Abstract
The susceptibility to several oligopeptide and amino acid antifungals of a Saccharomyces cerevisiae strain carrying multiple deletions in yeast multidrug resistance genes was compared to transformants containing the CDR1, CDR2, or MDR1 genes that encode the major Candida albicans drug efflux pumps. Recombinant yeast strains overexpressing Cdr1p and Cdr2p showed enhanced susceptibilities to all tested oligopeptide antifungals. The enhanced susceptibilities of multidrug-resistant yeast strains to oligopeptide antifungals corresponded to higher rates of oligopeptide uptake. Yeast cells overexpressing Cdr1p or Cdr2p effluxed protons at higher rates than the reference cells lacking these ABC transporters. An increased plasma membrane electrochemical gradient caused by the functional overexpression of Cdr1p or Cdr2p appeared to increase cellular susceptibility to oligopeptide antifungals by stimulating their uptake via oligopeptide permeases.
The frequency of fungal infections in immunocompromised patients and mortality due to invasive mycoses continue to be important clinical problems, and opportunistically pathogenic Candida species remain one of the leading causes of nosocomial bloodstream infections worldwide (13, 31, 35). Candida albicans comprises nearly half of the isolates of candidemia (44). The repertoire of antifungal chemotherapeutic agents is very limited, and although new antifungal drugs, such as voriconazole and caspofungin, have been introduced into clinical practice in recent years, the development of resistance, often multidrug resistance (MDR), has become increasingly significant (23).
The molecular mechanism underlying MDR is overexpression of membrane proteins belonging to members of a family of ATP-binding cassette (ABC) transporters or the major facilitator superfamily. In Candida albicans, Cdr1p and Cdr2p were identified as the major ABC drug transporters, while CaMdr1p and FLU1p are the main representatives of the major facilitator superfamily (36). The substrate specificity spectrum of fungal MDR transporters includes many therapeutically important antifungal drugs, including azole antifungals (40). The development of the MDR phenotype in C. albicans clinical strains isolated from patients subjected to antifungal chemotherapy with fluconazole is a well-documented phenomenon (1, 10, 11), and fluconazole is widely used for antifungal prophylaxis in immunocompromised patients (31). There is a need for new antifungals that circumvent MDR.
It was previously shown that recombinant yeast cells overexpressing the C. albicans Cdr1p drug efflux pump were paradoxically more susceptible to the action of oligopeptidic antifungal agents containing N3-(4-methoxyfumaroyl)-l-2,3-diaminopropanoic acid (FMDP). It was suggested that the observed hypersusceptibility might be due to the increased uptake of FMDP peptides mediated by oligopeptide permeases (27). In the present study, the effects of several structurally unrelated antifungal oligopeptides and amino acids were investigated in MDR clinical isolates of Candida albicans and genetically modified strains of the model yeast Saccharomyces cerevisiae that overexpress genes encoding C. albicans Cdr1p, Cdr2p, or Mdr1p drug efflux pumps.
MATERIALS AND METHODS
Chemicals.
Nva-FMDP and Lys-Nva-FMDP were synthesized by R. Andruszkiewicz (3). Methods for the synthesis of oligopeptides containing oxalysine (OLys), m-fluorophenylalanine (FPhe), or 5-fluoroorotic acid were described previously (4, 21, 43). Nikkomycin X/Z was a gift from H. Zähner, University of Tübingen, Germany. Fluconazole was from Pliva Krakow (Cracow, Poland). Histatin 5 (DSHAKRHHGYKRKFHEKHHSHRGY) and other chemicals were from Sigma or Calbiochem.
Yeast strains and growth conditions.
The yeast strains used in this investigation are presented in Table 1. S. cerevisiae AD12345678 (hereafter denoted strain AD) was kindly provided by A. Goffeau, Université Catholique de Louvain, Belgium. The AD-derived MDR cells were obtained by previously described methods (14, 37, 42). The AD cells were propagated in yeast-nitrogen base-glucose (YNBG) medium containing 0.67% YNB without amino acids and ammonium sulfate (Difco), 2% glucose, and 4 mg/ml of l-proline (or, if indicated, 1 mg/ml of l-glutamate) supplemented with l-histidine at 40 μg/ml and uracil at 30 μg/ml, while the AD-derived transformants were maintained in a similar medium lacking uracil. C. albicans cells were propagated in 2% glucose, 1% yeast extract, 1% Bacto peptone medium. C. albicans B3, B4, Gu4, and Gu5 clinical isolates were kindly provided by Joachim Morschhäuser, Würzburg, Germany. Gu4 and B3 are fluconazole-sensitive isolates obtained from early infection episodes, while Gu5 and B4 are the corresponding fluconazole-resistant isolates obtained from later episodes in the same patients treated with fluconazole (10). For each sensitive/resistant pair, comparison of CARE-2 fingerprint hybridization patterns confirmed that a single strain was responsible for the recurrent infection. Reverse transcription-PCR, Northern blotting, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the plasma membrane preparations indicated the molecular basis of the phenotypes ascribed in Table 1.
TABLE 1.
Microbial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| S. cerevisiae ATCC 9763 | Wild type | ATCC |
| S. cerevisiae AD12345678 | MATα pdr1-3 his ura3 pdr5Δ snq2Δ pdr10Δ pdr11Δ pdr15Δ yor1Δ ycf1Δ | 9 |
| S. cerevisiae ADCDR1 | AD12345678 transformed with CDR1 | 42 |
| S. cerevisiae ADCDR2 | AD12345678 transformed with CDR2 | 42 |
| S. cerevisiae ADMDR1 | AD12345678 transformed with MDR1 | 14 |
| C. albicans ATCC 10261 | Wild type | ATCC |
| C. albicans Gu4 | Clinical isolate (fluconazole sensitive) | 11 |
| C. albicans Gu5 | Clinical isolate (fluconazole resistant due to the overexpression of CDR1 and CDR2) | 11 |
| C. albicans B3 | Clinical isolate (fluconazole sensitive) | 11 |
| C. albicans B4 | Clinical isolate (fluconazole resistant due to the overexpression of MDR1) | 11 |
Susceptibility testing methods.
MICs of antifungals were determined by the microdilution methods using 96-well microtiter plates. MICs for C. albicans clinical isolates were determined in RPMI-1640 medium using the CLSI M27-A2 document recommendations (30). For S. cerevisiae recombinants, most conditions were the same, but an appropriately supplemented YNBG medium (see above) was used, and incubation was for 24 h at 30°C. Turbidity in individual wells was measured with a plate reader (Victor3; PerkinElmer). MIC was defined as a drug concentration that gave at least an 80% decrease in turbidity relative to that of the drug-free growth control.
Determination of peptide and amino acid uptake rates.
Initial velocities of oligopeptide or amino acid uptake were determined using a modification of the method of Payne and Nisbet (32). Yeast cells grown in the YNBG-proline medium were harvested, washed, and suspended in 50 mM phosphate-citrate buffer, pH 6.0, containing 1% glucose, to a final optical density at 660 nm (OD660) of 1.0. The cell suspension was preincubated for 10 min at 30°C, and a peptide or amino acid solution was added to give the final concentration of 50 μM. In some experiments, 100 μM fluconazole was added at the beginning of the preincubation. Suspensions supplemented with 10 mM NaN3 were used as negative controls to correct for the background passive binding of each oligopeptide/amino acid to the cells. A suspension without an oligopeptide/amino acid was used as a blank. Samples of 1 ml of cell suspension, withdrawn at zero time and at 5-min intervals, were filtered through Whatman GF/C filters. Samples of 200 μl from each filtrate were combined with 2-ml portions of 0.1 M tetraborate-HCl buffer, pH 6.2 (oligopeptide uptake determination) or pH 8.4 (amino acid uptake determination). Fluorescamine in acetone (0.15 mg/ml and 500 μl) was added, and the resulting fluorescence intensities were measured (excitation, 390 nm; emission, 485 nm). Initial uptake rates were determined from the plots of the oligopeptide/amino acid remaining versus time. Standard curves were generated for each oligopeptide and amino acid after dissolution in the appropriate tetraborate-HCl buffer and treatment with fluorescamine.
Efflux of oligopeptides and amino acids from yeast cells.
AD, ADCDR1, or ADCDR2 cells were grown in YNBG to the logarithmic phase of growth, harvested, washed three times in water, and resuspended to an OD660 of 1.0 in 50 mM phosphate buffer, pH 6.0, containing 5 mM 2-deoxy-d-glucose (2DG). Cell suspensions were incubated for 2 h at 30°C with gentle shaking, and the cells were harvested, washed three times in water, and resuspended in 50 mM phosphate-citrate buffer, pH 4.0. Amino acids or oligopeptides (200 μM) were added, and the cell suspensions were incubated for 60 min at 30°C with gentle shaking. Samples of the cell suspensions were collected at 10-min time intervals to monitor amino acid/oligopeptide uptake by the fluorescamine method (see above). Amino acid/oligopeptide-loaded cells were harvested, washed two times in water, and resuspended at an OD660 of 1.0 in prewarmed (30°C) 50 mM phosphate buffer, pH 6.0. After a 5-min preincubation, glucose (10 mM) was added and cell suspensions were incubated at 30°C with gentle shaking. Culture supernatants were collected at 5-min time intervals to monitor the appearance of effluxed amino acid/oligopeptide by the fluorescamine method. Yeast cell cultures treated as above, except for loading with an amino acid/oligopeptide, were used as a negative control to detect a background level of any fluorescamine-reactive compounds extruded by the cells. In separate experiments, cells deenergized by incubation with 2DG were loaded with rhodamine 6G (10 μM) instead of an amino acid/oligopeptide. An active glucose-dependent extrusion of the dye was measured as described previously (45).
Other methods.
Amino acids released from the oligopeptides by hydrolytic enzymes present in cell extracts from the yeast cells were quantified using the Cd-ninhydrin method (28). Intracellular pH was determined by the method of Bracey et al. with 5(6)-carboxyfluorescein diacetate-succinimidyl ester (CFDA-SE) as a fluorescent probe (7). The initial rates of proton efflux were determined as described previously (18).
RESULTS
Microorganisms used in the study.
This study used as a model system the recombinant S. cerevisiae AD strain, deleted of seven MDR transporters (9), and its transformants overexpressing either CDR1, CDR2, or MDR1 genes encoding the major drug effluxing pumps of C. albicans. In all transformants, each gene of interest was expressed from the centromeric pYEUra3 plasmid under the control of its natural promoter (14, 42). C. albicans clinical isolates that developed resistance to fluconazole during antifungal chemotherapy with this drug were also tested. The resistance of isolates Gu5 and B4 is due to the documented overexpression of CDR1 and/or CDR2 in the former and MDR1 in the latter (10, 11). Their matched fluconazole-sensitive isolates Gu4 and B3, respectively, exhibited basal expression of these resistance genes.
Growth inhibition of MDR yeast by oligopeptide and amino acid antifungals.
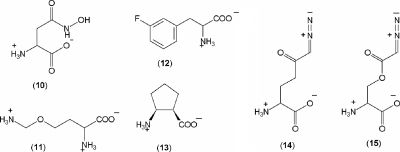
The in vitro susceptibilities of the strains described above to several antifungal oligopeptides and amino acids were determined. A set of the antifungal peptides included the following: 5-FO-l-leucyl-l-leucine 1, three oligopeptides incorporating OLys 2 to 4, a dipeptide and a tripeptide incorporating FPhe 5 and 6, two oligopeptides containing FMDP 7 and 8, the peptide-nucleoside antibiotic nikkomycin X/Z 9, and a human salivary polypeptide histatin 5. A set of the antifungal amino acids included the following: l-Asp-γ-hydroxamate 10, OLys 11, FPhe 12, cis-pentacin 13, 6-diazo-5-oxo-l-norleucine (DON) 14, and azaserine 15 (Fig. 1). Fluconazole was included as a reference to confirm the susceptibility/resistance of each strain.
FIG. 1.
Structures of amino acid antifungals 10 to 15 used in this study.
The results of the determination of antifungal in vitro activities of oligopeptidic antifungals against the S. cerevisiae strains are shown in Table 2. The yeast MDR genes disrupted in AD cells did not significantly change the strain's drug susceptibility profile, in comparison to the control ATCC 9763 strain, apart from a barely detectable increase in sensitivity to fluconazole. The ADCDR1, ADCDR2, and ADMDR1 strains were much less susceptible to fluconazole than AD. In contrast, the ADCDR1 and ADCDR2 strains showed enhanced susceptibilities to several oligopeptide antifungals. The strongest effects, reflected in the AD MIC/ADCDR1 MIC or AD MIC/ADCDR2 MIC ratios in the 64-fold/8-fold range, were observed for Leu-FPhe, Met-Met-FPhe, Nva-FMDP, Lys-Nva-FMDP, and nikkomycin. For other oligopeptides, the MIC ratios were in the two- to fourfold range. The ADMDR1 transformant showed an oligopeptide antifungal susceptibility comparable to that of the parent AD strain, despite a marked increase in resistance to fluconazole. A salivary polypeptide histatin 5 showed comparable activity against all strains.
TABLE 2.
In vitro fungistatic activities of oligopeptide antifungals and fluconazole against S. cerevisiae reference and recombinant strains
| Oligopeptide | MIC (μg/ml) ofa:
|
||||
|---|---|---|---|---|---|
| ATCC 9763 | AD | ADCDR1 | ADCDR2 | ADMDR1 | |
| 5FO-Leu-Leu (1) | 512 | 512 | 128 | 256 | 512 |
| OLys-Leu-Gly (2) | 8 | 4 | 1 | 1 | 4 |
| OLys-Leu-Leu-Gly (3) | 64 | 64 | 16 | 32 | 64 |
| OLys-Leu-Leu-Leu-Gly (4) | 256 | 256 | 128 | 128 | 256 |
| Leu-FPhe (5) | 256 | 256 | 8 | 4 | 256 |
| Met-Met-FPhe (6) | 256 | 512 | 32 | 32 | 256 |
| Nva-FMDP (7) | 4 | 4 | 0.5 | 1 | 4 |
| Lys-Nva-FMDP (8) | 32 | 32 | 4 | 8 | 32 |
| Nikkomycin (9) | 128 | 128 | 16 | 16 | 128 |
| Histatin 5 | 128 | 128 | 128 | 128 | 128 |
| Fluconazole | 2 | 1 | 32 | 16 | 32 |
MICs were determined as described in Materials and Methods by the microtiter serial twofold dilution method using the YNBG-proline medium containing 2% glucose. Inoculum size was 105 CFU/ml. Plates were incubated for 24 h at 30°C.
The results of the determination of growth inhibitory activities of antifungal amino acids presented in Table 3 showed a few cases of slightly different susceptibilities of the MDR strains to l-Asp-γ-hydroxamate, FPhe, OLys, DON, and azaserine. FPhe, OLys, DON, and azaserine were only slightly more active (two- to fourfold) against ADCDR1 and ADCDR2 cells than against the parent AD strain. All MDR transformants showed decreased susceptibilities to cis-pentacin.
TABLE 3.
In vitro fungistatic activities of amino acid antifungals against S. cerevisiae reference and recombinant strains
| Amino acid | MIC(s) (μg/ml) ofa:
|
||||
|---|---|---|---|---|---|
| ATCC 9763 | AD | ADCDR1 | ADCDR2 | ADMDR1 | |
| Asp-γ-hydroxamate (10) | 256 | 256 | 64 | 64 | 128 |
| OLys (11) | 8 | 8 | 2 | 4 | 16 |
| FPhe (12) | 32 | 32 | 16 | 16 | 32 |
| cis-pentacin (13) | 64, 4b | 128, 8b | 256, 16b | 256, 16b | 256, 16b |
| DON (14) | 0.5 | 0.5 | 0.125 | 0.25 | 0.5 |
| Azaserine (15) | 2 | 2 | 1 | 1 | 2 |
MIC(s) determined under conditions described in Table 2.
MICs determined by using the YNBG medium containing 1 mg/ml of l-glutamate instead of l-proline as the main nitrogen source.
MICs of antifungal amino acids, oligopeptides, and fluconazole against C. albicans clinical isolates Gu4 and B3, presented in Table 4, were similar to those found for the reference ATCC 10261 strain. Strains Gu5 and B4 were strongly resistant to fluconazole. Compared with Gu4, Gu5 cells overexpressing CDR1 and/or CDR2 showed enhanced susceptibilities to oligopeptides 2 and 5 to 9. Gu4 MIC/Gu5 MIC ratios were in the 4- to 16-fold range, and the highest values were found for OLys-Leu-Gly and Leu-FPhe. In B4, overexpression of MDR1 had little or no effect on the susceptibilities of C. albicans cells to oligopeptides 2 and 5 to 9 and amino acid antifungals 10 to 12, 14, and 15. All C. albicans strains were equally susceptible to histatin 5. Gu5 cells were slightly more susceptible to OLys 11 and DON 14 than the Gu4 cells. No differences were found for FPhe and azaserine. The MDR strains Gu5 and B4 were less susceptible to cis-pentacin 13 than their parental strains.
TABLE 4.
In vitro susceptibilities of Candida albicans reference strain and clinical isolates to oligopeptide and amino acid antifungals and fluconazole
| Compound | MIC (μg/ml) ofa:
|
||||
|---|---|---|---|---|---|
| ATCC 10261 | Gu4 | Gu5 (CDR1 and/or CDR2) | B3 | B4 (MDR1) | |
| OLys-Leu-Gly (2) | 128 | 256 | 16 | 64 | 32 |
| Leu-FPhe (5) | 64 | 128 | 8 | 64 | 64 |
| Met-Met-FPhe (6) | 128 | 128 | 32 | 128 | 128 |
| Nva-FMDP (7) | 2 | 2 | 0.125 | 4 | 4 |
| Lys-Nva-FMDP (8) | 8 | 4 | 1 | 8 | 8 |
| Nikkomycin (9) | 32 | 16 | 4 | 8 | 8 |
| Histatin 5 | 32 | 32 | 32 | 32 | 32 |
| Asp-γ-hydroxamate (10) | 256 | 256 | 128 | 256 | 256 |
| OLys (11) | 16 | 16 | 4 | 16 | 32 |
| FPhe (12) | 256 | 128 | 128 | 128 | 128 |
| cis-pentacin (13) | 1 | 1 | 4 | 2 | 8 |
| DON (14) | 4 | 4 | 1 | 4 | 4 |
| Azaserine (15) | 16 | 16 | 16 | 32 | 32 |
| Fluconazole | 4 | 8 | 256 | 4 | 64 |
MICs were determined by using RPMI-1640 buffered medium, as described in Materials and Methods. No growth inhibition was observed for oligopeptides 1, 3, and 4 at a concentration of ≤1,024 μg/ml.
Uptake of amino acid and oligopeptide antifungals by MDR yeast cells and intracellular processing of oligopeptides.
The antifungal action of all oligopeptide and amino acid antifungals used in this study is antagonized by proteinogenic oligopeptides or amino acids, respectively, which compete for uptake into the fungal cells via oligopeptide or amino acid permeases (4, 8, 21, 26, 28, 43, 46, 47). To test whether the presence and activity of drug-effluxing transporters affected oligopeptide or amino acid transport, the uptake rates of these compounds, alanine, Ala-Ala, and Ala-Ala-Ala, were determined.
The uptake rates of oligopeptides and amino acids tested were constant for at least 15 to 20 min and then gradually decreased. These properties allowed the determination of initial uptake velocities. The rates were corrected for passive binding of the compounds tested by measuring uptake in the presence of NaN3, accounting for less than 3% of the initial rate of oligopeptide/amino acid uptake.
The results of the determination of the initial uptake rates of compounds 1 to 9 and the model (Ala)2 and (Ala)3 oligopeptides are presented in Table 5. The initial uptake rates of compounds 1 to 9 into AD cells were in each case lower than those of the alanyl oligopeptides, but the ADCDR1 and ADCDR2 cells took up all the oligopeptides tested 1.5- to 3-fold faster than the AD strain. Uptake rates for the ADMDR1 strain were virtually identical to those of the AD strain. A 10-min pretreatment with fluconazole of ADCDR1 and ADCDR2 cells, but not AD or ADMDR1 cells, enhanced by up to 10% the uptake of Ala-Ala and the oligopeptide antifungals OLys-Leu-Gly, Leu-FPhe, and nikkomycin. Histatin 5 was taken up slowly (0.6 ± 0.05 nmol/min/mg dry weight) by all strains, but there was no substantial difference between AD and AD-derived cells. A comparison of Tables 5 and 2 suggests that all cases of increased transport rates of oligopeptide antifungals correlated with enhanced susceptibilities of MDR cells to these compounds. There was no direct correlation between susceptibilities (Table 3) and uptake rates (Table 6) for amino acid antifungals.
TABLE 5.
Initial rates of oligopeptide uptake by yeast mutants
| Oligopeptide | Initial transport rate (nmol/min/mg dry wt) ± SD ofa:
|
|||
|---|---|---|---|---|
| AD | ADCDR1 | ADCDR2 | ADMDR1 | |
| Ala-Ala | 2.55 ± 0.38b | 4.20 ± 0.47b | 3.94 ± 0.50b | 2.61 ± 0.35b |
| 2.58 ± 0.32 | 4.46 ± 0.51 | 4.08 ± 0.44 | 2.59 ± 0.33 | |
| Ala-Ala-Ala | 3.15 ± 0.45 | 5.30 ± 0.57 | 4.74 ± 0.60 | 2.91 ± 0.25 |
| OLys-Leu-Gly (2) | 0.70 ± 0.07b | 1.32 ± 0.06b | 1.26 ± 0.08b | 0.85 ± 0.08b |
| 0.69 ± 0.08 | 1.42 ± 0.09 | 1.31 ± 0.10 | 0.87 ± 0.06 | |
| OLys-Leu-Leu-Gly (3) | 0.51 ± 0.18 | 0.83 ± 0.16 | 0.96 ± 0.12 | 0.65 ± 0.08 |
| OLys-Leu-Leu-Leu-Gly (4) | 0.31 ± 0.12 | 0.49 ± 0.13 | 0.51 ± 0.11 | 0.28 ± 0.08 |
| Leu-FPhe (5) | 0.79 ± 0.08b | 1.82 ± 0.04b | 2.23 ± 0.12b | 0.87 ± 0.11b |
| 0.79 ± 0.07 | 1.95 ± 0.11 | 2.37 ± 0.21 | 0.84 ± 0.09 | |
| Met-Met-FPhe (6) | 0.64 ± 0.09 | 1.11 ± 0.14 | 1.23 ± 0.16 | 0.67 ± 0.10 |
| Nva-FMDP (7) | 0.96 ± 0.09 | 1.86 ± 0.11 | 1.56 ± 0.13 | 1.02 ± 0.10 |
| Lys-Nva-FMDP (8) | 0.79 ± 0.05 | 1.61 ± 0.12 | 1.18 ± 0.09 | 0.81 ± 0.09 |
| Nikkomycin (9) | 0.32 ± 0.02b | 0.98 ± 0.03b | 0.88 ± 0.06b | 0.34 ± 0.04b |
| 0.33 ± 0.05 | 1.08 ± 0.08 | 0.97 ± 0.08 | 0.32 ± 0.05 | |
Initial uptake rates were determined as described in Materials and Methods. Values are the means of three determinations ± the standard deviation (SD).
Cells were pretreated with fluconazole for 10 min.
TABLE 6.
Initial rates of amino acid uptake by yeast mutants
| Amino acid | Initial transport rate (nmol/min/mg dry wt) ± SD ofa:
|
|||
|---|---|---|---|---|
| AD | ADCDR1 | ADCDR2 | ADMDR1 | |
| Ala | 3.51 ± 0.56 | 4.16 ± 0.38 | 4.35 ± 0.41 | 3.33 ± 0.33 |
| Asp-γ-hydroxamate (10) | 1.54 ± 0.16 | 2.25 ± 0.42 | 1.69 ± 0.11 | 1.25 ± 0.13 |
| FPhe (11) | 1.48 ± 0.16 | 1.81 ± 0.18 | 1.44 ± 0.12 | 1.36 ± 0.15 |
| OLys (12) | 1.78 ± 0.11 | 2.57 ± 0.15 | 1.86 ± 0.06 | 1.63 ± 0.07 |
| Cispentacin (13) | 1.63 ± 0.20 | 2.11 ± 0.21 | 1.85 ± 0.16 | 1.67 ± 0.18 |
| DON (14) | 2.0 ± 0.22 | 2.32 ± 0.31 | 2.25 ± 0.16 | 1.98 ± 0.19 |
| Azaserine (15) | 1.75 ± 0.23 | 2.15 ± 0.17 | 1.92 ± 0.22 | 1.70 ± 0.17 |
Conditions of the determination were as described in Table 5. Values are the means of three determinations ± the standard deviation (SD).
Antifungal action of oligopeptides 1 to 8 requires intracellular cleavage to release an active enzyme inhibitor. The rate of this cleavage could affect the antifungal activity of oligopeptides. The cleavage rates obtained for cell extracts prepared from ATCC 9763, AD, ADCDR1, ADCDR2, and ADMDR1 cells were all within the 14 to 25 nmol/min/mg protein range (data not shown). There was no statistically significant difference between the cleavage rates for a particular oligopeptide determined in extracts from cells lacking or overexpressing MDR transporters. These measurements indicated that intracellular cleavage did not limit the activities of oligopeptide antifungals 1 to 8. Nikkomycin 9 and histatin 5 were very slowly cleaved (the rates were 0.4 to 0.5 and 0.1 to 0.15 nmol/min/mg protein, respectively); however, both of these agents act on the targets in an intact form (15, 46). The cleavage rates determined for C. albicans ATCC 10261, Gu4, Gu5, B3, and B4 cells were slightly higher (20 to 32 nmol/min/mg protein for compounds 1 to 8 and 0.3 to 0.8 nmol/min/mg protein for nikkomycin and histatin 5), and there was no significant difference between MDR and sensitive strains.
Changes in intra- and extracellular pH in MDR yeast strains.
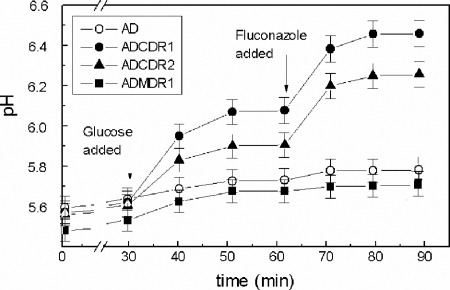
We previously found that yeast mutants overexpressing CDR1 had demonstrated enhanced proton-extruding activity (27). To test if the enhanced rates of uptake of oligopeptides and amino acids in cells overexpressing CDR1 and/or CDR2 might result from modifications of plasma membrane electrochemical potential, the intracellular pH and the rates of glucose-dependent proton efflux were measured. The intracellular pH was monitored using CFDA-SE as a fluorescent probe. The results of this experiment, presented in Fig. 2, show that probe-loaded, glucose-deprived yeast cells maintain an intracellular pH in the range of 5.4 to 5.6. This relatively low intracellular pH, compared with that determined in other studies (7, 20), may be due to the deactivation of the plasma membrane H+-ATPase, Pma1p (2), at a low pH in the absence of glucose (43). The glucose addition, while only slightly increasing the intracellular pH of the AD and ADMDR1 cells, markedly increased the intracellular pHs of ADCDR1 and ADCDR2 cells. Subsequent addition of fluconazole further alkalinized the cytoplasm of ADCDR1 and ADCDR2 cells, but the intracellular pHs of AD and ADMDR1 remained essentially unchanged. The addition of fluconazole without a prior glucose addition did not significantly increase the intracellular pH. Thus, metabolic energy is needed for fluconazole-induced enhancement of intracellular pH.
FIG. 2.
Changes in the intracellular pH of yeast cells caused by glucose and fluconazole addition. Yeast cells suspended in a buffer at pH 4.0 were starved of glucose and loaded with CFDA-SE. Cells were then suspended in buffer, pH 4.5, and treated with glucose and fluconazole. Samples were collected at time intervals, and fluorescence at excitation of 435 and 495 nm (F495 and F435) was measured at 525 nm. F495 and F435 ratios were used to calculate the intracellular pH values.
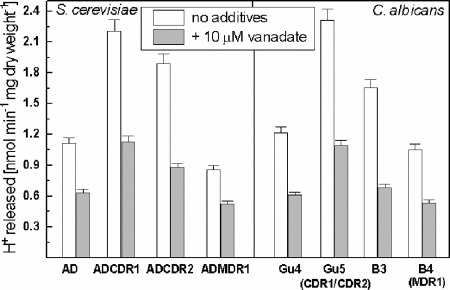
Glucose-dependent proton efflux was monitored in unbuffered cell suspensions adjusted to pH 5.2 ± 0.05 (Fig. 3). The ADCDR1 and ADCDR2 cells extruded protons 1.5 to 2-fold more rapidly than the AD or ADMDR1 cells. These results confirm previous observations made for the CDR1-containing transformants (27) and indicate that overexpression of Cdr1p or Cdr2p causes similar effects. The same phenomenon was observed for C. albicans clinical isolates overexpressing CDR1 and CDR2 but not MDR1. Vanadate, an inhibitor of the in vitro ATPase activity of the fungal Pma1p and ABC transporters (6), partially inhibited proton efflux in all types of cells tested.
FIG. 3.
Initial rates of proton efflux by yeast mutants. The rates were determined by monitoring pH changes of yeast cell suspensions. Cells were transferred from the minimal growth medium to unbuffered water, and pH changes were recorded after glucose addition. Each bar represents the mean of three independent determinations ± the standard deviation (SD).
The differences in glucose-dependent internal pH and glucose-dependent proton pumping between the cells overexpressing the ABC transporters and the control and ADMDR1 strains suggested that overexpression of ABC transporters increased the plasma membrane electrochemical potential. The susceptibilities of yeast cells to the protein synthesis inhibitor hygromycin B are pH dependent, and resistance has been correlated with depolarization of the plasma membrane in pma1 mutants (33). The antifungal activity of hygromycin B was determined for mutant yeast cells used in this study. The hygromycin MICs were as follows: 128, 64, 32, and 128 μg/ml for the AD, ADCDR1, ADCDR2, and ADMDR1 cells, respectively.
Oligopeptides and amino acids are not substrates for Cdr1p and Cdr2p.
Cells deenergized with 2DG and suspended in buffer (pH 4.0) were loaded with an amino acid or oligopeptide and then reenergized by a glucose addition at pH 6.0, thus triggering possible efflux. Cdr1p and Cdr2p functionality in the reenergized cells was confirmed using rhodamine 6G as a probe. During the 60-min loading, the cells accumulated 32 to 67% of each amino acid/oligopeptide. No fluorescamine-positive compounds were detected in spent medium samples collected from the AD, ADCDR1, and ADCDR2 cell suspensions loaded with most of the antifungal compounds tested and reenergized with glucose at pH 6.0. The only exception was the extrusion of cis-pentacin 13 from the ADCDR1 and ADCDR2 cells (5 ± 2 μM of the fluorescamine-cis-pentacin conjugate was detected after 30 min). These results show that all oligopeptides and amino acids tested, except cis-pentacin, are not extruded by Cdr1p and Cdr2p.
DISCUSSION
MDR yeast strains overexpressing CDR1 show strikingly increased susceptibilities to antifungal FMDP oligopeptides (27). The present work has shown that recombinant yeast and clinical C. albicans isolates overexpressing CDR1 and/or CDR2 genes are more susceptible to several different antifungal oligopeptides than their respective parental strains lacking these genes or expressing them at the basal level. On the other hand, yeast strains overexpressing the MDR1 gene showed susceptibilities to all oligopeptides similar to those of their parental strains. The enhanced susceptibilities to numerous oligopeptide antifungals, which represent a broad range of structures and mechanisms of antifungal action, appear to be a feature of MDR yeast strains overexpressing the Cdr1p and Cdr2p ABC drug transporters.
We suggest that the enhanced susceptibility of MDR yeast strains to oligopeptide antifungals is due to enhanced oligopeptide antifungal uptake mediated by oligopeptide permeases that are affected by an increased membrane potential induced by overexpression of the ABC drug efflux pumps Cdr1p and Cdr2p. In yeast strains, the inward transport of oligopeptides containing two to eight amino acid residues is exclusively mediated by active transport systems involving permeases acting as ligand/H+ symporters (16, 34, 39). In contrast, and consistent with our observations, relatively large antifungal polypeptides, like histatin 5, are internalized in an energy-dependent manner via cell wall-located receptors (24), while compounds known as the “cell-penetrating peptides” cross the membrane by free diffusion (17). The presence of the peptide permeases makes the yeast cells susceptible to the action of oligopeptide antifungals targeting intracellular enzymes. Some of these compounds, including nikkomycin 9, attack their targets in an intact form (46). Other oligopeptides, like compounds 1 to 8, are cleaved intracellularly by peptidases, and the released enzyme inhibitor is then able to reach its target (3, 4, 21, 28, 29, 43). The uptake of antifungal oligopeptides, rather than their intracellular cleavage, appears to be the rate-limiting step that determines their potency (25, 26). Thus, the relatively slow accumulation of oligopeptides 1 to 9 by the AD cells, a substantial enhancement of this uptake in AD-derived mutants overexpressing CDR1 or CDR2 and a good match between the enhanced transport rates and the increased antifungal activity, are consistent with the enhanced susceptibilities to oligopeptide antifungals being due to accelerated uptake.
The enhanced rates of glucose-dependent proton efflux and the fluconazole-stimulated alkalinization of the cytoplasm suggested that the overexpression of the ABC transporters Cdr1p and Cdr2p increased the membrane potential associated with the yeast plasma membrane. This concept was confirmed by demonstrating the enhanced susceptibilities of ADCDR1 and ADCDR2 cells to hygromycin B. Consistent with a mechanism in which the additional proton motive force drives proton gradient-dependent oligopeptide permeases, yeast isolate susceptibility to histatin 5 was not increased in cells overexpressing these two ABC transporters. The observed faster accumulation of amino acids by ADCDR1 and ADCDR2 was also most likely due to the increased membrane potential, since these compounds are transported into yeast cells by proton motive force-dependent permeases (5, 19, 38).
The molecular basis of pH changes observed in yeast cells expressing the ABC drug transporters has yet to be explained. They may result from a disturbance of physical properties of the cell membrane or be due to stimulation of proton export mediated by ABC drug transporters or other membrane proteins. The transmembrane proton gradient in yeast cells is normally maintained by the vanadate-sensitive plasma membrane proton pump Pma1p (2, 41). In this study, proton efflux was partially inhibited by vanadate in all cell types. Vanadate inhibits the ATP hydrolytic activity of both ABC transporters and Pma1p in plasma membrane preparations (22), but its effects on these activities in whole cells are not known. Nevertheless, stimulation of Pma1p activity due to the changes in membrane properties cannot be excluded. Another possibility is proton-effluxing activities of Cdr1p and Cdr2p, similar to those of human Mdr1p expressed in S. cerevisiae, found by Fritz et al. (12). This possibility may be supported by the fact that fluconazole efflux in ADCDR1 and ADCDR2 cells gave stronger alkalinization to the cytoplasm and a higher uptake rate of oligopeptide antifungals. It may be possible as well that the observed higher glucose-induced internal alkalinization of ADCDR1 and ADCDR2 cells was due to Cdr1p or Cdr2p proton-effluxing activity, since those cells were prestarved under conditions ensuring deactivation of Pma1p. Further studies are necessary to explore and exploit the possibilities. Nevertheless, we conclude that the phenomenon of the enhanced susceptibilities of yeast cells overexpressing the ABC-type MDR transporters to a broad range of oligopeptidic antifungals results from an accelerated uptake driven by an additional proton motive force created in these cells.
Acknowledgments
This work was supported by the Polish Ministry of Science and Higher Education Grant no. 3 P05F 023 24.
We thank Gillian Payne and Izabela Łącka for their help in some experiments. The generous gift of strains from Joachim Morschhäuser is gratefully acknowledged.
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Albertson, G. D., M. Niimi, and R. D. Cannon. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambesi, A., M. Miranda, V. V. Petrov, and C. W. Slayman. 2000. Biogenesis and function of the yeast plasma membrane H+-ATPase. J. Exp. Biol. 203:155-160. [DOI] [PubMed] [Google Scholar]
- 3.Andruszkiewicz, R., S. Milewski, T. Zieniawa, and E. Borowski. 1990. Anticandidal properties of N3-(4-methoxyfumaroyl)-l-2,3-diaminopropanoic acid oligopeptides. J. Med. Chem. 33:132-135. [DOI] [PubMed] [Google Scholar]
- 4.Basrai, M. A., H. L. Zhang, D. Miller, F. Naider, and J. M. Becker. 1992. Toxicity of oxalysine and oxalysine-containing peptides against Candida albicans: regulation of peptide transport by amino acids. J. Gen. Microbiol. 138:2353-2362. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, S., M. Roy, and A. Datta. 2003. N-acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149:2597-2608. [DOI] [PubMed] [Google Scholar]
- 6.Bowman, B. J., and C. W. Slayman. 1979. The effect of vanadate on the plasma membrane ATPase of Neurospora crassa. J. Biol. Chem. 254:2928-2934. [PubMed] [Google Scholar]
- 7.Bracey, D., C. D. Holyoak, G. Nebe-von Caron, and P. J. Coote. 1998. Determination of the intracellular pH (pHi) of growing cells of Saccharomyces cerevisiae: the effect of reduced-expression of the membrane H+-ATPase. J. Microbiol. Methods 31:113-125. [Google Scholar]
- 8.Capobianco, J. O., D. Zakula, M. N. Coen, and R. C. Goldman. 1993. Anti-Candida activity of cispentacin: the active transport by amino acid permeases and possible mechanisms of action. Biochem. Biophys. Res. Commun. 190:1037-1042. [DOI] [PubMed] [Google Scholar]
- 9.Decottignies, A., A. M. Grant, J. W. Nichols, H. de Wet, D. B. McIntosh, and A. Goffeau. 1998. ATP-ase and multidrug transport activities of overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273:12612-12622. [DOI] [PubMed] [Google Scholar]
- 10.Franz, R., M. Ruhnke, and J. Morschhäuser. 1999. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses 42:453-458. [DOI] [PubMed] [Google Scholar]
- 11.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhäuser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritz, F., E. M. Howard, M. M. Hoffman, and P. D. Roepe. 1999. Evidence for altered ion transport in Saccharomyces cerevisiae overexpressing human MDR1 protein. Biochemistry 38:4214-4226. [DOI] [PubMed] [Google Scholar]
- 13.Gudlaugsson, O., S. Gillespie, K. Lee, J. Van de Berg, J. Hu, S. Messer, L. Herwaldt, M. Pfaller, and D. Diekema. 2003. Attributable mortality of nosocomial candidemia. Clin. Infect. Dis. 37:1172-1177. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, V., A. Kohli, S. Krishnamurthy, N. Puri, S. A. Aalamgeer, S. Panwar, and R. Prasad. 1998. Identification of mutant alleles of CaMDR1, a major facilitator of C. albicans which confers multidrug resistance and its in vitro transcriptional activation. Curr. Genet. 34:192-199. [DOI] [PubMed] [Google Scholar]
- 15.Gyurko, C., U. Lendenmann, E. J. Helmerhorst, R. F. Troxler, and F. G. Oppenheim. 2001. Killing of Candida albicans by Histatin 5: cellular uptake and energy requirement. Antonie van Leeuwenhoek 79:297-309. [DOI] [PubMed] [Google Scholar]
- 16.Hauser, M., V. Narita, A. M. Donhardt, F. Naider, and J. Becker. 2001. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol. Microbiol. 18:105-112. [PubMed] [Google Scholar]
- 17.Henriques, S. T., M. N. Melo, and A. N. B. Canstanho. 2006. Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem. J. 399:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Höfer, M., and P. C. Misra. 1978. Evidence for a proton/sugar symport in the yeast Rhodotorula gracilis (glutinis). Biochem. J. 172:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horák, J. 1997. Yeast nutrient transporters. Biochim. Biophys. Acta 1331:41-79. [DOI] [PubMed] [Google Scholar]
- 20.Imai, T., and T. Ohno. 1995. Measurements of yeast intracellular pH by image processing and the changes it undergoes during growth phase. J. Biotechnol. 38:165-172. [DOI] [PubMed] [Google Scholar]
- 21.Kingsbury, W. D., J. C. Boehm, M. J. Mehta, and S. F. Grappel. 1983. Transport of antimicrobial agents using peptide carrier systems: anticandidal activity of m-fluorophenylalanine-peptide conjugates. J. Med. Chem. 26:1725-1729. [DOI] [PubMed] [Google Scholar]
- 22.Krishnamurthy, S., U. Chatterjee, V. Gupta, R. Prasad, P. Das, P. Snehlata, S. E. Hasmain, and R. Prasad. 1998. Deletion of transmembrane domain 12 of CDR1, a multidrug transporter from Candida albicans, leads to altered drug specificity: expression of a yeast multidrug transporter in baculovirus expression system. Yeast 14:535-550. [DOI] [PubMed] [Google Scholar]
- 23.Loeffler, J., and D. A. Stevens. 2003. Antifungal drug resistance. Clin. Infect. Dis. 36:31-41. [DOI] [PubMed] [Google Scholar]
- 24.Li, X. S., J. N. Sun, K. Okamoto-Shibayama, and M. Edgerton. 2006. Candida albicans cell wall Ssa proteins bind and facilitate import of human salivary histatin 5 required for toxicity. J. Biol. Chem. 281:22453-22463. [DOI] [PubMed] [Google Scholar]
- 25.Lichliter, W. D., F. Naider, and J. M. Becker. 1976. Basis for the design of anticandidal agents from studies of peptide utilization in Candida albicans. Antimicrob. Agents Chemother. 10:483-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy, P. J., D. J. Newman, L. J. Nisbet, and W. D. Kingsbury. 1985. Relative rates of transport of peptidyl drugs by Candida albicans. Antimicrob. Agents Chemother. 28:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milewski, S., F. Mignini, R. Prasad, and E. Borowski. 2001. Unusual susceptibility of a multidrug-resistant yeast strain to peptidic antifungals. Antimicrob. Agents Chemother. 45:223-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milewski, S., R. Andruszkiewicz, L. Kasprzak, J. Mazerski, F. Mignini, and E. Borowski. 1991. Mechanism of action of anticandidal dipeptides containing inhibitors of glucosamine-6-phosphate synthase. Antimicrob. Agents Chemother. 35:36-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moneton, P., P. Sarthou, and F. Le Goffic. 1986. Transport and hydrolysis of peptides in Saccharomyces cerevisiae. J. Gen. Microbiol. 132:2147-2153. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeast, 2nd ed., vol. 22. Approved standard M27-A2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 31.Patterson, T. F. 2005. Advances and challenges in management of invasive mycoses. Lancet 366:1013-1025. [DOI] [PubMed] [Google Scholar]
- 32.Payne, J. W., and T. M. Nisbet. 1981. Continuous monitoring of substrate uptake by microorganisms using fluorescamine: application to peptide transport by Saccharomyces cerevisiae and Streptococcus faecalis. J. Solid-Phase Biochem. 3:447-458. [Google Scholar]
- 33.Perlin, D. S., C. L. Brown, and J. E. Haber. 1988. Membrane potential defect in hygromycin B-resistant pma1 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 263:18118-18122. [PubMed] [Google Scholar]
- 34.Perry, J. R., M. A. Basrai, H.-Y. Steiner, F. Naider, and J. Becker. 1994. Isolation and characterization of a Saccharomyces cerevisiae peptide transporter. Mol. Cell. Biol. 14:104-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad, R., and K. Kapoor. 2005. Multidrug resistance in yeast Candida. Int. Rev. Cytol. 242:215-248. [DOI] [PubMed] [Google Scholar]
- 37.Prasad, R., P. De Wergifosse, and A. Goffeau. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1 conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 38.Regenberg, B., L. During-Olsen, M. C. Kielland-Brandt, and S. Holmberg. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36:317-328. [DOI] [PubMed] [Google Scholar]
- 39.Reuss, O., and J. Morschhäuser. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60:795-812. [DOI] [PubMed] [Google Scholar]
- 40.Sanglard, D., and J. Bille. 2002. Current understanding of the modes of action of and resistance mechanisms to conventional and emerging antifungal agents for treatment of Candida infections, p. 349-383. In R. A. Calderone (ed.), Candida and candidiasis, ASM Press, Washington, DC.
- 41.Serrano, R., M. C. Keilland-Brandt, and G. R. Fink. 1986. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+- ATPases. Nature (London) 319:689-693. [DOI] [PubMed] [Google Scholar]
- 42.Smriti, S. Krishnamurthy, B. L. Dixit, C. M. Gupta, S. Milewski, and R. Prasad. 2002. ABC transporters Cdr1p, Cdr2p and Cdr3p of a human pathogen Candida albicans are general phospholipid translocators. Yeast 19:303-318. [DOI] [PubMed] [Google Scholar]
- 43.Ti, J. S., A. S. Steinfeld, F. Naider, A. Gulumoglu, S. V. Lewis, and J. M. Becker. 1980. Anticandidal activity of pyrimidine-peptide conjugates. J. Med. Chem. 23:913-918. [DOI] [PubMed] [Google Scholar]
- 44.Viudes, A., J. Peman, E. Canton, P. Ubeda, J. L. Lopez-Ribot, and M. Gobernado. 2002. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur. J. Clin. Microbiol. Infect. Dis. 21:767-774. [DOI] [PubMed] [Google Scholar]
- 45.Wakieć, R., R. Prasad, J. Morschhäuser, F. Barchiesi, E. Borowski, and S. Milewski. 2007. Voriconazole and multidrug resistance in Candida albicans. Mycoses 50:109-115. [DOI] [PubMed] [Google Scholar]
- 46.Yadan, J. C., M. Gonneau, P. Sarthou, and F. Le Goffic. 1984. Sensitivity to nikkomycin Z in Candida albicans: role of peptide permeases. J. Gen. Microbiol. 160:884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng, H., H. L. Zhang, J. M. Becker, F. Naider, and W. R. Farkas. 1992. The lysine analog l-oxalysine is an inhibitor of RNA synthesis. Int. J. Biochem. 24:145-149. [DOI] [PubMed] [Google Scholar]