Abstract
Phospholipase C-ε (PLC-ε) is a highly elaborated PLC required for a diverse set of signaling pathways. Here we use a combination of cellular assays and studies with purified proteins to show that activated RhoA and Ras isoforms directly engage distinct regions of PLC-ε to stimulate its phospholipase activity. Purified PLC-ε was activated in a guanine nucleotide- and concentration-dependent fashion by purified lipidated K-Ras reconstituted in PtdIns(4,5)P2-containing phospholipid vesicles. Whereas mutation of two critical lysine residues within the second Ras-association domain of PLC-ε prevented K-Ras-dependent activation of the purified enzyme, guanine nucleotide-dependent activation by RhoA was retained. Deletion of a loop unique to PLC-ε eliminated its activation by RhoA but not H-Ras. In contrast, removal of the autoinhibitory X/Y-linker region of the catalytic core of PLC-ε markedly activates the enzyme (Hicks, S. N., Jezyk, M. R., Gershburg, S., Seifert, J. P., Harden, T. K., and Sondek, J. (2008) Mol. Cell, 31, 383–394), but PLC-ε lacking this regulatory region retained activation by both Rho and Ras GTPases. Additive activation of PLC-ε by RhoA and K- or H-Ras was observed in intact cell studies, and this additivity was recapitulated in experiments in which activation of purified PLC-ε was quantified with PtdIns(4,5)P2-containing phospholipid vesicles reconstituted with purified, isoprenylated GTPases. A maximally effective concentration of activated RhoA also increased the sensitivity of purified PLC-ε to activation by K-Ras. These results indicate that PLC-ε can be directly and concomitantly activated by both RhoA and individual Ras GTPases resulting in diverse upstream control of signaling cascades downstream of PLC-ε.
Activation of receptors by a variety of hormones, neurotransmitters, growth factors, and other extracellular signaling molecules promotes activation of inositol lipid-specific phospholipase C (PLC)2 enzymes to catalyze hydrolysis of PtdIns(4,5)P2 into diacylglycerol and inositol (1,4,5)-trisphosphate (1). These second messenger molecules are responsible for the stimulation of protein kinase C isozymes and the mobilization of intracellular Ca2+, respectively (2, 3). PLC-catalyzed depletion of cellular PtdIns(4,5)P2 also provides regulation of many additional cellular events. For example, a broad range of proteins contain structural domains, including PH, FERM, ENTH, and PX domains, that specifically bind inositol lipids necessary for their function (4, 5), and PtdIns(4,5)P2-binding proteins are implicated in a broad range of cell signaling events including membrane trafficking, changes in the actin cytoskeleton, and ion channel and transporter function (5).
The six PLC subfamilies (PLC-β, -γ, -δ, -ε, -ζ, and -η) all contain conserved X- and Y-regions, which fold cooperatively to form a catalytic triose-phosphate isomerase (TIM) barrel. Elaboration of the catalytic core with various protein binding and/or regulatory domains gives rise to multiple modes of regulation and different PLC subfamilies. PLC-β isozymes are activated by heterotrimeric Gα subunits of the Gq family (6–8), by Gβγ subunits (9–11), and by the small GTPase Rac (12–15). The region linking the catalytic X- and Y-boxes of PLC-γ isoforms encompass a split PH domain, two SH2 domains, and an SH3 domain, and is subject to activation by tyrosine phosphorylation (16–18). More recently, Rac GTPases have been shown to specifically activate PLC-γ2, but not PLC-γ1 (19). The recently identified PLC-η2 is directly activated by Gβγ subunits (20, 21). The regulation of PLC-δ (22, 23) and PLC-ζ (24, 25) is less well understood, although both are relatively sensitive to changes in cytosolic calcium concentration compared with other PLC isozymes.
The sixth family member of PLC isozymes, PLC-ε, was initially identified in Caenorhabditis elegans as a Ras-binding protein (26). Subsequent cloning of the mammalian homolog of PLC-ε confirmed the presence of conserved Ras-interacting domains, including tandem C-terminal RA domains and an N-terminal CDC25 domain that imparts guanine nucleotide exchange activity to the protein (27–29). Co-expression of constitutively active Ras family GTPases, including Ras and Rap family members, with PLC-ε results in an increased cellular accumulation of inositol phosphates (29–32). Co-expression of PLC-ε with Gα12, Gα13 (28, 33), or Gβγ (30) also promotes increased phospholipase activity in COS-7 cells. Finally, our studies with PLC-ε coexpressed with Rho family GTPases (33), as well as experiments with purified proteins (34), indicate that PLC-ε is a direct effector of RhoA, RhoB, and RhoC.
Although many effectors of Ras, Rho, and Rac GTPases have been identified, knowledge of signaling proteins directly regulated by multiple GTP-bound GTPases through interactions with distinct regulatory domains is limited. Therefore, the goals of this study were to establish incontrovertible evidence for direct and dual regulation of PLC-ε by Rho and Ras family GTPases, to confirm that Rho and Ras interact with distinct regions of PLC-ε, and to delineate the consequences of simultaneous activation of a single PLC isozyme by two GTP-bound activators. Dual, simultaneous, and additive regulation of PLC-ε is illustrated, and our biochemical studies have revealed that GTP-bound RhoA increases the sensitivity of PLC-ε to activation by K-Ras.
EXPERIMENTAL PROCEDURES
Materials—The open reading frame of rat PLC-ε in pCMV script was a kind gift from Grant Kelley, State University of New York, Syracuse, NY. The expression vector for GFP-RhoA was a kind gift from Keith Burridge, University of North Carolina, Chapel Hill, NC. Hemagglutinin A-tagged and 63L and 12V GTPase-deficient mutants of Ras-family GTPases were obtained from the Guthrie Institute (Sayre, PA). His6-tagged TEV protease was purified as previously described (35). K-Ras-4B was the form of K-Ras used in intact cell studies and for protein purification.
Construction of Mammalian PLC-ε Expression Vectors—PLC-ε fragments were constructed as previously described (33). Briefly, full-length rat PLC-ε cDNA was used as a template, and PCR was applied to amplify the selected regions following standard protocols. The resulting amplified products were ligated in-frame into pCMV-myc (Clontech). Constructs were confirmed by sequencing. The specific constructs used and ranges of amino acid residues included in each construct were as follows: CDC25-Ct, 509–2281; CDC25-RA1, 509–2113; EF-RA2, 1198–2215; and EF-Ct, 1198–2281; EF-Ct-ΔY, 1198–2281 lacking residues 1667–1728; EF-Ct-Δlinker 1198–2281 lacking residues 1532–1641.
Transfection of COS-7 Cells and Quantification of Inositol Phosphate Accumulation—COS-7 cells were maintained in high glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in an atmosphere of 90% air/10% CO2. Cells were seeded in 12-well culture dishes at a density of ∼65,000 cells per well 24 h prior to transfection. The indicated DNA vectors were transfected using FuGENE 6 (Roche Applied Science) transfection reagent according to the manufacturer's protocol. The total amount of transfected DNA was 700 ng and included empty vector to maintain an equal amount of DNA per well. The culture medium was changed ∼24 h after transfection to inositol-free Dulbecco's modified Eagle's medium (ICN Biomedicals) containing 1 μCi/well myo-[2-3H(N)]inositol (American Radiolabeled Chemicals). Metabolic labeling was allowed to proceed for 12–18 h. Accumulation of 3H-labeled inositol phosphates were quantified following a 60-min incubation in the presence of the 10 mm LiCl to inhibit inositol phosphate phosphatases. Incubations were terminated by aspiration of the culture medium and subsequent addition of 50 mm formic acid followed by neutralization with 150 mm NH4OH. [3H]Inositol phosphates were isolated and quantified using Dowex chromatography as described previously (36). Variability in cpm of [3H]inositol phosphate accumulation across experiments occurred due to differences in the amount of [3H]inositol used for labeling, duration of the prelabeling period, or different amounts of DNA used for transfection.
Coimmunoprecipitation of PLC-ε with Ras and Rho GTPases from COS-7 Cells—Cells were seeded in 10-cm culture dishes at a density of 1 million cells per dish 24 h prior to transfection. The indicated DNA vectors were transfected using FuGENE 6 transfection reagent according to the manufacturer's protocol. The total amount of transfected DNA was 14 μg and included empty vector to maintain an equal amount of DNA per dish. The culture medium was changed ∼24 h after transfection to serum-free Dulbecco's modified Eagle's medium (Invitrogen). 15 h after serum starvation, cells were washed twice with ice-cold PBS and once with 10 mm Hepes, pH 7.4. Cells were incubated in 1 ml of hypotonic lysis buffer (10 mm Hepes, pH 7.4, 10 mm NaCl, 5 mm MgCl2, 1 mm EDTA, and protease inhibitor mixture) for 15 min on ice with rocking. Cells were collected with a cell scraper, Dounce-homogenized (20 strokes), and passed through a 27-gauge needle (3 times). Crude membrane preparations were isolated by centrifuging whole cell lysates (40,000 × g, 1 h, 4 °C) using a table-top ultracentrifuge (TLA 55 rotor). The membrane pellet was resuspended in 500 μl of lysis buffer (above) + 1% Triton X-100 + protease inhibitor mixture, and Dounce-homogenized (30 strokes). Membrane samples were precleared with protein A/G beads at 4 °C for 1 h. Precleared samples were incubated with 2.5 μg anti-Myc 9E10 antibody (Roche Applied Science) and protein A/G beads (Santa Cruz Biotechnology) at 4 °C for 3 h. Beads were collected by centrifugation at 13,800 × g for 1 min and washed three times with hypotonic lysis buffer + protease inhibitor mixture. Immunoprecipitated samples were eluted by incubating with 2× Lammeli buffer at room temperature for 30 min, and all samples were boiled prior to SDS-PAGE and immunoblot analysis.
Purification of PLC Constructs—PLC-ε-EF-RA2 (amino acids 1258–2215) was amplified from full-length rat PLC-ε and subcloned into NcoI/XhoI-digested pFastBacHT, which incorporates an N-terminal hexahistidine tag and TEV protease site for cleavage of the affinity tag. Site-directed mutagenesis was used to generate PLC-ε-EF-RA2(K>E), a construct with mutations of lysine residues 2150 and 2152 to glutamic acid. Baculoviruses encoding each PLC-ε construct were generated using the Bac-to-Bac method per the manufacturer's instructions (Invitrogen).
PLC-ε-EF-RA2 and PLC-ε-EF-RA2(K>E) were purified as follows. HighFive cells were grown in 4 liter shaker flasks at 27 °C to a density of ∼2.0 × 106 cells/ml and infected with virus encoding PLC-ε-EF-RA2 or PLC-ε-EF-RA2(K>E) at a multiplicity of infection of 1.0. Cells were harvested 48 h after infection by low speed centrifugation and resuspended to a final volume of 150 ml in buffer A (20 mm HEPES, pH 8.0, 300 mm NaCl, 1 mm CaCl2, 10% glycerol, and EDTA-free Complete protease inhibitor tablets (Roche Applied Science)). The resuspended cells were lysed by passage through an EmulsiFlex C5 homogenizer (Avestin). Intact cells and nuclei were removed from the lysate by low speed centrifugation at 500 × g for 15 min. The supernatant from the low-speed centrifugation was clarified by centrifugation at 100,000 × g for 1 h. The resulting supernatant was diluted to a final volume of 150 ml and loaded onto a 5-ml HisTrap metal chelate column (GE Healthcare) charged with Ni2+. The loaded column was washed with 10 column volumes of buffer A, followed by a 10 column volumes, 3% buffer B wash (buffer A + 1 m imidazole). Recombinant protein was eluted using a 3–50% buffer B linear gradient over 20 column volumes collecting 5-ml fractions. Fractions containing recombinant protein were pooled, and hexahistidine-tagged TEV protease was added to the sample to cleave the hexahistidine tag from the PLC-ε fragment. The sample was dialyzed overnight against buffer A to remove the imidazole. The cleaved PLC-ε fragment was subjected to a second passage over a Ni2+-charged 1-ml HisTrap column, and the protein was eluted with buffer containing 5 mm imidazole, which separates the cleaved PLC-ε fragment from the hexahistidine tag and TEV protease. Fractions containing the recombinant protein were pooled and concentrated using a 50,000 MWCO PES centrifugal filtering device (Sartorius). Typical yields for PLC-ε-EF-RA2 and PLC-ε-EF-RA2(K>E) were ∼1 mg per liter of infected cells.
Purification of Monomeric GTPases—Coding sequences for full-length human monomeric GTPases were amplified by PCR and ligated into BamHI/XhoI-digested pFastBacHT, which incorporates an N-terminal hexahistidine tag. Baculoviruses of each GTPase were produced using the Bac-to-Bac system per the manufacturer's protocol. HighFive cells at a density of ∼2.0 × 106 cells/ml were infected with baculovirus at a multiplicity of infection of ∼1.0. Cells were harvested 48 h after infection by low speed centrifugation and resuspended in a final volume of 100 ml of buffer A (20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm MgCl2, 5% glycerol, 1 mm dithiothreitol, one EDTA-free Complete protease inhibitor tablet). Cells were lysed using an Emulsiflex C5 homogenizer. Cell lysates were cleared of intact cells and nuclei by low speed centrifugation at 500 × g for 15 min. Membranes of virus-infected cells were harvested from the resulting supernatant by high speed centrifugation at 100,000 × g for 1 h. Post-translationally modified, membrane-bound GTPases were extracted from the harvested membranes after resuspension in buffer A containing 1% sodium cholate and one EDTA-free Complete protease inhibitor at a final protein concentration of 5 mg/ml. Resuspended membranes were subjected to rapid stirring for 1 h at 4 °C. Solubilized membrane proteins were separated from the particulate fraction by centrifugation at 100,000 × g for 1 h.
Ni-NTA agarose (Qiagen) was added to the resulting supernatant and incubated for 1 h at 4 °C. The resulting slurry mixture was added to a gravity flow column and washed with 10 column volumes (CV) of buffer A containing 500 mm NaCl. The column was then equilibrated with buffer A containing 10 mm NaCl, and recombinant protein was eluted in 10 CV of buffer B (20 mm HEPES, pH 8.0, 10 mm NaCl, 1 mm MgCl2, 400 mm imidazole, 5% glycerol, 0.5% sodium cholate). Sample containing the eluted protein was diluted to 50 ml in buffer B and applied to a 1 ml HiTrap Q column (GE Healthcare). The loaded column was washed with 10 CV of buffer B, and protein was eluted using a linear NaCl gradient from 10 to 500 mm over 25 CV. Fractions containing the recombinant protein were pooled and applied directly to a 1-ml HisTrap column (GE Healthcare) charged with Ni2+. The loaded column was washed with 10 CV of buffer C (20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm MgCl2, 0.5% sodium cholate, 5% glycerol) and 10 CV of 5% buffer D (buffer C + 1 m imidazole). Protein specifically bound to the column was eluted with a 0 to 50% buffer D gradient over 20 CV collecting 1 ml fractions. Fractions containing recombinant GTPase were pooled and concentrated using a 10,000 MWCO PES centrifugal filtering device (Sartorius). All steps were completed within 1 day with no freezing of the sample between column purifications. The concentrations of the individual GTPases were determined by quantifying the binding of [35S]GTPγS under identical buffer and assay conditions described below for reconstitution of the GTPases with phospholipid vesicles. The ability of purified, lipidated GTPases to associate with phospholipid vesicles was tested as previously described (37).
Phospholipase C Reconstitution Assay—Experiments to determine regulation of PLC activity by G proteins were performed as previously described with minor modifications (34, 38). Briefly, phospholipid vesicles were created by combining 600 μm PE (Avanti Polar Lipids), 50 μm PtdIns(4,5)P2 (Perkin Elmer), and ∼10,000 cpm of [3H]PtdIns(4,5)P2 (Perkin Elmer) per assay into a glass borosilicate tube and drying the mixture under a stream of nitrogen. Vesicles were created by resuspension of the dried lipids in 20 mm HEPES, pH 7.4, followed by probe sonication. Assays were performed in a final volume of 60 μl in a reaction mixture containing 20 mm HEPES, pH 7.4, 70 mm KCl, 3 mm EGTA, 2 mm dithiothreitol, 0.16 mg/ml fatty acid-free bovine serum albumin, 10 μm free calcium, and 0.03% sodium cholate. Purified GTPases were diluted 6-fold into the assay. The GTPases were loaded with nucleotide in the absence of PLC by incubating the G protein/lipid suspension for 30 min at 30 °C with 10 μm GDP or 10 μm GTPγS. Reactions were initiated by the addition of purified PLC and incubations were at 30 °C for 5–10 min as indicated in the figure legends. Reactions were terminated by the addition of 200 μl of 10% trichloroacetic acid and 100 μl of 10 mg/ml fatty acid-free bovine serum albumin. Soluble [3H]inositol phosphates were quantified by liquid scintillation counting of the soluble fraction after centrifugation of the reaction mixture.
Data Analysis—All experiments were carried out at least three times with duplicate or triplicate samples in each experiment. Results are presented as representative or pooled data sets. Dose-response curves were generated using GRAPHPAD PRISM software (Graphpad Software Inc., San Diego, CA). Statistical significance was assessed with a Student's t test.
RESULTS
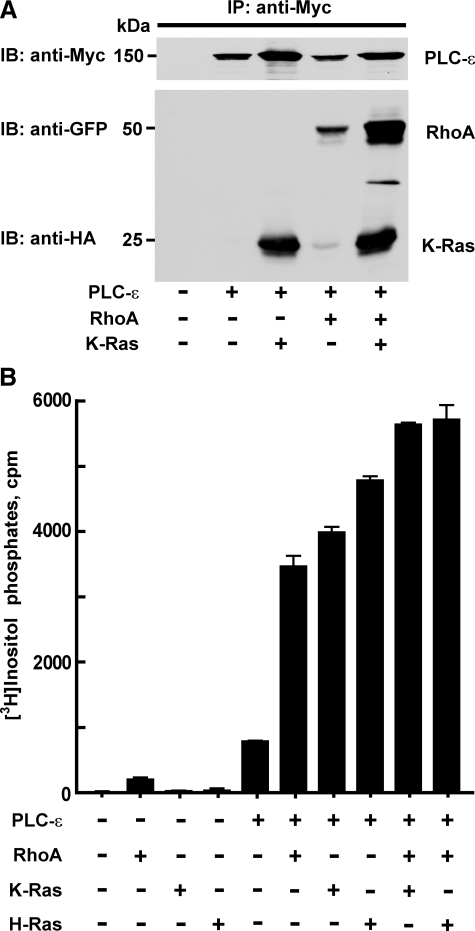
Activation of PLC-ε by Rho and Ras—To establish evidence for dual and simultaneous regulation of PLC-ε by Rho and Ras GTPases, the capacity of GTPases to interact with PLC-ε in intact cells was tested in pull-down assays using antibody directed against a c-Myc tag on the N terminus of PLC-ε. Expression of PLC-ε with either GTPase-deficient RhoA or K-Ras resulted in PLC-ε-dependent pull-down of the GTPase from membrane extracts of these cells (Fig. 1A). Moreover, both RhoA and K-Ras were pulled down from membrane extracts of cells expressing both GTPases with PLC-ε. The amount of RhoA interacting with PLC-ε in cells expressing RhoA and K-Ras simultaneously was greater than that observed in samples from cells expressing only RhoA with PLC-ε, and the amount of K-Ras pulled down in cells expressing RhoA and K-Ras simultaneously was at least as great as that observed in samples from cells expressing only K-Ras with PLC-ε (Fig. 1A).
FIGURE 1.
G protein activation of PLC-ε in cells overexpressing GTPases. A, COS-7 cells were transfected with an expression vector for Myc-PLC-ε-EF-Ct (6 μg) in the absence or presence of constitutively active HA-K-Ras (4 μg), GFP-RhoA (4 μg), or with both HA-K-Ras (4 μg) and GFP-RhoA (4 μg). Samples were immunoprecipitated using anti-Myc 9E10 antibody as described in “Experimental Procedures,” and the samples were immunoblotted with anti-Myc 9E10 (Roche Applied Science) for PLC-ε, anti-GFP-HRP (Santa Cruz Biotechnology) for RhoA, and anti-HA-HRP 3F10 (Roche Applied Science) for K-Ras. Data shown are representative of three individual experiments. B, full-length Myc-PLC-ε (50 ng) was expressed alone in COS-7 cells, or expressed with different combinations of RhoA (30 ng), H-Ras (30 ng), or K-Ras (30 ng), as indicated. [3H]Inositol phosphate accumulation was quantified as described under “Experimental Procedures.” [3H]Inositol phosphate accumulation observed with empty vector alone was subtracted from each value. The data shown are the mean ± S.D. for triplicate samples and are representative of results from three or more experiments in each case.
The relative capacity of both Ras and Rho GTPases to activate PLC-ε also was examined in COS-7 cells transiently transfected with DNA encoding GTPase-deficient G-proteins and PLC-ε. Whereas expression of H-Ras or K-Ras (Fig. 1B) or N-Ras (data not shown) alone had no effect on [3H]inositol phosphate accumulation, marked accumulation occurred when these GTPases were coexpressed with PLC-ε, and data for the effects of maximally effective amounts of Ras GTPases are illustrated in Fig. 1B. We previously demonstrated that RhoA directly activates PLC-ε (33, 34), and the amount of PLC-ε-dependent accumulation of [3H]inositol phosphates was similar with maximally effective amounts of GTPase-deficient RhoA and K-Ras or H-Ras (Fig. 1B). Maximally activating amounts of RhoA and K-Ras or H-Ras also were simultaneously expressed with PLC-ε in COS-7 cells. Co-expression of RhoA with either K-Ras or H-Ras resulted in PLC-ε-dependent inositol phosphate accumulation significantly greater than that observed after expression of either GTPase individually. The effect of coexpression of PLC-ε with both RhoA and K-Ras (or H-Ras) ranged from a partially additive effect as shown in Fig. 1B to a fully additive effect (data not shown). In contrast, co-expression of maximally effective concentrations of H-Ras with K-Ras resulted in no additivity of PLC-ε-dependent response (data not shown). Western blots confirmed that the levels of GTPase or PLC-ε expression from the indicated amounts of DNA were not changed by co-expression with various other expression vectors (data not shown).
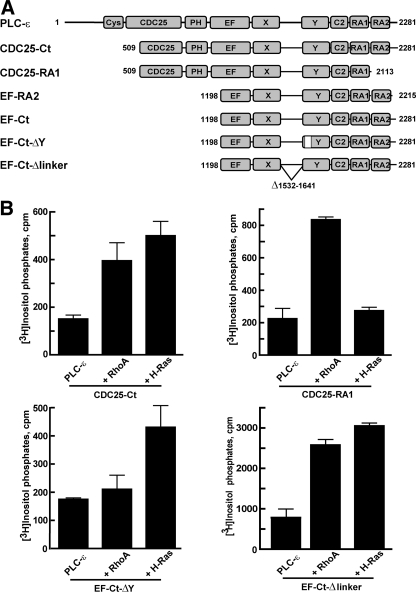
Our previous studies suggested that Rho and Ras GTPases regulate the activity of PLC-ε through specific and separate regions of the enzyme (33). These observations were extended by a series of experiments in COS-7 cells transiently transfected with DNA encoding GTPase-deficient RhoA or H-Ras with various constructs of PLC-ε (Fig. 2A). Activity observed with PLC-ε lacking the N-terminal region (CDC25-Ct) was markedly increased by co-expression with either RhoA or H-Ras (Fig. 2B). As previously illustrated (32), removal of the second RA domain of PLC-ε resulted in an isozyme (CDC25-RA1) that was not activated by H-Ras; however, this construct retained capacity for RhoA-dependent activation (Fig. 2B). PLC-ε lacking an insertion within the Y-box (EF-Ct-ΔY) that is unique to PLC-ε isozymes (30) was not activated by co-expression with RhoA; however, removal of the Y-box insert did not affect the ability of PLC-ε to be regulated by H-Ras (Fig. 2B).
FIGURE 2.
Activation of PLC-ε by RhoA and H-Ras requires unique regions of interaction. A, specific PLC-ε constructs used and amino acid ranges were as follows: CDC25-Ct, 509–2281; CDC25-RA1, 509–2113; EF-RA2, 1198–2215; EF-Ct, 1198–2281; EF-Ct-ΔY, 1198–2281 lacking residues 1667–1728 (white box); and EF-Ct-Δlinker, 1198–2281 lacking residues 1532–1641. Note that the two mutants, CDC25-Ct and CDC25-RA1, only contain part of the sequences of the CDC25 domain; and similarly, the four mutants, EF-RA2, EF-Ct, EF-Ct-ΔY, EF-Ct-Δlinker, only contain part of the sequences of the EF-hand domains. B, expression vectors (30 ng) for constitutively active mutants of RhoA(63L) or H-Ras(61L) were expressed in COS-7 cells in the absence or presence of the indicated PLC-ε constructs (50 ng each of vectors for CDC25-Ct, CDC25-RA1, and EF-Ct-ΔY; and 0.1 ng of vector for EF-Ct-Δlinker). [3H]Inositol phosphate accumulation was quantified as described under “Experimental Procedures.” [3H]Inositol phosphate accumulation observed with empty vector alone was subtracted from the corresponding samples expressing PLC-ε. The data shown are the mean ± S.D. for triplicate samples and are representative of results from three or more experiments in each case.
Our recent structural and biochemical studies exploring the mechanism whereby Rac1 activates PLC-β2 have implicated the poorly conserved X/Y-linker region of the catalytic core of PLC isozymes in regulation of their activity (15, 39). These studies indicated that the X/Y-linker region of PLC-β2 and potentially other PLC isozymes subserves an autoinhibitory function, and removal of this region results in activation of PLC-β2 as well as other isozymes. For example, removal of this region of PLC-ε results in a 10–100-fold increase in basal enzyme activity as compared with wild-type PLC-ε when expressed in COS-7 cells (39). To determine whether PLC-ε lacking the X/Y-linker region retains activation by Rho and Ras GTPases, a fragment of PLC-ε lacking this region (PLC-ε-EF-Ct-Δlinker) was co-expressed with RhoA and H-Ras in COS-7 cells. As illustrated in Fig. 2B, PLC-ε-EF-Ct-Δlinker retained capacity for activation by RhoA and H-Ras when co-expressed with either of these GTPases.
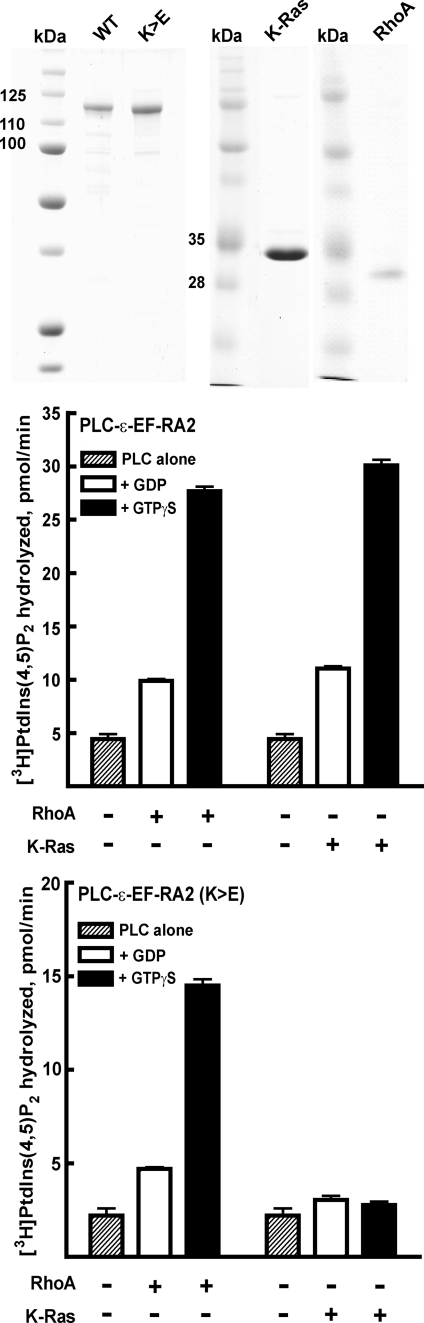
Selective G Protein-mediated Regulation of Purified PLC-ε—Experiments also were carried out to begin to address unequivocally the idea that PLC-ε is a direct effector of both Rho and Ras GTPases independently regulating the isozyme through separate binding surfaces. Ras superfamily G proteins were overexpressed in HiFive insect cells, detergent-extracted from isolated membranes, and purified to homogeneity as described under “Experimental Procedures.” K-Ras was used as the model Ras GTPase in these studies because, as was the case with RhoA, it was readily purified (see Fig. 4, top panel) in ample amounts in a form that bound [35S]GTPγS stoichiometrically and associated with reconstituted substrate-containing phospholipid vesicles as described in “Experimental Procedures.” In contrast, attempts to generate pure, active H-Ras in a form that reproducibly and quantitatively associated with phospholipid vesicles were unsuccessful.
FIGURE 4.
Distinct regions of PLC-ε are required for its activation by RhoA and K-Ras. Purified proteins (top panel) corresponding to PLC-ε-EF-RA2, PLC-ε-EF-RA2(K>E), RhoA, and K-Ras were resolved by SDS-PAGE, and gels were stained with Coomassie Blue. Wild-type PLC-ε-EF-RA2 (middle panel) or PLC-ε-EF-RA2(K>E) (bottom panel) were incubated with phospholipid vesicles containing [3H]PtdIns(4,5)P2 substrate. Assays were performed with vesicles in the absence of G-proteins (dashed bars) or in the presence of GDP-bound (open bars) or GTPγS-bound (black bars) RhoA (300 nm) or K-Ras (300 nm). G-proteins were preincubated in the presence of 10 μm GDP or 10 μm GTPγS for 30 min at 30 °C to allow nucleotide exchange. Assays were initiated by the addition of PLC enzyme (2 ng) and incubations were for 8 min at 30 °C. Data are presented as the mean ± S.E. of an individual experiment, and the results are representative of those observed in three individual experiments.
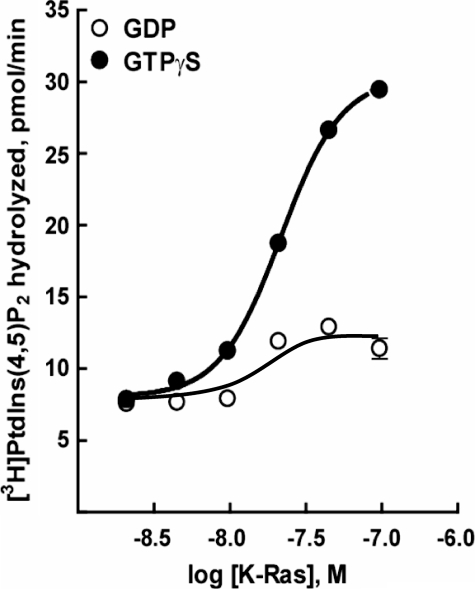
Various concentrations of purified K-Ras were reconstituted in [3H]PtdIns(4,5)P2 substrate-containing phospholipid vesicles, and the capacity of this GTPase to activate purified PLC-ε was quantified. Whereas little effect of K-Ras was observed in the presence of GDP, marked concentration-dependent stimulation occurred with K-Ras activated with GTPγS (Fig. 3). The extent of GTPγS-dependent activation (2–6 times greater than basal) of purified PLC-ε-EF-RA2 by a maximally effective concentration of purified lipidated K-Ras varied with different protein preparations and experiments but was similar to that observed with a maximally effective concentration of purified lipidated RhoA (Fig. 4, middle panel) tested under the same assay conditions. Increases in enzyme activity also were observed in some experiments with GDP-bound K-Ras or RhoA. However, this activity was always much less than that observed in the presence of GTPγS, and also occurred with purified lipidated Rac1, although no GTPγS-dependent activation of PLC-ε is observed with this GTPase (34). In contrast to results obtained with lipidated RhoA or K-Ras, soluble forms of these GTPases purified from bacteria did not activate PLC-ε-EF-RA2 (data not shown).
FIGURE 3.
Direct activation of PLC-ε by K-Ras. K-Ras was purified from detergent-extracted HighFive insect cell membranes as described under “Experimental Procedures,” was reconstituted at various concentrations in phospholipid vesicles containing [3H]PtdIns(4,5)P2 and was incubated with 10 μm GDP (open circles) or 10 μm GTPγS (closed circles) for 30 min at 30 °C. The assay was initiated by the addition of purified PLC-ε-EF-RA2 (2 ng) and incubations were continued for 8 min at 30 °C. The results are the mean ± S.E. of a representative experiment performed six times.
RhoA and K-Ras Interact with Distinct Regions of PLC-ε for Its Regulation—The interaction and regulation in intact cells of PLC-ε with Ras family GTPases (27–30, Fig. 2), but not that observed with Rho (33, 34), is dependent on the second RA domain near the C terminus of PLC-ε. To illustrate that PLC-ε is directly regulated through distinct interactions of the enzyme with K-Ras and RhoA, a double mutation in the second RA2 domain of PLC-ε-EF-RA2(K>E), previously shown to inhibit activation of PLC-ε by Ras GTPases in intact cells (29, 30, 33), was introduced into the wild-type PLC-ε-EF-RA2, and this construct was purified to homogeneity after expression from a baculovirus in insect cells (Fig. 4, top panel). This double mutation in the RA domain of PLC-ε results in a charge reversal of lysine residues 2150 and 2152, creating a negatively charged surface potential expected to repel the negatively charged potential of Ras family switch regions. Additionally, Lys-2150 recently was shown to form contacts with switch I of H-Ras (40).
Basal phospholipase C activity observed with purified PLC-ε-EF-RA2(K>E) was similar to that observed with wild-type PLC-ε-EF-RA2 within the range of error over three individual experiments (Fig. 4, middle and bottom panel). In contrast to wild-type PLC-ε-EF-RA2, PLC-ε-EF-RA2(K>E) retained stimulation by GTPγS-bound RhoA, but enzyme activity was not increased by a concentration of GTPγS-bound K-Ras that maximally stimulated wild-type PLC-ε-EF-RA2 (Fig. 4, bottom panel). Moreover, a wide range of concentrations of GTPγS-bound K-Ras failed to activate PLC-ε-EF-RA2(K>E) (data not shown).
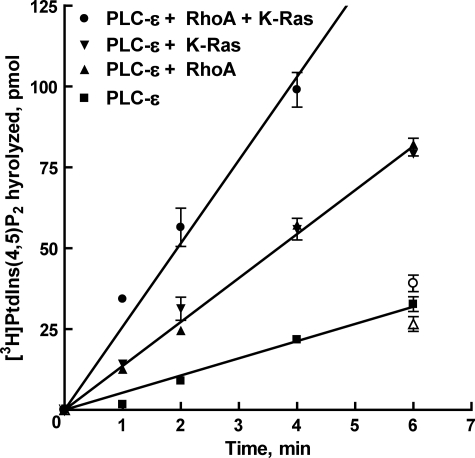
Dual Regulation of PLC-ε by RhoA and K-Ras—Results from experiments described above confirm that PLC-ε is directly regulated by both RhoA and K-Ras in a GTPγS-dependent manner through interactions with distinct regions of the isozyme. To test whether a combination of the two GTPases increases the activity of PLC-ε above that observed with a single GTPase, RhoA, and K-Ras were co-reconstituted in the same phospholipid vesicle preparation, and the phospholipase activity of purified PLC-ε-EF-RA2 was assessed. Maximally activating concentrations (300 nm) of either RhoA or K-Ras preloaded with GTPγS resulted in ∼2.5-fold increases in enzyme activity over that observed with PLC-ε alone (Fig. 5, Table 1). When maximally activating concentrations of RhoA (300 nm) and K-Ras (300 nm) were combined in the same vesicles, the enzymatic rate of PLC-ε-EF-RA2 was approximately additive relative to the stimulation observed with RhoA or K-Ras individually (Fig. 5). Inositol lipid hydrolysis was linear for at least 6 min under the assay conditions utilized.
FIGURE 5.
Time course of PtdIns(4,5)P2 hydrolysis by purified PLC-ε in the combined presence of RhoA and K-Ras. The time-dependent stimulation of purified, wild-type PLC-ε-EF-RA2 (1 ng) was tested in phospholipid vesicles reconstituted with 300 nm RhoA (closed upward triangles) or K-Ras (closed downward triangles), or vesicles containing 300 nm of both RhoA and K-Ras (closed circles). GTPases reconstituted in phospholipid vesicles were preincubated in the presence of 10 μm GTPγS for 30 min at 30 °C to allow nucleotide exchange. The activity of PLC-ε-EF-RA2 in the absence of G proteins also is shown (closed squares). The ability of GDP-bound RhoA (open upward triangles), K-Ras (open downward triangles), or RhoA and K-Ras (open circles) to regulate PLC-ε-EF-RA2 activity are indicated at the 6 min time point. These results are representative of results from five individual experiments and are presented as the mean ± S.E.
TABLE 1.
RhoA and K-Ras regulation of PLC-ε-EF-RA2 activity
| Rate | EC50 | |
|---|---|---|
| pmol/min/ng | nm | |
| PLC-ε-EF-RA2 | 5.3 ± 0.2 | |
| +RhoAa | 13.7 ± 0.2 | 21.3 ± 3.4b |
| +RhoA + K-Rasc | 21.1 ± 1.0 | 19.3 ± 0.5b |
| +K-Rasd | 13.6 ± 0.3 | 14.9 ± 2.6e |
| +K-Ras + RhoAf | 21.1 ± 1.0 | 5.6 ± 0.7e |
A concentration effect curve was generated for RhoA alone. The rate indicated is that observed for a maximally effective concentration K-Ras (300 nm) plus 300 nm RhoA.
An EC50 value was determined for RhoA in the absence or presence of 300 nm K-Ras. No statistically significant difference (p > 0.05) was observed.
A concentration effect curve was generated for RhoA in the presence of 300 nm K-Ras. The rate indicated is that observed for a maximally effective concentration of RhoA (300 nm) plus 300 nm K-Ras.
A concentration effect curve was generated for K-Ras alone. The rate indicated is that observed for a maximally effective concentration of K-Ras (300 nm).
An EC50 value was determined for K-Ras in the absence or presence of 300 nm RhoA. An approximately three-fold increase in potency of K-Ras was observed in the presence of RhoA (p < 0.01).
A concentration effect curve was generated for K-Ras in the presence of 300 nm RhoA. The rate indicated is that observed for a maximally effective concentration of K-Ras (300 nm) plus 300 nm RhoA.
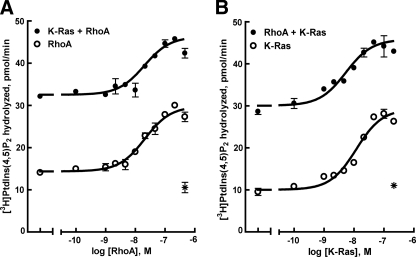
To further assess the regulation of PLC-ε by both RhoA and K-Ras, each GTPase was reconstituted in combination over a wide range of concentrations in phospholipid vesicles (Fig. 6). Enzyme activity of purified PLC-ε was increased in a concentration-dependent manner by GTPγS-bound RhoA or K-Ras. Under these assay conditions, RhoA (∼20 nm) and K-Ras (∼15 nm) exhibited similar EC50 values for activation of PLC-ε-EF-RA2. The same concentration range of RhoA was examined in the presence of a maximally activating concentration of K-Ras. The resulting activity was essentially additive and little or no change was observed in the EC50 of RhoA (Fig. 6A). In contrast to the lack of effect of K-Ras on the RhoA activation curve, the presence of 300 nm RhoA resulted in a 3-fold decrease (p < 0.01) in the EC50 observed for K-Ras (Fig. 6B and Table 1). Thus, dual, independent, and additive activation of PLC-ε occurs in response to purified RhoA and K-Ras. Activation of PLC-ε by RhoA also results in an increase in sensitivity to activation by K-Ras.
FIGURE 6.
Dual and independent regulation of PLC-ε by purified RhoA and K-Ras. A, increasing concentrations of RhoA were reconstituted in phospholipid vesicles containing [3H]PtdIns(4,5)P2 in the absence (open circles) or presence of 300 nm K-Ras (closed circles). After a 30-min preincubation of RhoA or RhoA and K-Ras containing vesicles in the presence of 10 μm GTPγS, PLC-ε EF-RA2 (2 ng) was added, and the incubation was continued for 8 min at 30 °C. B, increasing concentrations of K-Ras were reconstituted in phospholipid vesicles in the absence (open circles) or presence of 300 nm RhoA (closed circles). Assay was performed for 8 min at 30 °C. Purified PLC-ε EF-RA2 also was added to phospholipid vesicles containing maximally activating concentrations of RhoA and K-Ras preloaded with GDP (asterisk). The results are representative of those from three individual experiments and are presented as the mean ± S.E.
DISCUSSION
More than 150 Ras superfamily GTPases promote multifarious cell signaling pathways, but few examples exist of regulation of single effector proteins by GTPases from multiple GTPase subfamilies. The current work confirms that PLC-ε is a unique G protein-regulated effector that is stimulated by both Ras and Rho GTPases acting by direct interaction with distinct regions of the isozyme.
This work together with previous studies places PLC-ε at a unique point of convergence for the remarkably broad range of signaling pathways that promote Rho and Ras GTPase-mediated signaling. For example, receptor-mediated activation of heterotrimeric G proteins results in activation of Rho GTPases by both Gα12/13-activated (41) (e.g. p115RhoGEF) and Gαq-activated (42, 43, 44) (e.g. p63RhoGEF) RhoGEFs, and at least 70 other RhoGEFs, regulated by many different upstream regulators, exist (45). The activating proteins and pathways upstream of Ras also are legion (46). Our data obviously do not establish whether Rho and Ras simultaneously exist in an activated state in a cellular membrane microenvironment where PLC-ε is commonly and simultaneously activated by both GTPases. However, these results illustrate that spatial and temporal convergence of activated Rho and Ras would result in lipase activity that is essentially additive over that occurring with either GTPase alone. It also will be important to place our observation of Rho-promoted increases in sensitivity of PLC-ε to activation by K-Ras in a physiologically relevant context.
Our studies also did not address the activity of the CDC25 guanine nucleotide exchange domain of the N terminus of PLC-ε, and the function of this domain remains unclear. Perhaps activation of PLC-ε by Rho causes unique downstream engagement of Ras GTPases or promotes a feed-forward loop and additional activation of the phospholipase C by enhanced activation of Ras, Rap, or other Ras subfamily GTPases, which in turn associate with PLC-ε through the RA2 domain. Alternatively, PLC-ε may be sequentially trafficked to specific membrane environments as a consequence of Rho-promoted activation of Ras GTPases. Citro et al. (47) recently illustrated that PLC-ε functions as a nexus in Rho- and Rap-promoted astrocyte proliferation. Thus, activation of a thrombin receptor results in activation of Rho, presumably through activation of a Gα12/13-activated RhoGEF. Rho in turn stimulates both the lipase activity of PLC-ε and the function of the enzyme as a guanine nucleotide exchange factor for Rap1. ERK is consequentially activated, and astrocyte mitogenesis ensues.
The mechanism(s) by which Rho and Ras activate PLC-ε is unclear. Our studies indicate that lipidation of these GTPases is essential for activation of PLC-ε since increases in activity of the isozyme were not observed with bacterially expressed soluble GTPases. These observations in turn suggest that activated GTPases recruit PLC-ε to phosphoinositide substrate-containing membrane surfaces and therein promote increased enzyme activity. Other potential mechanisms of PLC-ε regulation by Ras superfamily GTPases include promotion of conformational changes within the enzyme upon binding of activated GTPases. Bunney et al. (40) have proposed a mechanism of regulation of PLC-ε whereby the RA domains fold back onto the catalytic regions of the enzyme to effect autoinhibition, which is relieved upon binding to Ras and translocation to the plasma membrane. In addition, we previously identified a conserved stretch of ∼70 amino acids found in the Y-region of the catalytic core of PLC-ε of all species but not found in other PLC isozymes (30, 34). The unique regulation of PLC-ε by Rho suggested that this PLC-ε-specific region is important for Rho-mediated activation, which indeed has proven to be the case. Thus, Rho binds in the catalytic core of the isozyme. Whether this simply results in membrane recruitment of PLC-ε, or rather results in a conformational change of the active site is yet to be resolved.
The aforementioned possibilities should be considered in face of recent structural and mechanistic insight into GTP-dependent activation of PLC-β2 by Rac. Rac binds PLC-β2 through interaction with the PH domain (14, 15), and the structure of holo-PLC-β2 superimposes with the structure of PLC-β2 in a GTP γS-promoted complex with Rac1 (39). Thus, binding to Rac1 has no effect on the catalytic core of PLC-β2, but rather, results in membrane localization and orientation of the catalytic region of PLC-β2 in a position that facilitates increased access to substrate. These structures of PLC-β2 also revealed that an ordered region of the otherwise disordered linker that separates the two halves, i.e. the X- and Y-regions, of the TIM barrel, occludes the active site and therein autoinhibits the isozyme. Existence of dense negative charge in the X/Y-linker region of this and other PLC isozymes suggests a unifying mechanism of regulation of catalysis whereby autoinhibition is removed when the negatively charged X/Y-linker encounters the negative surface of the plasma membrane. This model predicts that removal of the X/Y-linker should activate the enzyme, as we recently illustrated for PLC-β2 as well as for PLC-δ1 and PLC-ε (39). We show here that PLC-ε lacking the X/Y-linker region retains regulation by RhoA and K-Ras, and therefore, binding of neither GTPase requires this region of the enzyme.
The large size of PLC-ε allows the enzyme to bind various GTPases through distinct regions and implies that the binding of either GTPase alone would not affect the binding of a complimentary GTPase at a more distant site. Enzyme activity in the simultaneous presence of both GTPases is additive, but activation of PLC-ε by RhoA also increases the sensitivity to activation by K-Ras. Taken together, our data suggest that RhoA binding to PLC-ε orients the enzyme at the membrane in a favorable position for K-Ras to bind the RA2 domain of PLC-ε. Alternatively, binding of RhoA within the catalytic core of PLC-ε relieves an inhibitory constraint provided by a potential interaction of the RA domains with the catalytic core of the enzyme as suggested by Bunney et al. (40). Our data clearly illustrate that the binding of Rho or Ras GTPases to PLC-ε are mutually independent events, and we favor the idea that activation occurs as a consequence of GTPase-promoted increases in membrane association and producti veorientation for polyphosphoinositide hydrolysis.
Two decades ago, two groups independently suggested a role for Ras family proteins in the regulation of phosphoinositide metabolism (48, 49). The identification of PLC-ε as a direct and dual effector of Rho and Ras adds a new level of complexity to the role of Ras superfamily proteins in phospholipid signaling and metabolism. Appreciation of the importance of PLC-ε in cardiac development and function has followed from the work of Smrcka (50, 51) and Kataoka (52, 53) and their colleagues. Hildebrandt and co-workers (54) recently discovered that epithelial cell PLC-ε plays a crucial role in human kidney podocyte and glomeruli development, and early onset nephrotic syndrome occurs in children with truncating mutations of the PLC-ε gene. It will be important to establish to what extent coordinate, and perhaps cooperative, regulation of PLC-ε by Rho and Ras occurs in development and function of the heart, kidney, and other tissues. Future experiments also will elucidate the spatial and temporal regulation of PLC-ε by multiple GTPase signaling pathways, and will provide structurally-based mechanistic understanding of the regulation of this isozyme by Ras superfamily GTPases.
Acknowledgments
We thank Rhonda Carter and Svetlana Gershburg for excellent technical assistance and Len Stephens, Gary Waldo, and Rhonda Carter for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM57391 and GM65533. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PLC, phospholipase C; RA, Ras-associating; GTPγS, guanosine 5′-O-(3-thiotriphosphate); PtdIns, phosphatidylinositol; PtdIns(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PE, phosphatidylethanolamine; PH, pleckstrin homology; GEF, guanine nucleotide exchange factor; EF, elongation factor; TIM, triose-phosphate isomerase; HA, hemagglutinin A; CV, column volume.
References
- 1.Berridge, M. J. and Irvine, R. F. (1989) Nature 341 197–205 [DOI] [PubMed] [Google Scholar]
- 2.Kishimoto, A., Takai, Y., Mori, T., Kikkawa, U., and Nishizuka, Y. (1980) J. Biol. Chem. 255 2273–2276 [PubMed] [Google Scholar]
- 3.Streb, H., Irvine, R. F., Berridge, M. J., and Schulz, I. (1983) Nature 306 67–69 [DOI] [PubMed] [Google Scholar]
- 4.Lemmon, M. A. (2003) Traffic. 4 201–213 [DOI] [PubMed] [Google Scholar]
- 5.Niggli, V. (2005) Annu. Rev. Cell Dev. Biol. 21 57–79 [DOI] [PubMed] [Google Scholar]
- 6.Smrcka, A. V., Hepler, J. R., Brown, K. O., and Sternweis, P. C. (1991) Science 251 804–807 [DOI] [PubMed] [Google Scholar]
- 7.Taylor, S. J., Chae, H. Z., Rhee, S. G., and Exton, J. H. (1991) Nature 350 516–518 [DOI] [PubMed] [Google Scholar]
- 8.Waldo, G. L., Boyer, J. L., Morris, A. J., and Harden, T. K. (1991) J. Biol. Chem. 266 14217–14225 [PubMed] [Google Scholar]
- 9.Camps, M., Carozzi, A., Schnabel, P., Scheer, A., Parker, P. J., and Gierschik, P. (1992) Nature 360 684–686 [DOI] [PubMed] [Google Scholar]
- 10.Boyer, J. L., Waldo, G. L., and Harden, T. K. (1992) J. Biol. Chem. 267 25451–25456 [PubMed] [Google Scholar]
- 11.Blank, J. L., Brattain, K. A., and Exton, J. H. (1992) J. Biol. Chem. 267 23069–23075 [PubMed] [Google Scholar]
- 12.Illenberger, D., Schwald, F., Pimmer, D., Binder, W., Maier, G., Dietrich, A., and Gierschik, P. (1998) EMBO J. 17 6241–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Illenberger, D., Walliser, C., Nurnberg, B., Diaz, L. M., and Gierschik, P. (2003) J. Biol. Chem. 278 3006–3014 [DOI] [PubMed] [Google Scholar]
- 14.Snyder, J. T., Singer, A. U., Wing, M. R., Harden, T. K., and Sondek, J. (2003) J. Biol. Chem. 278 21099–21104 [DOI] [PubMed] [Google Scholar]
- 15.Jezyk, M. R., Snyder, J. T., Gershberg, S., Worthylake, D. K., Harden, T. K., and Sondek, J. (2006) Nat. Struct. Mol. Biol. 13 1135–1140 [DOI] [PubMed] [Google Scholar]
- 16.Wahl, M. I., Nishibe, S., Suh, P. G., Rhee, S. G., and Carpenter, G. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 1568–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meisenhelder, J., Suh, P. G., Rhee, S. G., and Hunter, T. (1989) Cell 57 1109–1122 [DOI] [PubMed] [Google Scholar]
- 18.Rhee, S. G. and Choi, K. D. (1992) J. Biol. Chem. 267 12393–12396 [PubMed] [Google Scholar]
- 19.Piechulek, T., Rehlen, T., Walliser, C., Vatter, P., Moepps, B., and Gierschik, P. (2005) J. Biol. Chem. 280 38923–38931 [DOI] [PubMed] [Google Scholar]
- 20.Zhou, Y., Wing, M. R., Sondek, J., and Harden, T. K. (2005) Biochem. J. 391 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou, Y., Sondek, J., and Harden, T. K. (2008) Biochemistry 47 4410–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, Y. H., Park, T. J., Lee, Y. H., Baek, K. J., Suh, P. G., Ryu, S. H., and Kim, K. T. (1999) J. Biol. Chem. 274 26127–26134 [DOI] [PubMed] [Google Scholar]
- 23.Baek, K. J., Kang, S., Damron, D., and Im, M. (2001) J. Biol. Chem. 276 5591–5597 [DOI] [PubMed] [Google Scholar]
- 24.Saunders, C. M., Larman, M. G., Parrington, J., Cox, L. J., Royse, J., Blayney, L. M., Swann, K., and Lai, F. A. (2002) Development 129 3533–3544 [DOI] [PubMed] [Google Scholar]
- 25.Yoda, A., Oda, S., Shikano, T., Kouchi, Z., Awaji, T., Shirakawa, H., Kinoshita, K., and Miyazaki, S. (2004) Dev. Biol. 268 245–257 [DOI] [PubMed] [Google Scholar]
- 26.Shibatohge, M., Kariya, K., Liao, Y., Hu, C. D., Watari, Y., Goshima, M., Shima, F., and Kataoka, T. (1998) J. Biol. Chem. 273 6218–6222 [DOI] [PubMed] [Google Scholar]
- 27.Song, C., Hu, C. D., Masago, M., Kariyai, K., Yamawaki-Kataoka, Y., Shibatohge, M., Wu, D., Satoh, T., and Kataoka, T. (2001) J. Biol. Chem. 276 2752–2757 [DOI] [PubMed] [Google Scholar]
- 28.Lopez, I., Mak, E. C., Ding, J., Hamm, H. E., and Lomasney, J. W. (2001) J. Biol. Chem. 276 2758–2765 [DOI] [PubMed] [Google Scholar]
- 29.Kelley, G. G., Reks, S. E., Ondrako, J. M., and Smrcka, A. V. (2001) EMBO J. 20 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wing, M. R., Houston, D., Kelley, G. G., Der, C. J., Siderovski, D. P., and Harden, T. K. (2001) J. Biol. Chem. 276 48257–48261 [DOI] [PubMed] [Google Scholar]
- 31.Kelley, G. G., Reks, S. E., and Smrcka, A. V. (2004) Biochem. J. 378 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunney, T. D., and Katan, M. (2006) Trends Cell Biol. 16 640–648 [DOI] [PubMed] [Google Scholar]
- 33.Wing, M. R., Snyder, J. T., Sondek, J., and Harden, T. K. (2003) J. Biol. Chem. 278 41253–41258 [DOI] [PubMed] [Google Scholar]
- 34.Seifert, J. P., Wing, M. R., Snyder, J. T., Gershburg, S., Sondek, J., and Harden, T. K. (2004) J. Biol. Chem. 279 47992–47997 [DOI] [PubMed] [Google Scholar]
- 35.Kapust, R. B., Tozser, J., Fox, J. D., Anderson, D. E., Cherry, S., Copeland, T. D., and Waugh, D. S. (2001) Protein Eng. 14 993–1000 [DOI] [PubMed] [Google Scholar]
- 36.Brown, H. A., Lazarowski, E. R., Boucher, R. C., and Harden, T. K. (1991) Mol. Pharmacol. 40 648–655 [PubMed] [Google Scholar]
- 37.Suire, S., Hawkins, P., and Stephens, L. (2002) Curr. Biol. 12 1068–1075 [DOI] [PubMed] [Google Scholar]
- 38.Seifert, J. P., Snyder, J. T., Sondek, J., and Harden, T. K. (2006) Methods Enzymol. 406 260–271 [DOI] [PubMed] [Google Scholar]
- 39.Hicks, S. N., Jezyk, M. R., Gershburg, S., Seifert, J. P., Harden, T. K., and Sondek, J. (2008) Mol. Cell 31 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunney, T. D., Harris, R., Gandarillas, N. L., Josephs, M. B., Roe, S. M., Sorli, S. C., Paterson, H. F., Rodrigues-Lima, F., Esposito, D., Ponting, C. P., Gierschik, P., Pearl, L. H., Driscoll, P. C., and Katan, M. (2006) Mol. Cell 21 495–507 [DOI] [PubMed] [Google Scholar]
- 41.Kozasa, T., Jiang, X., Hart, M. J., Sternweis, P. M., Singer, W. D., Gilman, A. G., Bollag, G., and Sternweis, P. C. (1998) Science 280 2109–2111 [DOI] [PubMed] [Google Scholar]
- 42.Lutz, S., Freichel-Blomquist, A., Yang, Y., Rumenapp, U., Jakobs, K. H., Schmidt, M., and Wieland, T. (2005) J. Biol. Chem. 280 11134–11139 [DOI] [PubMed] [Google Scholar]
- 43.Rojas, R. J., Yohe, M. E., Gershburg, S., Kawano, T., Kozasa, T., and Sondek, J. (2007) J. Biol. Chem. 282 29201–29210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lutz, S., Shankaranarayanan, A., Coco, C., Ridilla, M., Nance, M. R., Vettel, C., Baltus, D., Evelyn, C. R., Neubig, R. R., Wieland, T., and Tesmer, J. J. (2007) Science 318 1923–1927 [DOI] [PubMed] [Google Scholar]
- 45.Rossman, K. L., Der, C. J., and Sondek, J. (2005) Nat. Rev. Mol. Cell Biol. 6 167–180 [DOI] [PubMed] [Google Scholar]
- 46.Mitin, N., Rossman, K. L., and Der, C. J. (2005) Curr. Biol. 15 R563-R574 [DOI] [PubMed] [Google Scholar]
- 47.Citro, S., Malik, S., Oestreich, E. A., Radeff-Huang, J., Kelley, G. G., Smrcka, A. V., and Brown, J. H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15543–15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cantley, L. C., Whitman, M., Chahwala, S., Fleischman, L., Kaplan, D. R., Schaffhausen, B. S., and Roberts, T. M. (1986) Ann. N. Y. Acad. Sci. 488 481–490 [DOI] [PubMed] [Google Scholar]
- 49.Wakelam, M. J., Davies, S. A., Houslay, M. D., McKay, I., Marshall, C. J., and Hall, A. (1986) Nature 323 173–176 [DOI] [PubMed] [Google Scholar]
- 50.Wang, H., Oestreich, E. A., Maekawa, N., Bullard, T. A., Vikstrom, K. L., Dirksen, R. T., Kelley, G. G., Blaxall, B. C., and Smrcka, A. V. (2005) Circ. Res. 97 1305–1313 [DOI] [PubMed] [Google Scholar]
- 51.Oestreich, E. A., Wang, H., Malik, S., Kaproth-Joslin, K. A., Blaxall, B. C., Kelley, G. G., Dirksen, R. T., and Smrcka, A. V. (2007) J. Biol. Chem. 282 5488–5495 [DOI] [PubMed] [Google Scholar]
- 52.Bai, Y., Edamatsu, H., Maeda, S., Saito, H., Suzuki, N., Satoh, T., and Kataoka, T. (2004) Cancer Res. 64 8808–8810 [DOI] [PubMed] [Google Scholar]
- 53.Tadano, M., Edamatsu, H., Minamisawa, S., Yokoyama, U., Ishikawa, Y., Suzuki, N., Saito, H., Wu, D., Masago-Toda, M., Yamawaki-Kataoka, Y., Setsu, T., Terashima, T., Maeda, S., Satoh, T., and Kataoka, T. (2005) Mol. Cell Biol. 25 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinkes, B., Wiggins, R. C., Gbadegesin, R., Vlangos, C. N., Seelow, D., Nurnberg, G., Garg, P., Verma, R., Chaib, H., Hoskins, B. E., Ashraf, S., Becker, C., Hennies, H. C., Goyal, M., Wharram, B. L., Schachter, A. D., Mudumana, S., Drummond, I., Kerjaschki, D., Waldherr, R., Dietrich, A., Ozaltin, F., Bakkaloglu, A., Cleper, R., Basel-Vanagaite, L., Pohl, M., Griebel, M., Tsygin, A. N., Soylu, A., Muller, D., Sorli, C. S., Bunney, T. D., Katan, M., Liu, J., Attanasio, M., O'toole, J. F., Hasselbacher, K., Mucha, B., Otto, E. A., Airik, R., Kispert, A., Kelley, G. G., Smrcka, A. V., Gudermann, T., Holzman, L. B., Nurnberg, P., and Hildebrandt, F. (2006) Nat. Genet. 38 1397–1405 [DOI] [PubMed] [Google Scholar]