Abstract
The aim of this study was to evaluate the relationship between the virological response to darunavir-based salvage antiretroviral therapy and the darunavir genotypic and virtual inhibitory quotients (gIQ and vIQ, respectively). Thirty-seven HIV-infected patients failing protease inhibitor-based antiretroviral regimens who started salvage therapy containing darunavir-ritonavir were prospectively studied. The primary outcome of the study was a viral load (VL) of <50 copies/ml at week 48. The trough concentrations of darunavir in plasma, the number of darunavir resistance mutations, the change in the 50% inhibitory concentration (IC50) of darunavir in the virtual phenotype, and the darunavir gIQ and vIQ were correlated with the virological outcome in regression analyses adjusted by the number of active drugs in the background regimen. The VL was <50 copies/ml in 56.8% of patients at week 48. Changes in the VL were not significantly associated with the darunavir concentration (P = 0.304), the number of darunavir resistance mutations (P = 0.695), or the change in the IC50 (P = 0.750). However, patients with darunavir vIQs of ≥1.5 had a 12-fold greater chance of achieving a ≥1 log10 reduction in the VL (odds ratio [OR], 12.7; 95% confidence interval [95% CI], 1.9 to 81.6; P = 0.007), and a 5-fold greater chance of achieving a VL of <50 copies/ml (OR, 5.4; 95% CI, 1.2 to 24.5; P = 0.028), at week 48 than patients with darunavir vIQs of <1.5. The positive and negative predictive values of this darunavir vIQ cutoff for achieving a VL of <50 copies/ml at week 48 were 70% and 69%, respectively. The darunavir vIQ predicts virological response to darunavir-based salvage therapy better than the darunavir trough concentration or resistance mutations alone. We suggest targeting a darunavir vIQ of 1.5 for achieving long-term viral suppression.
Current guidelines for the management of antiretroviral therapy for patients infected with human immunodeficiency virus (HIV) (19) focus on maintaining maximal suppression of viral replication over time, regardless of whether the patients have prior experience with antiretroviral drugs or not. To achieve this goal, the inclusion of at least two active drugs in the therapeutic regimen is strongly recommended.
Darunavir is a recently licensed HIV protease inhibitor that exerts potent antiretroviral activity against both wild-type and mutant viral strains with reduced susceptibility to other protease inhibitors (5). The clinical effectiveness of darunavir for patients who had experienced triple-drug-class virological failure has been shown in different clinical trials (3, 12, 13, 17) where patients treated with darunavir had a fourfold greater chance of attaining undetectable viral loads than patients treated with an investigator-selected comparator protease inhibitor (3). However, a significant proportion of patients still did not achieve durable control of viral replication despite receiving darunavir-based therapy (3, 12, 13, 17).
The number of active drugs included in the antiretroviral regimen and the degree of viral resistance to darunavir are factors associated with virological and immunological responses in patients undergoing darunavir-based salvage therapy (3, 6, 8, 17; V. Sekar, S. De Meyer, T. Vangeneugden, E. Lefebvre, M. De Pauw, B. van Baelen, E. De Paepe, M. P. de Béthune, R. Hoetelmans, and W. Parys, presented at the 13th Conference on Retroviruses and Opportunistic Infections, 2006; V. Sekar, S. De Meyer, T. Vangeneugden, E. Lefebvre, M. De Pauw, B. van Baelen, E. De Paepe, M. P. de Béthune, D. Miralles, and R. Hoetelmans, presented at the 16th International AIDS Conference, 2006). Conversely, only a weak correlation between the darunavir trough concentration (Ctrough) and virological response has been reported (V. Sekar et al., posters presented at the 13th Conference on Retroviruses and Opportunistic Infections and at the 16th International AIDS Conference, 2006).
The inhibitory quotient (IQ), which is the ratio between the concentration of a drug in plasma and a measurement of viral susceptibility to that drug, has been shown to be a better predictor of virological response to salvage antiretroviral therapy than drug concentration or resistance data considered separately (10). In the case of darunavir, Sekar et al. (posters presented at the 13th Conference on Retroviruses and Opportunistic Infections and at the 16th International AIDS Conference, 2006) showed that the ratio between the Ctrough and the 50% inhibitory concentration (IC50) of darunavir, estimated through phenotypic resistance assays and designated the phenotypic inhibitory quotient (pIQ), was strongly related to the virological response to darunavir-based salvage therapy. However, despite the predictive value of pIQ, its use is limited in clinical practice by the complexity and cost inherent to phenotypic resistance testing (19). As a consequence, combining pharmacological data with alternative measurements of drug resistance that are simpler to implement in the clinical setting, such as the number of mutations associated with resistance to protease inhibitors in the viral genotype (genotypic inhibitory quotient [gIQ]) or the IC50 for the virus estimated through the virtual phenotype (virtual inhibitory quotient [vIQ]), would be cheaper, simpler, and faster to undertake than using the pIQ in routine clinical practice (10). The objective of this study was to evaluate the relationship between the virological response to darunavir-based salvage antiretroviral therapy and the darunavir gIQ or vIQ for HIV-infected patients who failed to respond to protease inhibitor-based antiretroviral regimens.
MATERIALS AND METHODS
Study design.
This was an open-label, prospective study including HIV-infected patients aged ≥18 years who were failing on a protease inhibitor-based antiretroviral regimen excluding darunavir. Virological failure was defined as a plasma HIV type 1 (HIV-1) RNA level of >50 copies/ml after at least 24 weeks of therapy or after the viral load had become undetectable. Patients gave informed consent before enrollment, and the protocol was approved by the ethics committee of our hospital.
Salvage antiretroviral therapy consisted of darunavir-ritonavir (600 and 100 mg, respectively, twice daily) with at least two other drugs (optimized background regimen), which were selected by the investigators according to treatment history and according to genotype and virtual-phenotype testing, performed at screening. Patients were visited at baseline and 4, 12, 24, 36, and 48 weeks after darunavir was initiated. The plasma HIV-1 RNA load and CD4+ T-cell count were determined at every visit, and the concentration of darunavir in plasma was measured at weeks 4, 12, 24, 36, and 48.
Study end points.
The primary end point of the study was the percentage of patients with plasma HIV-1 RNA loads of <50 copies/ml after 48 weeks of follow-up (virological response). Secondary end points included the magnitude of viral load reduction and the variability in the Ctrough of darunavir in plasma during follow-up.
Calculation of the OBS.
The genetic sequences of the HIV protease and reverse transcriptase genes and the virtual phenotype were obtained within the 4 weeks before the start of darunavir therapy by using the TruGene HIV-1 genotyping kit (Siemens Medical Solutions, Barcelona, Spain) and vircoTYPE HIV-1, version 3 (Virco BVBA, Mechelen, Belgium), respectively. A genotypic sensitivity score was calculated for each drug included in the background regimen. The drugs included in the background regimen to which the virus was not susceptible in the virtual phenotype were scored as 0, while the drugs to which the virus was partially or fully susceptible in the virtual phenotype were scored as 0.5 or 1, respectively. The optimized background score (OBS) was defined as the sum of the genotypic sensitivity scores for all drugs included in the regimen.
Enfuvirtide and raltegravir were considered to be active when they were used for the first time or if the patients had not previously failed to respond to antiretroviral regimens containing these agents. Resistance to etravirine was considered when three or more mutations related to resistance to nonnucleoside reverse transcriptase inhibitors were present in the reverse transcriptase gene.
Pharmacokinetic analysis.
For darunavir Ctrough determinations, blood samples were collected into potassium- and EDTA-containing 10-ml tubes before the morning dose of darunavir. Plasma was isolated by centrifugation (at 3,200 × g for 15 min) and stored at −20°C until analysis (4). Darunavir concentrations were determined by using high-performance liquid chromatography with a photo diode array detector (HPLC-PDA 2996; Waters, Barcelona, Spain) according to a validated method. A NovaPak C18 analytical column (3.9 by 150 mm) was used, with a NovaPak C18 guard column (Waters). The method involved liquid-liquid extraction of the drug from plasma with tert-butyl methyl ether after basification and a second wash with hexane. The mobile phase consisted of a gradient elution with phosphate buffer-acetonitrile (pH 6.70). The method was linear over the range of 0.05 to 10.0 mg/liter, and the intra- and interday variations were <10%.
Darunavir IQs.
The darunavir gIQ and vIQ were calculated for each participant. The darunavir gIQ was calculated as the median darunavir Ctrough during the follow-up divided by the number of darunavir resistance-associated mutations (RAM), which included the 11I, 32I, 33F, 47V, 50V, 54M/L, 73S, 76V, 84V, and 89V mutations in the protease gene (11). The darunavir vIQ was calculated as the median darunavir Ctrough during the follow-up divided by the change (n-fold) in the darunavir IC50 in the virtual phenotype and then multiplied by the protein-binding-corrected darunavir IC50 for protease inhibitor-resistant viruses (550 ng/ml) (5). For the calculation of both IQs, the darunavir Ctrough was expressed in nanograms per milliliter.
Statistical analysis.
Statistical analysis was performed using SPSS statistical software (version 15.0; SPSS Inc., Chicago, IL). The relationships between virological end points and different predictors were assessed by linear, nonlinear, or logistic regression analysis as appropriate and adjusted for the OBS.
Receiving operator characteristic (ROC) curves were used to explore possible cutoff values of resistance, pharmacological, and IQ data in order to properly classify patients as responders or nonresponders. Percentages were compared with the chi-square test or the Fisher exact test, as appropriate, and survival analyses using the Kaplan-Meier and the log rank test were performed to compare the lengths of time for patients to achieve HIV-1 RNA loads of <50 copies/ml.
Interindividual and intraindividual variabilities in the darunavir Ctrough were assessed at each visit and for those patients who had more than one darunavir determination available during follow-up, respectively. In both cases, variability was expressed as the coefficient of variation, which was calculated by dividing the standard deviation by the mean darunavir Ctrough.
RESULTS
Study population characteristics.
A total of 37 patients were included in the study. Patients had received a median (interquartile range) of 14 (12 to 15) antiretroviral regimens, including 5.0 (5.0 to 6.5) protease inhibitors, prior to the introduction of darunavir. Moreover, 81.1% of the patients had received five or more protease inhibitors before enrollment.
At the time of study entry, lopinavir and tipranavir were the most frequently used protease inhibitors (35% and 27%, respectively), and the median (interquartile range) viral load and CD4+ T-cell count were 4.6 (3.8 to 4.9) log10 copies/ml and 210 (95 to 310) cells/mm3, respectively. Enfuvirtide was included in the optimized background regimen for 17 (45.9%) patients. Six (16.2%) patients received etravirine and four (10.8%) patients received raltegravir during the study. The median (interquartile range) number of active drugs in the optimized background regimen was 1.5 (0.75 to 2.0). Table 1 summarizes other characteristics of the patients at baseline.
TABLE 1.
Demographic and main clinical characteristics of the study populationa
| Characteristic | Value for:
|
Pb | ||
|---|---|---|---|---|
| All patients | Virological responders | Virological nonresponders | ||
| No. (%) of patients | 37 | 21 (56.8) | 16 (43.2) | |
| HIV-1 RNA load (log10 copies/ml) | 4.6 (3.8-4.9) | 4.4 (3.8-4.9) | 4.8 (3.9-5.1) | 0.490 |
| CD4+ T-cell count (cells/mm3) | 210 (95-303) | 238 (170-295) | 174 (51-333) | 0.265 |
| OBS | 1.5 (0.7-2.0) | 1.5 (0.7-2.2) | 1.5 (0.6-2.0) | 0.565 |
| No. of RAM in the PRO gene | 8 (6-11) | 9 (7-11) | 7 (4-10) | 0.918 |
| No. of DRV RAM | 1 (0-2) | 1 (0-2) | 1 (0-2) | 0.918 |
| Fold change in the DRV IC50 | 1.4 (0.6-3.5) | 1.2 (0.6-2.4) | 2.0 (0.6-9.3) | 0.651 |
| DRV Ctrough (ng/ml) | 3,518 (2,612-5,085) | 3,389 (2,961-4,656) | 3,361 (1,593-4,813) | 0.377 |
| DRV gIQ | 2,780 (1,937-3,396) | 3,144 (2,250-3,514) | 2,314 (1,459-3,382) | 0.353 |
| DRV vIQ | 4.1 (0.9-9.7) | 6.6 (1.9-10.5) | 1.4 (0.8-9.6) | 0.159 |
| No. (%) of patients with a DRV gIQ of >2,400 | 22 (59.5) | 15 (71.4) | 7 (43.8) | 0.105 |
| No. (%) of patients with a DRV vIQ of >1.5 | 24 (64.8) | 17 (70.8) | 7 (29.2) | 0.028 |
Data are expressed as the median (interquartile range), except where otherwise noted. PRO, protease; DRV, darunavir.
For difference between virological responders and nonresponders, determined by logistic regression analysis adjusted for the OBS.
Virological and immunological outcomes.
The median (interquartile range) increases in the CD4+ T-cell count were 85 (14 to 181) cells/mm3 and 69 (10 to 218) cells/mm3 at weeks 24 and 48, respectively. The median (interquartile range) decreases in the viral load were −2.3 (−1.0 to −2.8) log10 copies/ml at week 24 and −2.1 (−0.8 to −2.9) log10 copies/ml at week 48. The percentage of patients with HIV-1 RNA loads of <50 copies/ml was 59.5% at week 24 and 56.8% at week 48.
Pharmacological and resistance analyses.
A median (interquartile range) of 3 (1 to 4) blood samples were drawn from each patient during the study, and the median darunavir Ctrough was 3,518 (2,612 to 5,085) ng/ml. The median inter- and intraindividual coefficients of variation in the darunavir Ctrough determinations were 57% (49% to 63%) and 27% (19% to 44%), respectively.
Patients showed a median of 8 (6 to 11) RAM in the protease gene, including 1 (0 to 2) darunavir RAM. The L33F mutation was observed in 19 patients (51.3%), followed by the I84V mutation (in 12 patients [32.4%]), the G73S mutation (in 5 patients [13.5%]), and the I54L and V32I mutations (in 3 patients [8.1%] each). Two patients showed the N88S mutation, and one patient had the I50L mutation, in the protease gene; both are related to increased susceptibility to darunavir (N. Parkin, E. Stawiski, C. Chappey, and E. Coakley, presented at the 14th Conference on Retroviruses and Opportunistic Infections, 2007).
The median (interquartile range) change in the darunavir IC50 in the virtual phenotype was 1.4-fold (0.6- to 3.5-fold). Fourteen patients (40%) showed a darunavir IC50 change of <1-fold, and six patients (16.2%) had a darunavir IC50 change of >10-fold at baseline.
Determinants of virological response.
By regression analysis, changes in the plasma HIV-1 RNA load were not significantly associated with the median darunavir Ctrough (r = 0.27 and P = 0.112 by the sigmoid Emax model), the number of darunavir RAM (r = 0.36 and P = 0.695 by the linear model), or the change in the IC50 of darunavir in the virtual phenotype (r = 0.16 and P = 0.344 by the sigmoid Emax model) when they were considered separately. Likewise, ROC curves and logistic regression analysis did not identify any cutoff in these parameters that was significantly associated with virological response at week 48 (Table 1).
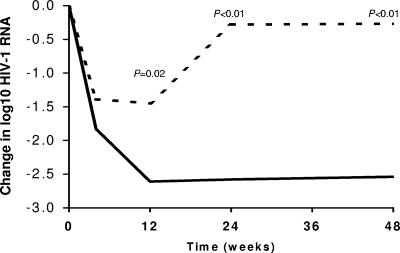
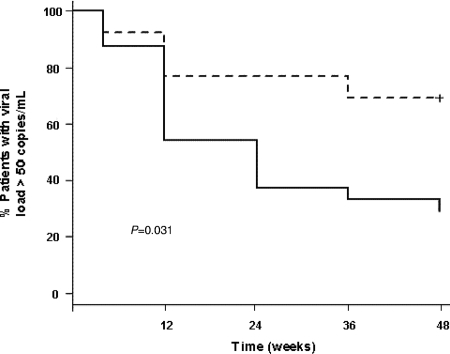
When darunavir pharmacological and resistance data were combined, changes in the plasma viral load were not associated with the darunavir gIQ (r = 0.27 and P = 0.100 by the sigmoid Emax model) but were associated with the darunavir vIQ (r = 0.40 and P = 0.014 by the sigmoid Emax model) by regression analysis. ROC curves identified cutoff values for the darunavir gIQ and vIQ that were related to virological response at week 48. There was a trend toward a better virological response in patients with darunavir gIQs of ≥2,400. Patients who had darunavir gIQs of ≥2,400 had a median (interquartile range) decrease in the HIV-1 RNA load of −2.5 (−1.4 to −3.1) log10 copies/ml at week 48, compared with −1.6 (−0.1 to −2.7) log10 copies/ml for patients who had darunavir gIQs of <2,400 (P = 0.139). Similarly, the chance of achieving a reduction in the viral load of >1 log10 unit or a viral load of <50 copies/ml at week 48 of follow-up was fourfold (odds ratio [OR], 4.0; 95% confidence interval [95% CI], 8.0 to 20.1; P = 0.089) or threefold greater, respectively, for patients with darunavir gIQs of >2,400 than for those with darunavir gIQs of <2,400 (Table 1). On the other hand, patients who had darunavir vIQs of ≥1.5 showed a median (interquartile range) decrease in the HIV-1 RNA load of −2.5 (−3.1 to −1.6) log10 copies/ml at week 48, compared with only −0.27 (−2.58 to 0.25) log10 copies/ml for patients who had vIQs of <1.5 (P = 0.004) (Fig. 1). Moreover, patients with darunavir vIQs of ≥1.5 had a 12-fold greater chance of achieving a reduction in the viral load of >1 log10 unit (OR, 12.7; 95% CI, 1.9 to 81.6; P = 0.007) and a 5-fold greater chance of achieving a viral load of <50 copies/ml (OR, 5.4; 95% CI, 1.2 to 24.5; P = 0.028) at week 48 than patients whose darunavir vIQs were <1.5 (Table 1; Fig. 2). The positive and negative predictive values of this darunavir vIQ cutoff for predicting the virological response at week 48 were 70% and 69%, respectively.
FIG. 1.
Changes in HIV-1 RNA load during follow-up. The dashed line represents patients with darunavir vIQs of <1.5, and the solid line represents patients with darunavir vIQs of ≥1.5.
FIG. 2.
Percentages of patients with HIV-1 RNA load of >50 copies/ml during follow-up. The dashed line represents patients with darunavir vIQs of <1.5, and the solid line represents patients with darunavir vIQs of ≥1.5.
DISCUSSION
In our population of HIV-infected patients who had been heavily treated previously, the darunavir IQ was shown to be a better predictor of long-term virological response to darunavir-based salvage antiretroviral therapy than either the darunavir Ctrough or darunavir RAM considered separately. Moreover, we were able to define a vIQ cutoff that correlated with a greater chance of maintaining complete long-term viral suppression.
In agreement with reports for other protease inhibitors as well as for darunavir itself (1, 9, 10, 14-16; Sekar et al., posters presented at the 13th Conference on Retroviruses and Opportunistic Infections and at the 16th International AIDS Conference, 2006), we did not observe a close relationship between virological response and darunavir exposure, although the interindividual variability in darunavir concentrations among our patients was notable. Sekar et al. (posters presented at the 13th Conference on Retroviruses and Opportunistic Infections and at the 16th International AIDS Conference, 2006) also pointed out the limited value of considering pharmacological data separately for the prediction of the virological response to salvage antiretroviral therapy with darunavir, noting the importance of integrating this information with resistance data in this setting. In those studies, the virological outcome of darunavir-based salvage therapy was much better predicted by the darunavir pIQ than by either the darunavir concentration or the change in the darunavir IC50 alone. However, phenotypic resistance testing is complex and expensive in the clinical setting, and pIQ data may therefore not be available for a large proportion of patients outside clinical trials. The gIQ and vIQ are simpler to obtain than the pIQ and have been shown to predict virological response to salvage antiretroviral therapy with several protease inhibitors (1, 9, 10, 14-16).
In our study, in which darunavir resistance was assessed by genotypic assays, the darunavir vIQ was the best predictor of virological response after 48 weeks of follow-up. According to our results, the chances of attaining complete viral suppression were fivefold greater when patients had darunavir vIQs of ≥1.5. Using this cutoff value, we were able to correctly classify 70% of the patients as responders or nonresponders, allowing us to predict virological response in an individualized fashion. Consequently, this vIQ cutoff could be used to guide possible dose adjustments of darunavir in the context of therapeutic drug monitoring programs aimed at individualizing darunavir dosing in order to overcome partial resistance to darunavir and maximize the chances of virological response in the long term.
We were unable to confirm the relationship between the degree of viral resistance to darunavir and the virological response to darunavir-based salvage therapy that was observed in the POWER studies (3, 6). This discrepancy between our results and those reported earlier may be explained by a lesser extent of resistance to darunavir in our population than in the POWER trials, where 22% of the patients who participated had three or more darunavir RAM at baseline and the median change in the IC50 of darunavir was approximately fourfold (3). Conversely, only two patients (5%) in our study had three or more darunavir RAM at baseline; the median change in the darunavir IC50 was only 1.4-fold in our population; and only six patients (16.2%) had a >10-fold change in the IC50 at baseline (four of these patients were responders at week 48 even when their darunavir vIQs were <1.5). Similarly, the low number of darunavir RAM may also explain the lack of a significant relationship between the darunavir gIQ and virological response in our study. In addition, although darunavir RAM are supposed to affect drug susceptibility to different degrees, all mutations were weighted equally in the calculation of the darunavir gIQ in the present study. In order to overcome this limitation, different RAM scoring systems that assign different weights to each mutation can be applied, or the virtual phenotype can be used, as in our calculation of the vIQ.
The number of active drugs in the background regimen (indicated by the OBS) is another relevant factor that has been related to the virological response to salvage antiretroviral therapy in different studies (2, 3, 7, 8, 12, 13, 18). We were not able to identify such a relationship in our study, probably because of the low OBS in our population. A quarter of the patients in our study were treated with less than one active drug in the background regimen, and the median OBS was <2. Nonetheless, we are aware of the relevance of the OBS and of its influence on virological response, and linear and logistic analyses were therefore adjusted for the OBS.
In conclusion, the darunavir vIQ was related to the chance of maintaining complete long-term viral suppression in our treatment-experienced HIV-infected patients on darunavir-based salvage antiretroviral therapy. This tool allows virological response to be predicted at the individual level and could be helpful in targeting darunavir concentrations in this setting.
Acknowledgments
We acknowledge the contribution of Mary Ellen Kerans, who assisted with English language expression.
The authors have no conflicts of interest that are directly relevant to the context of this study.
This study was funded by a grant from the Lluita Contra La SIDA Foundation and by ISCIII-RETIC RD06/006. Supplemental funding was obtained from a United Kingdom Department of Health New and Emerging Applications of Technology award (CO52). M. Valle is supported by FIS trough grant CP04/00121 from the Spanish Health Department in collaboration with the Institut de Recerca de l'Hospital de la Santa Creu i Sant Pau, Barcelona, Spain, and is a member of CIBERSAM (supported by the Spanish Ministry of Health, Instituto de Salud Carlos III).
Footnotes
Published ahead of print on 25 August 2008.
REFERENCES
- 1.Barrios, A., A. L. Rendon, O. Gallego, L. Martin-Carbonero, L. Valer, P. Rios, I. Maida, T. Garcia-Benayas, I. Jimenez-Nacher, J. Gonzalez-Lahoz, and V. Soriano. 2004. Predictors of virological response to atazanavir in protease inhibitor-experienced patients. HIV Clin. Trials 5:201-205. [DOI] [PubMed] [Google Scholar]
- 2.Cahn, P., J. Villacian, C. Katlama, B. Grinsztejn, K. Arasteh, P. Lopez, N. Clumeck, J. Gerstoft, N. Stavrineas, S. Moreno, F. Antunes, D. Neubacher, and D. Mayers. 2006. Ritonavir-boosted tipranavir demonstrates superior efficacy to ritonavir-boosted protease inhibitors in treatment-experienced HIV-infected patients: 24-week results of the Resist-2 trial. Clin. Infect. Dis. 43:1347-1356. [DOI] [PubMed] [Google Scholar]
- 3.Clotet, B., N. Bellos, J. M. Molina, D. Cooper, J. C. Goffard, A. Lazzarin, A. Wöhrmann, C. Katlama, T. Wilkin, R. Haubrich, C. Cohen, C. Farthing, D. Jayaweera, M. Markowitz, P. Ruane, S. Spinosa-Guzman, E. Lefebvre, and the POWER 1 and 2 study groups. 2007. Efficacy and safety of darunavir-ritonavir at week 48 in treatment-experienced patients with HIV-1 infection in POWER 1 and 2: a pooled subgroup analysis of data from two randomized trials. Lancet 369:1169-1178. [DOI] [PubMed] [Google Scholar]
- 4.D'Avolio, A., M. Siccardi, M. Sciandra, L. Baietto, S. Bonora, L. Trentini, and G. Di Perri. 2007. HPLC-MS method for the simultaneous quantification of the new HIV protease inhibitor darunavir, and 11 other antiretroviral agents in plasma of HIV-infected patients. J. Chromatogr. B 859:234-240. [DOI] [PubMed] [Google Scholar]
- 5.De Meyer, S., H. Azijn, D. Surleraux, D. Jochmans, A. Thari, R. Pawels, P. Wiquerinck, and M. P. de Bethune. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Meyer, S., T. Vangeneugden, E. Lefebvre, H. Van Mack, H. Azjin, I. De Baere, B. van Baelen, and M. P. de Béthune. 2006. Phenotypic and genotypic determinants of TMC114 (darunavir) resistance: POWER 1, 2, and 3 pooled analysis, abstr. P196. Prog. Abstr. 8th Int. Cong. Drug Ther. HIV Infect.
- 7.Gathe, J., D. A. Cooper, C. Farthing, D. Jayaweera, D. Norris, G. Pierone, C. R. Steinhart, B. Trottier, S. L. Walmsley, C. Workman, G. Mukwaya, V. Kohlbrenner, C. Dohnanyi, S. McCallister, and D. Mayers for the RESIST-1 Study Group. 2006. Efficacy of the protease inhibitors tipranavir plus ritonavir in treatment-experienced patients: 24-week analysis from the RESIST-1 trial. Clin. Infect. Dis. 43:1337-1346. [DOI] [PubMed] [Google Scholar]
- 8.Gathe, J., Jr., E. de Jesus, R. Falcon, S. Spinosa-Guzman, and T. Vangeneugden. 2006. Examination of factors influencing response to darunavir combined with low-dose ritonavir in POWER 1, 2 and 3: pooled 48-week analysis, abstr. 66, p. 72-73. HIV DART 2006: Front. Drug Dev. Antiretrovir. Ther. http://www.ihlpress.com/pdf%20files/AbstractBookFINAL_DART06.pdf.
- 9.Gonzalez de Requena, D., S. Bonora, A. Calcagno, A. D'Avolio, M. Siccardi, S. Fontana, M. G. Milia, M. Sciandra, S. Garazzino, A. Di Garbo, L. Baietto, L. Trentini, and G. Di Perri. 2008. Tipranavir (TPV) genotypic inhibitory quotient predicts virological response at 48 weeks to TPV-based salvage regimens. Antimicrob. Agents Chemother. 52:1066-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoefnagel, J. G. M., P. P. Koopmans, D. M. Burger, R. Schuurman, and J. M. D. Galama. 2005. Role of the inhibitory quotient in HIV therapy. Antivir. Ther. 10:879-892. [PubMed] [Google Scholar]
- 11.Johnson, V. A., F. Brun-Vézinet, B. Clotet, H. F. Günthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2007. Update of the drug resistance mutations in HIV-1: 2007. Top. HIV Med. 15:119-125. [PubMed] [Google Scholar]
- 12.Lazzarin, A., T. Campbell, B. Clotet, M. Johnson, C. Katlama, A. Moll, W. Towner, B. Trottier, M. Peeters, J. Vingerhoets, G. de Smedt, B. Baeten, G. Beets, R. Sinha, B. Woodfall, and the DUET-2 Study Group. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1 infected patients in DUET-2: 24-week results from a randomized, double-blind, placebo-controlled trial. Lancet 370:39-48. [DOI] [PubMed] [Google Scholar]
- 13.Madruga, J. V., P. Cahn, B. Grinsztejn, R. Haubrich. J. Lalezari, A. Mills, G. Pialoux, T. Wilkin, M. Peeters, J. Vingerhoets, G. de Smedt, L. Leopold, R. Trefiglio, B. Woodfall, and the DUET-1 Study Group. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1 infected patients in DUET-1: 24-week results from a randomized, double-blind, placebo-controlled trial. Lancet 370:29-38. [DOI] [PubMed] [Google Scholar]
- 14.Marcelin, A. G., I. Cohen-Codar, M. S. King, P. Colson, E. Guillevic, D. Descamps, C. Lamotte, V. Schneider, J. Ritter, M. Segondy, H. Peigue-Lafeuille, L. Morand-Joubert, A. Schmuck, A. Ruffault, P. Palmer, M. L. Chaix, V. Mackiewicz, V. Brodard, J. Izopet, J. Cottalorda, E. Kohli, J. P. Chauvin, D. J. Kempf, G. Peytavin, and V. Calvez. 2005. Virological and pharmacological parameters predicting the response to lopinavir-ritonavir in heavily protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 49:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcelin, A. G., C. Dalban, G. Peytavin, C. Lamotte, R. Agher, C. Delaugerre, M. Wirden, F. Conan, S. Dantin, C. Katlama, D. Costagliola, and V. Calvez. 2004. Clinically relevant interpretation of genotype and relationship to plasma drug concentrations for resistance to saquinavir-ritonavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 48:4687-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcelin, A. G., C. Lamotte, C. Delaugerre, N. Ktorza, H. A. Mohand, R. Cacace, M. Bonmarchand, M. Wirden, A. Simon, P. Bossi, F. Bricaire, D. Costagliola, C. Katlama, G. Peytavin, V. Calvez, and the Genophar Study Group. 2003. Genotypic inhibitory quotient as predictor of virological response to ritonavir-amprenavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 47:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina, J. M., C. Cohen, C. Katlama, B. Grinsztejn, A. Timerman, R. J. Pedro, T. Vangeneugden, D. Miralles, S. De Meyer, W. Parys, and E. Lefebvre on Behalf of the TMC114-C208 and -C215 Study Groups. 2007. Safety and efficacy of darunavir (TMC114) with low-dose ritonavir in treatment-experienced patients. 24-week results of POWER 3. J. Acquir. Immune Defic. Syndr. 46:24-31. [DOI] [PubMed] [Google Scholar]
- 18.Nelson, M., K. Arastéh, B. Clotet, D. A. Cooper, K. Henry, C. Katlama, J. P. Lalezari, A. Lazzarin, J. S. G. Montaner, M. O'Hearn, P. J. Piliero, J. Reynes, B. Trottier, S. L. Walmsley, C. Cohen, J. J. Eron, Jr., D. R. Kuritzkes, J. Lange, H. J. Stellbrink, J. F. Delfraissy, N. E. Buss, L. Donatacci, C. Wat, L. Smiley, M. Wilkinson, A. Valentine, D. Guimaraes, R. DeMasi, J. Chung, and M. P. Salgo. 2005. Durable efficacy of enfuvirtide over 48 weeks in heavily treatment-experienced HIV-1-infected patients in the T-20 versus optimized background regimen only 1 and 2 clinical trials. J. Acquir. Immune Defic. Syndr. 40:404-412. [DOI] [PubMed] [Google Scholar]
- 19.Panel on Antiretroviral Guidelines for Adults and Adolescents. 2008. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services, Washington, DC. http://aidsinfo.nih.gov/contentfiles/AdultandAdolescentGL.pdf.