Aβ oligomers have been referred to as amorphous aggregates, micelles, protofibrils, prefibrillar aggregates, ADDLs, Aβ*56, globulomers, amylospheroids, “tAβ” (toxic soluble Aβ), “paranuclei,” and annular protofibrils (11, 13, 15–17, 19–26). A similar spectrum of soluble oligomers has been observed for many types of amyloids such as α-synuclein (27), islet amyloid (28), and non-disease-associated “neoamyloids” (21). Although these oligomers have been formed under different conditions and display different toxic activities, sizes, and morphologies, it is not yet clear whether they represent the same or distinct structures.
Immunological Classification of Amyloid Oligomers
If the structures of these amyloid oligomers were known, it would be obvious what their relationships are and how many different and unique structures they represent. Only the structures of a few of the amyloid fibrils are known (29–33), and many of these structures are not known at a resolution sufficient to evaluate how many structural variants exist. These structural studies have revealed that fibrils are composed of parallel, in-register, hydrogen-bonded peptide strands, and a recent x-ray crystallographic analysis of a large number fibrils formed by small, 6–7-residue peptides confirms that parallel in-register structure is a common motif and provides additional details of the fibril structures at atomic resolution (34).
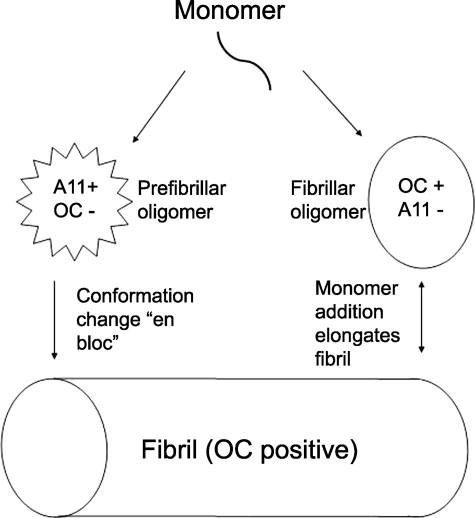
In the absence of high resolution oligomer structures, conformation-dependent antibodies can distinguish different types of amyloid structures. Conformation-dependent antibodies and antisera that specifically recognize amyloid fibrils (35–38) or prefibrillar oligomers (Fig. 1) (39) have been reported. Many of these antibodies are also interesting because they have the unusual property of recognizing generic epitopes that are associated with specific aggregation states regardless of their amino acid sequence. The fact that these epitopes are widely distributed yet distinct and non-overlapping between fibrils and prefibrillar oligomers suggests that there is a fundamental structural difference in the organization of the polypeptide backbone between these two classes of amyloid structures that is shared by many different types of amyloids. Although fibrils have often been operationally defined as insoluble material that sediments at 100,000 × g, the fact that small soluble oligomers also react with fibril-specific antibodies indicates that oligomers with the same type of structural organization as the insoluble fibril also exist (Fig. 1) (37). Although fibrillar oligomers may sound like an oxymoron, the fact that fibril assembly is known to be a nucleation-dependent process indicates that the existence of these small “seed” aggregates, in which the peptide is organized in the same lattice structure of the fibrils, is to be expected.
FIGURE 1.
Schematic representation of the distinct types of amyloid oligomers and fibrils. The aggregation pathway begins with a misfolded amyloidogenic monomer (top) and can diverge into two paths depending on which conformation it adopts. Monomers can aggregate to form prefibrillar oligomers that are A11-positive and OC-negative (left pathway). These prefibrillar oligomers may then align to form protofibrils (not shown) and undergo a concerted conformation change “en bloc” to form fibrils. They are termed prefibrillar oligomers because they are transient intermediates that ultimately become fibrils. In the other pathway, amyloidogenic monomers aggregate to form a fibrillar conformation or lattice that is OC-positive and A11-negative (right pathway). These fibrillar oligomers may represent fibril nuclei or seeds that are aggregates capable of elongating by recruiting additional monomers at their ends. Addition of monomers ultimately elongates the fibrils to a size that satisfies an arbitrary definition of insolubility and would be recognized as fibrillar under the electron or atomic force microscope, although no conformational difference is apparent by antibody reactivity. Fibrils may be distinct from fibrillar oligomers on the basis of their content of multiple protofilaments (not shown), but this does not imply that a fundamental conformation change in their integral peptide-building blocks is necessary for fibrillar oligomers to convert to fibrils. They may simply coalesce or grow by monomer addition to form fibrils.
What Are the Generic Epitopes, and Why Are They Different in Fibrillar and Prefibrillar Oligomers?
It is rather remarkable that the polyclonal immune response to fibrillar and prefibrillar amyloid antigens is largely independent of protein sequence and conformation-specific (37). Why would an antibody raised against aggregates from one sequence recognize the same aggregation state of another protein sequence yet be conformationally specific for fibrillar or prefibrillar states? The structure of amyloid fibrils and oligomers may provide clues for the molecular basis for this peculiar specificity. Most of the fibril structures are relatively simple: parallel, in-register, intermolecularly hydrogen-bonded strands. This motif gives rise to side chain “steric zippers” composed of a single amino acid side chain that runs up and down the sheet (34, 40). These structures were first observed in β-helix proteins where a repeating sequence strand is interspersed with a helical spacer, giving rise to a parallel, in-register β-sheet that has stacks of identical amino acid side chains aligned up and down the sheet (41). Because many amyloid fibril structures have these side chain zipper tracts and because specific amino acid side chains are widely distributed among amyloid-forming sequences (42), most fibrils would be expected to share these steric zippers, and they may constitute the fundamental structures recognized as generic epitopes. This hypothesis suggests that there are many different potential epitopes, at least one per side chain. If the epitope recognizes two adjacent zippers, the number of potential epitopes could be exponentially greater. Whether a given amyloid structure displays a particular steric zipper would depend on the primary sequence and folded structure of the β-sheet.
Although steric zippers can explain the widespread generic nature of the epitopes, they do not explain why the antibodies distinguish between fibrillar and prefibrillar amyloid structures. The fact that the recognition of these generic structures by antibodies is distinct and mutually exclusive suggests that the difference is a fundamental difference in the organization of the peptide backbone. One possibility that has been suggested is that prefibrillar oligomers may be α-extended sheets (43), whereas fibrillar structures are β-sheets. Another possibility is that the difference is due to differences in the twist, tilt, or hydrogen-bonding pattern of the individual β-strands within the sheet. Whether these hypothetical inferences are valid must await structural characterization of the prefibrillar oligomeric state.
Does Size Matter?
For a long time, the aggregation state of amyloids has been judged by the solubility or the size of the oligomer according to sedimentation, size exclusion chromatography, and gel electrophoresis. Although this has been useful, size measurements may lack the structural resolution needed to distinguish alternative conformations and may not reflect the aggregation state under more physiological circumstances. On Western blots, Aβ aggregates ranging from the size of a dimer all the way up to aggregates >500 kDa that stain with the fibril-specific antibody OC are observed (Fig. 1) (37). In contrast, the prefibrillar oligomer-specific antibody A11 stains bands ranging from approximately tetrameric up to ∼75 kDa (Fig. 1). Similar results were obtained for fibrillar oligomers under nondenaturing conditions, indicating that the small size of fibrillar oligomers is not an artifact of SDS-induced dissociation. The size distributions of soluble fibrillar and prefibrillar Aβ oligomers broadly overlap, indicating that size is not a good indicator of their immunologically defined conformation and that distinct oligomeric conformations of the same size exist. If the oligomer structures are intermolecularly hydrogen-bonded extended structures as their β-sheet character would indicate, then the difference between a dimer and a trimer or a nonamer and a decamer would be no more significant that adding a single coin to the stack. Of course, size could matter for the activity of small oligomers in the 10–100-kDa range and those that are insoluble. Small oligomers would be expected to have a higher rate of diffusion, and if the growing ends of the β-sheets are important for pathogenesis, then small size provides a larger number of ends per unit mass of peptide.
Are There Structural Polymorphisms within a Class of Amyloid Aggregates?
In addition to prefibrillar oligomers and fibrils, structural variants or polymorphisms appear to exist within a class. This variation may be analogous to the well known phenomenon of strain variation in prions. Prion protein appears to be capable of forming aggregates that display subtle differences in their patterns of transmissibility and protease resistance (44). Recent evidence indicates that structural variants also exist within Aβ fibrils (45) and prefibrillar Aβ oligomers (46). For Aβ, distinct fibril morphologies have subtle differences in the underlying structures determined by solid-state NMR. These distinct variants can be propagated by seeding the assembly of monomers (45). For prefibrillar Aβ oligomers, a polymorphism at the N-terminal 6E10 epitope that is displayed at low pH but hidden or absent at neutral pH was noted (46). Yeast prion strains also display distinct conformations that arise by differences in the extent and overlap of the amyloid core region (47). In human prions, variation in fibril structure that is detected by differences in antibody binding has been observed to occur in alternating fashion within a single amyloid fibril (48). In view of the potential for structural heterogeneity within a fibril and the difficulty of determining the structure of amyloids, specific antibody binding may provide a more facile means of detecting and characterizing structural polymorphisms. The structural variability already reported in Aβ and prion structures suggests that different amyloid oligomers prepared under different conditions could have subtle differences in structure.
Structural Basis for the Classification of Amyloid Oligomers
Which oligomers are fibrillar or prefibrillar, and are there any other unique structures? Some data are beginning to emerge on which types of oligomers are formed in vivo and in vitro. ADDLs are formed in vitro by dissolving dry Aβ42 in Me2SO and diluting it in F12 cell culture medium to 100 μm, followed by incubation at 4 °C for 24 h (20). ADDLs range in size from trimer and tetramer to approximately dodecamer in vitro and in vivo (49, 50). Although it is not yet clear whether ADDLs represent fibrillar or prefibrillar oligomers or some other unique type of aggregates, results with conformation-dependent anti-ADDL antibodies are consistent with ADDLs representing fibril-type oligomers because the antibodies stain plaques (51, 52). ADDLs bind preferentially to synapses on neurons (50, 51), and their level is elevated in AD brain (50) and cerebrospinal fluid (53).
Prefibrillar oligomers recognized by antibody A11 also occur in human AD brain, although they are only rarely associated with plaque deposits (39). On Western blots, prefibrillar Aβ oligomers range in size from approximately tetramers to ∼20-mers (37). Aβ*56 is a dodecamer that is correlated with cognitive deficits in transgenic animal models, and it is a prefibrillar type of oligomer because it stains with antibody A11 on Western blots (22). Prefibrillar oligomers are also preferentially associated with axon terminals in AD and transgenic mouse brains (54, 55). Prefibrillar oligomers also bind preferentially to synapses on human neurons, suggesting that the conformational differences between fibrillar and prefibrillar oligomers are not important for synapse binding (56).
“Globulomers” are prepared in vitro by dissolving Aβ42 in hexafluoroisopropyl alcohol, drying, resuspending in Me2SO at a concentration of 5 mm, and diluting to 400 μm in phosphate-buffered saline containing 0.2% SDS. After incubation for 6 h at 37 °C, the solution is diluted 3-fold with water and incubated for an additional 18 h at 37 °C (23). Globulomers run at ∼38–48 kDa on SDS gels (23). Globulomers bind in a punctate distribution to neurons in an age-dependent fashion, and they alter synaptic activity (57). Anti-globulomer conformation-dependent antibodies do not react with monomeric Aβ but stain fibrillar plaque deposits, suggesting that they are also specific for fibril-related epitopes. The reactivity of globulomers with fibril-specific antibodies suggests that they are fibril-type oligomers. The conformation of other types of Aβ oligomers in not yet known, but it should be relatively easy to determine their status using the conformation-dependent antibodies that are currently available.
Naturally secreted Aβ oligomers that run predominantly as dimers and trimers on SDS gels have been shown to inhibit long-term potentiation in vitro, but the conformation of these low molecular mass oligomers is not yet clear (58–60). Aβ monomers and fibrils did not inhibit long-term potentiation in the same paradigm, indicating that the inhibition is conformation-dependent. These low molecular mass oligomers run at a size that is not recognized by antibody A11, suggesting that they may be fibrillar oligomers. Whether there are other, more subtle strain variants within these classes of oligomers that uniquely define different strains of oligomers awaits to be determined by the development of monoclonal antibodies that can detect these differences or by determination of their actual structures.
In the absence of more detailed structural information, conformation-dependent antibodies may provide a more rational means of classifying amyloid oligomers based on their underlying structural organization rather than on differences in size or sample preparation. This structure-based classification scheme may be more useful in comparing the toxic activity and pathological significance of oligomers. The availability of novel conformation-dependent antibodies that specifically recognize these assembly states also affords us a unique opportunity to clarify their roles in Aβ fibril assembly and cellular toxicity, and they may ultimately be useful as therapeutic agents.
Supplementary Material
This work was supported, in part or in whole, by National Institutes of Health Grants NS31230 and AG00538 and grants from the Cure Alzheimer Fund and the Larry L. Hillblom Foundation. This is the fifth article of eleven in the Thematic Minireview Series on the Molecular Basis of Alzheimer Disease. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: AD, Alzheimer disease; Aβ, amyloid-β; ADDLs, Aβ-derived diffusible ligands.
References
- 1.Kirschner, D. A., Abraham, C., and Selkoe, D. J. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wisniewski, H. M., and Terry, R. D. (1973) Prog. Neuropathol. 2 1–26 [Google Scholar]
- 3.Hardy, J., and Selkoe, D. J. (2002) Science 297 353–356 [DOI] [PubMed] [Google Scholar]
- 4.Terry, R. D. (1996) J. Neuropathol. Exp. Neurol. 55 1023–1025 [PubMed] [Google Scholar]
- 5.Westerman, M. A., Cooper-Blacketer, D., Mariash, A., Kotilinek, L., Kawarabayashi, T., Younkin, L. H., Carlson, G. A., Younkin, S. G., and Ashe, K. H. (2002) J. Neurosci. 22 1858–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billings, L. M., Oddo, S., Green, K. N., McGaugh, J. L., and Laferla, F. M. (2005) Neuron 45 675–688 [DOI] [PubMed] [Google Scholar]
- 7.McLean, C. A., Cherny, R. A., Fraser, F. W., Fuller, S. J., Smith, M. J., Beyreuther, K., Bush, A. I., and Masters, C. L. (1999) Ann. Neurol. 46 860–866 [DOI] [PubMed] [Google Scholar]
- 8.Lue, L. F., Kuo, Y. M., Roher, A. E., Brachova, L., Shen, Y., Sue, L., Beach, T., Kurth, J. H., Rydel, R. E., and Rogers, J. (1999) Am. J. Pathol. 155 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baglioni, S., Casamenti, F., Bucciantini, M., Luheshi, L. M., Taddei, N., Chiti, F., Dobson, C. M., and Stefani, M. (2006) J. Neurosci. 26 8160–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haass, C., and Selkoe, D. J. (2007) Nat. Rev. Mol. Cell Biol. 8 101–112 [DOI] [PubMed] [Google Scholar]
- 11.Burdick, D., Soreghan, B., Kwon, M., Kosmoski, J., Knauer, M., Henschen, A., Yates, J., Cotman, C., and Glabe, C. (1992) J. Biol. Chem. 267 546–554 [PubMed] [Google Scholar]
- 12.Hilbich, C., Kisters-Woike, B., Reed, J., Masters, C. L., and Beyreuther, K. (1991) J. Mol. Biol. 218 149–163 [DOI] [PubMed] [Google Scholar]
- 13.Soreghan, B., Kosmoski, J., and Glabe, C. (1994) J. Biol. Chem. 269 28551–28554 [PubMed] [Google Scholar]
- 14.Garzon-Rodriguez, W., Sepulveda-Becerra, M., Milton, S., and Glabe, C. G. (1997) J. Biol. Chem. 272 21037–21044 [DOI] [PubMed] [Google Scholar]
- 15.Walsh, D. M., Lomakin, A., Benedek, G. B., Condron, M. M., and Teplow, D. B. (1997) J. Biol. Chem. 272 22364–22372 [DOI] [PubMed] [Google Scholar]
- 16.Harper, J. D., Wong, S. S., Lieber, C. M., and Lansbury, P. T. (1997) Chem. Biol. 4 119–125 [DOI] [PubMed] [Google Scholar]
- 17.Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T., and Lansbury, P. T., Jr. (2002) Nature 418 291. [DOI] [PubMed] [Google Scholar]
- 18.Anguiano, M., Nowak, R. J., and Lansbury, P. T., Jr. (2002) Biochemistry 41 11338–11343 [DOI] [PubMed] [Google Scholar]
- 19.Lomakin, A., Teplow, D. B., Kirschner, D. A., and Benedek, G. B. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7942–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., Wals, P., Zhang, C., Finch, C. E., Krafft, G. A., and Klein, W. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. M., and Stefani, M. (2002) Nature 416 507–511 [DOI] [PubMed] [Google Scholar]
- 22.Lesne, S., Koh, M. T., Kotilinek, L., Kayed, R., Glabe, C. G., Yang, A., Gallagher, M., and Ashe, K. H. (2006) Nature 440 352–357 [DOI] [PubMed] [Google Scholar]
- 23.Barghorn, S., Nimmrich, V., Striebinger, A., Krantz, C., Keller, P., Janson, B., Bahr, M., Schmidt, M., Bitner, R. S., Harlan, J., Barlow, E., Ebert, U., and Hillen, H. (2005) J. Neurochem. 95 834–847 [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto, N., Matsubara, E., Maeda, S., Minagawa, H., Takashima, A., Maruyama, W., Michikawa, M., and Yanagisawa, K. (2007) J. Biol. Chem. 282 2646–2655 [DOI] [PubMed] [Google Scholar]
- 25.Hoshi, M., Sato, M., Matsumoto, S., Noguchi, A., Yasutake, K., Yoshida, N., and Sato, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6370–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bitan, G., Kirkitadze, M. D., Lomakin, A., Vollers, S. S., Benedek, G. B., and Teplow, D. B. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway, K. A., Harper, J. D., and Lansbury, P. T., Jr. (2000) Biochemistry 39 2552–2563 [DOI] [PubMed] [Google Scholar]
- 28.Janson, J., Ashley, R. H., Harrison, D., McIntyre, S., and Butler, P. C. (1999) Diabetes 48 491–498 [DOI] [PubMed] [Google Scholar]
- 29.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F., and Tycko, R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16742–16747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luhrs, T., Ritter, C., Adrian, M., Riek-Loher, D., Bohrmann, B., Dobeli, H., Schubert, D., and Riek, R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 17342–17347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Der-Sarkissian, A., Jao, C. C., Chen, J., and Langen, R. (2003) J. Biol. Chem. 278 37530–37535 [DOI] [PubMed] [Google Scholar]
- 32.Margittai, M., and Langen, R. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jayasinghe, S. A., and Langen, R. (2004) J. Biol. Chem. 279 48420–48425 [DOI] [PubMed] [Google Scholar]
- 34.Sawaya, M. R., Sambashivan, S., Nelson, R., Ivanova, M. I., Sievers, S. A., Apostol, M. I., Thompson, M. J., Balbirnie, M., Wiltzius, J. J., McFarlane, H. T., Madsen, A. O., Riekel, C., and Eisenberg, D. (2007) Nature 447 453–457 [DOI] [PubMed] [Google Scholar]
- 35.Hrncic, R., Wall, J., Wolfenbarger, D. A., Murphy, C. L., Schell, M., Weiss, D. T., and Solomon, A. (2000) Am J. Pathol. 157 1239–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Nuallain, B., and Wetzel, R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kayed, R., Head, E., Sarsoza, F., Saing, T., Cotman, C. W., Necula, M., Margol, L., Wu, J., Breydo, L., Thompson, J. L., Rasool, S., Gurlo, T., Butler, P., and Glabe, C. G. (2007) Mol. Neurodegener. 2 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moretto, N., Bolchi, A., Rivetti, C., Imbimbo, B. P., Villetti, G., Pietrini, V., Polonelli, L., Del Signore, S., Smith, K. M., Ferrante, R. J., and Ottonello, S. (2007) J. Biol. Chem. 282 11436–11445 [DOI] [PubMed] [Google Scholar]
- 39.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W., and Glabe, C. G. (2003) Science 300 486–489 [DOI] [PubMed] [Google Scholar]
- 40.Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. O., Riekel, C., Grothe, R., and Eisenberg, D. (2005) Nature 435 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurnak, F., Yoder, M. D., Pickersgill, R., and Jenkins, J. (1994) Curr. Opin. Struct. Biol. 4 802–806 [DOI] [PubMed] [Google Scholar]
- 42.Chiti, F., Stefani, M., Taddei, N., Ramponi, G., and Dobson, C. M. (2003) Nature 424 805–808 [DOI] [PubMed] [Google Scholar]
- 43.Armen, R. S., DeMarco, M. L., Alonso, D. O., and Daggett, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11622–11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morales, R., Abid, K., and Soto, C. (2007) Biochim. Biophys. Acta 1772 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petkova, A. T., Leapman, R. D., Guo, Z., Yau, W. M., Mattson, M. P., and Tycko, R. (2005) Science 307 262–265 [DOI] [PubMed] [Google Scholar]
- 46.Necula, M., Kayed, R., Milton, S., and Glabe, C. G. (2007) J. Biol. Chem. 282 10311–10324 [DOI] [PubMed] [Google Scholar]
- 47.Toyama, B. H., Kelly, M. J., Gross, J. D., and Weissman, J. S. (2007) Nature 449 233–237 [DOI] [PubMed] [Google Scholar]
- 48.Novitskaya, V., Makarava, N., Bellon, A., Bocharova, O. V., Bronstein, I. B., Williamson, R. A., and Baskakov, I. V. (2006) J. Biol. Chem. 281 15536–15545 [DOI] [PubMed] [Google Scholar]
- 49.Lambert, M. P., Viola, K. L., Chromy, B. A., Chang, L., Morgan, T. E., Yu, J., Venton, D. L., Krafft, G. A., Finch, C. E., and Klein, W. L. (2001) J. Neurochem. 79 595–605 [DOI] [PubMed] [Google Scholar]
- 50.Gong, Y., Chang, L., Viola, K. L., Lacor, P. N., Lambert, M. P., Finch, C. E., Krafft, G. A., and Klein, W. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10417–10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lacor, P. N., Buniel, M. C., Chang, L., Fernandez, S. J., Gong, Y., Viola, K. L., Lambert, M. P., Velasco, P. T., Bigio, E. H., Finch, C. E., Krafft, G. A., and Klein, W. L. (2004) J. Neurosci. 24 10191–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambert, M. P., Velasco, P. T., Chang, L., Viola, K. L., Fernandez, S., Lacor, P. N., Khuon, D., Gong, Y., Bigio, E. H., Shaw, P., De Felice, F. G., Krafft, G. A., and Klein, W. L. (2007) J. Neurochem. 100 23–35 [DOI] [PubMed] [Google Scholar]
- 53.Georganopoulou, D. G., Chang, L., Nam, J. M., Thaxton, C. S., Mufson, E. J., Klein, W. L., and Mirkin, C. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2273–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kokubo, H., Kayed, R., Glabe, C. G., Saido, T. C., Iwata, N., Helms, J. B., and Yamaguchi, H. (2005) Brain Res. 1045 224–228 [DOI] [PubMed] [Google Scholar]
- 55.Kokubo, H., Kayed, R., Glabe, C. G., and Yamaguchi, H. (2005) Brain Res. 1031 222–228 [DOI] [PubMed] [Google Scholar]
- 56.Deshpande, A., Mina, E., Glabe, C., and Busciglio, J. (2006) J. Neurosci. 26 6011–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nimmrich, V., Grimm, C., Draguhn, A., Barghorn, S., Lehmann, A., Schoemaker, H., Hillen, H., Gross, G., Ebert, U., and Bruehl, C. (2008) J. Neurosci. 28 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J., and Selkoe, D. J. (2002) Nature 416 535–539 [DOI] [PubMed] [Google Scholar]
- 59.Cleary, J. P., Walsh, D. M., Hofmeister, J. J., Shankar, G. M., Kuskowski, M. A., Selkoe, D. J., and Ashe, K. H. (2005) Nat. Neurosci. 8 79–84 [DOI] [PubMed] [Google Scholar]
- 60.Klyubin, I., Walsh, D. M., Lemere, C. A., Cullen, W. K., Shankar, G. M., Betts, V., Spooner, E. T., Jiang, L., Anwyl, R., Selkoe, D. J., and Rowan, M. J. (2005) Nat. Med. 11 556–561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.