Abstract
Conventional myosin II plays a fundamental role in the process of cytokinesis where, in the form of bipolar thick filaments, it is thought to be the molecular motor that generates the force necessary to divide the cell. In Dictyostelium, the formation of thick filaments is regulated by the phosphorylation of three threonine residues in the tail region of the myosin heavy chain. We report here on the effects of this regulation on the localization of myosin in live cells undergoing cytokinesis. We imaged fusion proteins of the green-fluorescent protein with wild-type myosin and with myosins where the three critical threonines had been changed to either alanine or aspartic acid. We provide evidence that thick filament formation is required for the accumulation of myosin in the cleavage furrow and that if thick filaments are overproduced, this accumulation is markedly enhanced. This suggests that myosin localization in dividing cells is regulated by myosin heavy chain phosphorylation.
INTRODUCTION
The process of cytokinesis is undoubtedly the most dramatic morphological event in the cell cycle. As such, it has been the object of much study over the past decades (Wolpert, 1960; Satterwhite and Pollard, 1992). Although it is known that the actin-based motor protein myosin II is important for this process, little is understood about how myosin becomes organized in the dividing cell and how it generates the morphological changes so characteristic of cytokinesis.
In most animal cells, cell division is thought to be powered by the constriction of a contractile ring in the cleavage furrow (Schroeder, 1968). This transient organelle has been studied extensively at the ultrastructural level and contains a thin (100–200 nm) band of actin filaments (Schroeder, 1968). The actin filaments are spaced ∼15 nm apart and are thought to adhere to the overlying membrane by interacting with accessory proteins in that location.
Myosin II (referred to as myosin in this paper) is present in the contractile ring (Fujiwara and Pollard, 1976; Yumura and Fukui, 1985; Maupin and Pollard, 1986). This myosin is unique among actin-based motors in that the carboxy-terminal domain of the heavy chain forms a long α-helical coiled coil that allows nonmuscle myosin (Clarke and Spudich, 1974), like muscle myosin (Huxley, 1963), to form bipolar thick filaments. Dictyostelium filaments are thought to contain ∼70 monomers that are oriented with their force-generating heads pointing in opposite directions at the ends of the filament (Clarke and Spudich, 1974; Mahajan and Pardee, 1996). In the contractile ring, myosin filaments are thought to bind to neighboring actin filaments and pull them together so that the ring shrinks as myosin moves along the actin.
The role of myosin in cytokinesis has been investigated by both genetic and biochemical methods in a number of organisms. Injection of antimyosin antibodies into starfish eggs inhibits cytokinesis (Mabuchi and Okuno, 1977; Kiehart et al., 1982). In Drosophila, mutations of the myosin-regulatory light chain result in defective cytokinesis (Karess et al., 1991). In Dictyostelium, myosin function has been disrupted genetically either by replacing the single myosin heavy chain gene with a truncated form or by removing it entirely (De Lozanne and Spudich, 1987; Manstein et al., 1989). Alternatively, myosin function can be reduced significantly by expressing antisense RNA or by expressing the carboxy-terminal rod domain of the protein (Knecht and Loomis, 1987; Burns et al., 1995). Regardless of the method of disrupting myosin function, the phenotype is similar: cells lacking myosin cannot undergo cytokinesis when grown in suspension culture. They can, however, divide on a surface by an event that morphologically resembles normal cytokinesis (Fukui et al., 1990; Neujahr et al., 1997; Zang et al., 1997). Wild-type behavior can be restored in the myosin null cell by expression of the myosin heavy chain gene from an extrachromosomal plasmid (Egelhoff et al., 1990).
Evidence that myosin plays a role in cytokinesis only when it is in bipolar thick filaments has come from studies of mutant myosins that either cannot form thick filaments or form them constitutively. In Dictyostelium, thick filament formation is regulated by the phosphorylation of three threonine residues in the tail region of the protein (Vaillancourt et al., 1988; Luck-Vielmetter et al., 1990; Egelhoff et al., 1993). When these three threonines are phosphorylated, filament formation in vitro is inhibited (Kuczmarski and Spudich, 1980; Pasternak et al., 1989; Ravid and Spudich, 1989; Egelhoff et al., 1993). The role of thick filament formation in vivo has been studied by making mutant myosins where each of the three threonines has been converted to either alanine (3XALA-myosin) or aspartic acid (3XASP-myosin). The 3XALA-myosin cannot be phosphorylated on those residues, and it constitutively forms thick filaments that are found almost exclusively in a Triton-insoluble cytoskeletal pellet (Egelhoff et al., 1993). Conversely, 3XASP-myosin mimics the phosphorylated state, cannot form bipolar thick filaments in vitro, and fails to assemble into a Triton-insoluble pellet (Egelhoff et al., 1993). Like cells with no myosin, cells expressing 3XASP-myosin cannot undergo cytokinesis when grown in suspension. This strongly suggests that thick filament formation is required for proper cytokinesis and that the regulation of the phosphorylation of these three threonine residues is crucial for proper localization or function of myosin in the contractile ring.
This requirement for myosin to be in the form of thick filaments to support cytokinesis could arise for two reasons. First, it is possible that monomeric myosin does not localize properly to the contractile ring and hence cannot power its contraction. Alternatively, monomeric myosin could localize correctly but fail to act as an effective motor because it cannot cross-link adjacent actin filaments to cause the ring to contract. We address this question directly by visualizing the location of various myosins in live cells undergoing cytokinesis.
To do this we have fused the green-fluorescent protein (GFP)1 to the amino terminus of the wild-type Dictyostelium myosin heavy chain (GFP-myosin), to 3XALA-myosin (GFP-3XALA-myosin), and to 3XASP-myosin (GFP-3XASP-myosin). GFP is a protein from Aequorea victoria that forms a fluorophore by an internal cyclization and oxidation reaction (Cody et al., 1993). By expressing these constructs in a Dictyostelium strain lacking endogenous myosin, we ensure that every myosin molecule in the cell is labeled with the fluorescent probe.
We found that GFP-myosin accumulates in the cleavage furrow during cytokinesis just as the cleavage furrow forms and that the rates of furrow constriction and cell migration are strictly linear. In cells expressing GFP-3XASP-myosin, a myosin that cannot form thick filaments, the myosin accumulation in the furrow is drastically reduced such that virtually no accumulation is apparent. Conversely, in cells expressing GFP-3XALA-myosin, a myosin that constitutively assembles into filaments, the accumulation in the furrow is dramatically increased relative to wild-type myosin.
These data suggest that the spatial organization of myosin during cytokinesis is controlled by heavy chain phosphorylation. Furthermore, the data provide evidence that thick filament formation is required to localize myosin to the cleavage furrow. Thus, it is likely that the formation of thick filaments is important not only for the productive interaction with neighboring actin filaments to constrict the dividing cell but also for the localization of myosin to the equatorial region of the cell before active furrowing occurs.
MATERIALS AND METHODS
Cell Culture
Dictyostelium cells were cultured on plastic Petri dishes or in suspension in HL-5 medium supplemented with 10% FM (Life Technologies, Paisley, United Kingdom), 60 units/ml penicillin, 60 μg/ml streptomycin, and 5 μg/ml G418 (Sussman, 1987). The cells used for all the studies of wild-type myosin were myosin heavy chain null cells (strain HS1) expressing a fusion of wild-type GFP to the amino terminus of the Dictyostelium myosin heavy chain (GFP-myosin). This construct was inserted into a high-copy extrachromosomal vector, and the expression was driven by the actin-15 promoter. The initial characterization of this construct and these cells has been reported elsewhere (Moores et al., 1996).
The GFP-3XASP-myosin heavy chain protein is identical with the above wild-type GFP-myosin with the exception that T1823, T1833, and T2029 are all mutated to aspartic acid residues (Egelhoff et al., 1993). The GFP-3XALA myosin heavy chain has these threonines mutated to alanines. These constructs were also expressed in the HS1 myosin null strain.
Cells were maintained with repeated passaging and replaced every 6–8 wk from fresh stocks of cells frozen with 5% dimethyl sulfoxide in the above medium.
Protein Purification
GFP-myosin was purified from stably transformed Dictyostelium cells grown in suspension culture. The purification protocol was identical with that previously described for wild-type myosin (Ruppel et al., 1994). Briefly, a whole cell lysate was made in the presence of 50 mM NaCl. Under these conditions, myosin forms thick filaments that interact with actin and can be separated from soluble proteins by centrifugation. The pellet is resuspended in the presence of ATP and 300 mM NaCl, thus solubilizing the myosin which is separated from most of the actin and other insoluble materials by centrifugation. Myosin filaments are then reformed by an overnight dialysis of the supernatant back into 50 mM NaCl, and the resultant filaments are collected by another centrifugation step.
Western Blot
Duplicate lanes of whole cell lysates were chromatographed using 7.5% SDS-PAGE. One lane was processed for Coomassie staining, and the others were electroblotted to nitrocellulose. The filters were probed separately with a primary monoclonal antibody to the myosin heavy chain (a kind gift of Dr. Thomas Pollard) and a polyclonal antibody to GFP (Clontech Laboratories, Palo Alto, CA). These were followed by horseradish peroxidase conjugated secondary antibodies (Bio-Rad Laboratories, Richmond, CA). The signal was visualized by enhanced chemiluminescence (Amersham Life Sciences, Arlington Heights, IL).
Fluorimetry
Fluorometric analysis of purified GFP-myosin was carried out with an Aminco-Bowman series 2 spectrofluorometer outfitted with a 150-W xenon lamp (SLM Aminco, Urbana, IL). Recombinant GFP was obtained as a purified protein (Clontech). All proteins were assayed in an 0.8-ml cuvette at a concentration of 33 μg/ml, as assayed by the Bradford method, using rabbit skeletal myosin as a standard (Bradford, 1976). The fluorophore was excited with 488 nm monochromatized light, and the emission was integrated over a band pass from 495 to 520 nm.
Immunofluorescence
Cells were grown on a clean No. 1 glass coverslip, fixed in −8°C methanol, and processed for immunofluorescence. After fixation and rinsing, they were incubated with a 1:2000 dilution of a primary polyclonal antibody raised against the Dictyostelium myosin heavy chain. The primary antibody was visualized using a rhodamine-conjugated donkey anti-rabbit secondary antibody (Jackson Immunoresearch Laboratories, West Grove, PA).
Imaging and Data Analysis
Dictyostelium amebae stably expressing GFP-myosin were plated on chambered No. 1 coverglasses (Nunc, Naperville, IL). They were submerged in imaging buffer containing 20 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.8, 2 mM MgSO4, and 0.2 mM CaCl2. The cells were imaged with a Zeiss Axiovert 100 microscope outfitted with a 63/1.4 Planapochromat objective and a 1.6× Optovar lens (Zeiss, Thornwood, NY). The GFP was illuminated using a High Q fluorescein isothiocyanate filter set (Chroma Technologies, Brattleboro, VT).
Images were collected on a cooled charge-coupled device camera configured on the basement port of the microscope. The charge-coupled device chip was a thinned, back illuminated device cooled to −30°C and used at a readout rate of 100 kHz (TK512D chip, Princeton Instruments, Trenton, NJ).
The cells were illuminated with a 100-W Atto-arc mercury lamp (Atto Instruments, Rockville, MD) attenuated to 50% output. Illumination was temporally controlled with a Uniblitz shutter (Vincent Associates, Rochester, NY).
Each image was 30 ms in duration. Images were collected every 5 s using Image1/MetaMorph software (Universal Imaging Corp., West Chester, PA) which controlled the shutter and the camera. All image analysis was carried out using Image1/MetaMorph and StatView (Abacus Concepts, Berkeley, CA).
To measure relative fluorescent intensity, regions of interest were drawn (using a freehand tool in MetaMorph) on the cells based on the shape of the cells. The extent of the furrow was defined simply by shape (see Figure 3). The jagged nature of the lines on this image reflects the fact that full pixel intensities were counted and the lines did not bisect individual pixels. These intensity values measure the amount of fluorophore imaged onto those pixels and do not evaluate the absolute concentration of fluorophore because this would require a measure of cell thickness. However, given that the thickness and shape of all the cells imaged were similar, the pixel intensity gives a good measure of relative concentration.
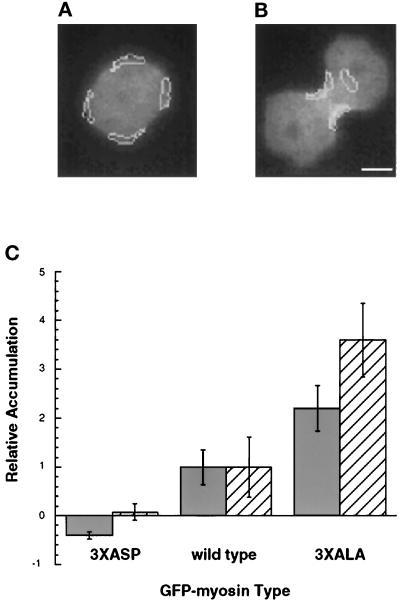
Figure 3.
Quantitation of myosin accumulation. The average fluorescence intensity within the indicated regions was calculated. (A) Four regions, shown by the white outline, were drawn on early mitosis cells, and their average intensity was taken as a measure of cortical myosin concentration. (B) In furrowing cells, the average intensity of two regions defining the furrow was taken as a measure of cortical furrow myosin concentration, and a region of perinuclear cytoplasm was used as a measure of cytoplasmic myosin concentration. Bar, 5 μm. (C) The amount of accumulation is shown normalized to those levels seen in GFP-myosin cells. Solid bars indicate the relative increase in myosin concentration in the furrow versus the adjacent cytoplasm as shown in Figure 4B. Striped bars indicate the relative increase in myosin concentration in the furrow (B) versus the cortex of the same cell in early mitosis as shown in Figure 4A. Bars, standard deviations.
RESULTS
Dictyostelium cells lacking the myosin heavy chain gene fail to undergo cytokinesis when grown in suspension culture and fail to complete the Dictyostelium developmental cycle (Manstein et al., 1989). Previous studies showed that GFP-myosin expression in these cells complements the null phenotype for both development and cytokinesis (Moores et al., 1996). In addition, in vitro biochemical studies showed that purified GFP-myosin has solution ATPase and motility properties that are similar to those of nontagged myosin (Moores et al., 1996). These results suggest that the addition of GFP to the amino terminus of the myosin heavy chain does not compromise myosin function in vitro or in vivo.
GFP-Myosin Is a Reliable Marker of Myosin Localization
To use GFP-myosin as a reliable quantitative marker of where myosin is localized in the cell, it is crucial to determine that the fluorescence imaged in the cell is caused by GFP and that the GFP in the cell is in the form of an intact GFP-myosin fusion protein.
To address this question, whole cell lysates from GFP-myosin-containing cells were analyzed by SDS-PAGE (Figure 1A). Although the lysate is a complex mixture of many proteins as shown by Coomassie staining (Figure 1, lane 1), only one band reacts with an antibody against GFP (Figure 1, lane 3). This band is of the same electrophoretic mobility as the major high molecular weight band that reacts with an antibody against the myosin heavy chain (Figure 1, lane 2). Whole cell lysates from wild-type cells or from myosin heavy chain null cells show an equally complex Coomassie pattern on SDS-PAGE but do not react with the anti-GFP antibody (our unpublished observations). This suggests that in intact cells, all of the GFP is associated with an intact GFP-myosin fusion protein.
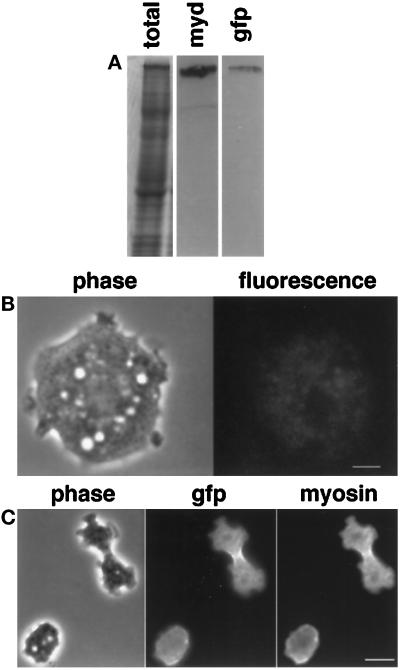
Figure 1.
(A) Expression of GFP-myosin in myosin-null Dictyostelium cells. Whole-cell lysates were run on SDS-PAGE. Lane 1, Coomassie stained; lane 2, antimyosin antibody; lane 3, anti-GFP antibody. A single band was identified by both antibodies, suggesting that the only GFP-labeled protein in the cell was myosin. The lighter band in the myosin blot is an antigen recognized by the secondary antibody and is not a myosin. (B) Minimal background fluorescence is seen in the myosin-null parent cell line. Live myosin-null cells were imaged with the same exposures used to image the GFP-myosin-containing cells. The cell shown in phase on the left has only minimal fluorescence as shown on the right. Bar, 3 μm. (C) GFP and myosin colocalize in vivo. A dividing GFP-myosin cell was fixed and stained with an antibody against myosin. The immunofluorescence localization of myosin mirrors that of the GFP fluorescence, implying that fluorescence localization is a reasonable marker of GFP-myosin localization. Bar, 10 μm.
However, in intact cells, fluorescence can arise from sources other than GFP. When the parent cell line that was used to make the GFP-myosin-expressing cells was imaged, a low intensity fluorescent signal was seen when the cell was imaged with the same exposure and light intensity that was used with GFP-myosin cells (Figure 1B). This background fluorescence was found mainly in vesicular structures. The intensity of this fluorescence was ∼10% of that seen in GFP-myosin cells.
Further evidence that fluorescence from sources other than GFP-myosin is minimal came from comparing the location of myosin and the GFP fluorescence in fixed cells (Figure 1C). A cell in the act of cytokinesis is shown in the phase image (Figure 1C, left). The GFP signal is highest in the cleavage furrow, but the fluorophore is obviously present throughout the cell (Figure 1C, center). Myosin localization, as detected with an antibody against Dictyostelium myosin, mirrors that of GFP (Figure 1C, right).
These data suggest that the vast majority of the fluorescence in these imaging experiments is attributable to intact GFP-myosin. Furthermore, the colocalization experiment shows that the GFP is active as a fluorophore on myosin throughout the cell and that the fluorescence localization is an accurate marker of GFP-myosin localization.
However, to use this protein-fluorophore as a quantitative marker of GFP-myosin localization, it is also necessary to show that the fluorescence intensity of the protein does not change when myosin interacts with other proteins in the cell. Although it is impossible at this time to test combinatorially every myosin-binding protein, we do know that myosin forms bipolar thick filaments and binds to actin.
To examine the effect of these protein-protein interactions on the fluorescence efficiency of the GFP-myosin, we purified GFP-myosin and asked whether the fluorescence changed when GFP-myosin was assembled into bipolar thick filaments or when it was bound to actin.
Thick filaments of Dictyostelium myosin form when the NaCl concentration is between 50 and 100 mM NaCl. They disassemble into soluble myosin molecules at 0 and 200 mM NaCl (Kuczmarski and Spudich, 1980; Kuczmarski et al., 1987). GFP-myosin formed thick filaments with the same dependence on ionic conditions as untagged Dictyostelium myosin (our unpublished observations). The GFP fluorescence intensity was not affected by filament formation. At 0 and 200 mM NaCl, when GFP-myosin was primarily soluble, the fluorescence intensity of 33 μg/ml myosin was 57.3 ± 7.7 and 33.5 ± 1.3 U, respectively (mean ± SD). At 50 and 100 mM NaCl, when GFP-myosin was in the form of filaments, the fluorescence intensity at the same myosin concentration was 53.6 ± 2.4 and 45.4 ± 6.8 U, respectively.
GFP-myosin bound to actin in a rigor state in the absence of ATP (our unpublished observations). When 33 μg/ml monomeric GFP-myosin were bound to actin in this way at 200 mM NaCl, the fluorescence intensity was 30.4 ± 3.1 U. In the absence of actin, monomeric myosin had a fluorescence intensity of 31.0 ± 1.4. Similarly, filamentous myosin also bound to actin in a rigor state in the absence of ATP and that binding did not affect fluorescence. At 50 mM NaCl, 33 μg/ml filamentous GFP-myosin bound to actin had a fluorescence intensity of 31.1 ± 1.4 U. In the absence of actin, GFP-myosin under these conditions had a fluorescence intensity of 30.4 ± 3.1.
GFP-Myosin Localizes to the Cleavage Furrow in Live Cells
Dictyostelium amebae expressing GFP fused to the wild-type myosin heavy chain (GFP-myosin) as their only myosin grow both on a surface and in suspension culture (Moores et al., 1996). For imaging, cells were transferred onto clean chambered coverglasses. We imaged 57 GFP-myosin-expressing cells during cytokinesis and saw no evidence of phototoxicity according to the imaging protocol outlined in MATERIALS AND METHODS. We also did not detect any fluorophore photobleaching or photoactivation. Furthermore, the total amount of fluorescence did not change during the course of imaging. If longer or more frequent exposures were used, phototoxicity was seen as evidenced by the cells rounding up and the GFP-myosin forming a serpentine filamentous aggregate.
A representative GFP-myosin cell going through cytokinesis is shown in Figure 2, WT. In all mitotic and interphase cells, fluorescence was present throughout the cytoplasm but was somewhat concentrated at the cortex (Figure 2, WT, 0 s). Dictyostelium cells do not undergo nuclear envelope breakdown during mitosis (Moens, 1976). This was confirmed in these cells by the observation that GFP-myosin was excluded from the nuclei, which therefore could be identified as regions of low fluorescence. A dividing nucleus can be seen in Figure 2, WT (0 and 35 s).
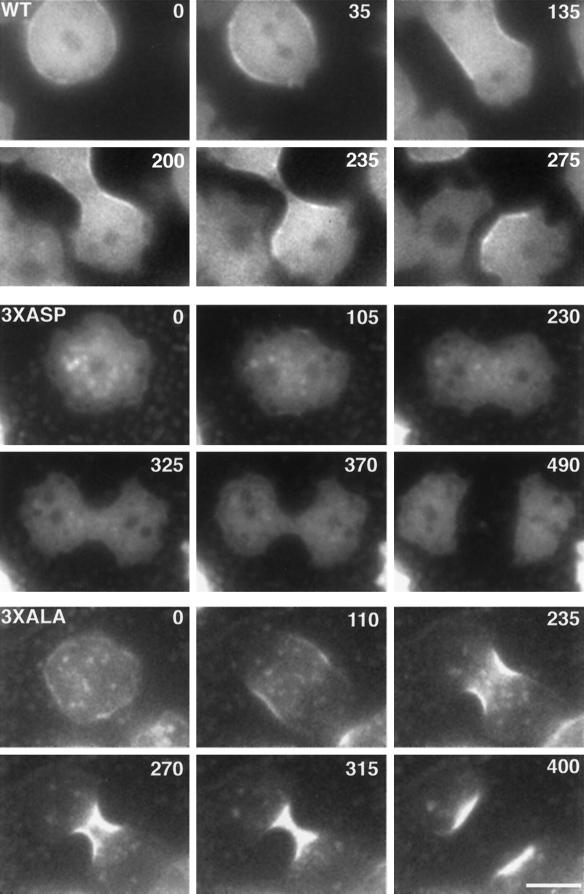
Figure 2.
GFP-myosin-expressing cells undergoing cell division. The elapsed time for each cell is shown in seconds. WT, a cell expressing GFP-myosin. Note that myosin accumulates in the central region of the cell early in mitosis before the cleavage furrow forms (35 s). Myosin is found in the contracting furrow as a band underneath the plasma membrane (135, 200, and 235 s). After cell separation, myosin is localized to the posterior ends of the daughter cells, which migrate away from one another (275 s). 3XASP, a cell expressing GFP-3XASP-myosin. Note that little myosin accumulates in the cleavage furrow (325 and 370 s). 3XALA, a cell expressing GFP-3XALA-myosin. GFP-3XALA-myosin is depleted from the distal ends of the cell (270 and 315 s) and accumulates to a dramatic degree in the cleavage furrow (315 s). Bar, 5 μm.
The accumulation of myosin in the region of the cell destined to furrow occurred just after the nuclei were beginning to separate (Figure 2, WT, 35 s). At this time, no cleavage furrow was morphologically apparent. However, one could detect the asymmetrical distribution of pseudopodia, which were present at the ends of the cell, but not in the central region where myosin was accumulating. Changes in the shape of the dividing cell were evident from the moment myosin accumulation was noticed (Figure 2, WT, 135 s).
By the end of telophase, a cleavage furrow was clearly seen with myosin accumulating as a band of fluorescence at the cell edge in the furrow (Figure 2, WT, 135 s). No myosin accumulation was seen at the pseudopod-rich ends of the dividing cell. As the cleavage furrow progressed, the intensity of myosin accumulation did not change (Figure 2, WT, 200 s and 235 s). After cell division, myosin accumulations were found at the posterior ends of each migrating daughter cell (Figure 2, WT 275 s). Both daughter cells continued to migrate away from each other along an axis perpendicular to the plane of cell division. As in the dividing cell, daughter cells showed pseudopodial activity only in the region that showed no myosin accumulation.
Although the specific cells shown in Figure 2 had different rates of cell division, the population mean values for cell division were not significantly different for the different cell types, as discussed below and shown in Table 1.
Table 1.
Cell division kinetics
| GFP-myosin expressed | N | Rates
(μm/s, mean ±
SD)
|
||
|---|---|---|---|---|
| Furrow constriction | Cell migration | Nuclear movement | ||
| Wild-type | 57 | 0.043 ± 0.011 | 0.043 ± 0.021 | 0.040 ± 0.017 |
| 3XASP | 13 | 0.043 ± 0.013 | 0.031 ± 0.011 | 0.027 ± 0.09 |
| 3XALA | 21 | 0.055 ± 0.010 | ND | ND |
Accumulation of GFP-Myosin in the Cleavage Furrow Is Regulated by Heavy Chain Phosphorylation
The above observations suggest that the accumulation of myosin in the cleavage furrow is closely linked to furrow contraction. We would very much like to understand the molecular basis of the spatial and temporal control of the formation of this actin-myosin-containing contractile ring. To pursue this issue, we took advantage of our earlier work regarding three threonine residues in the tail of myosin that are critical for assembly of myosin into thick filaments.
There are three critical threonine residues in the tail portion of myosin (residues T1823, T1833, and T2029), which are phosphorylated by a myosin heavy chain kinase both in vivo and in vitro. Phosphorylation of those three threonines inhibits myosin thick filament formation. When the three threonines were changed to alanine residues (3XALA-myosin), the myosin constitutively formed thick filaments (Egelhoff et al., 1993). Conversely, when those residues were changed to aspartate residues (3XASP-myosin), which mimic phosphorylated threonines, the myosin was unable to form thick filaments. We examined the dynamics of GFP fused to the two mutant myosins, 3XALA-myosin and 3XASP-myosin. These mutant myosins were expressed in cells lacking endogenous myosin. The expression levels of GFP-3XALA-myosin and GFP-3XASP-myosin, like that of GFP-myosin, were similar to the expression level of wild-type myosin. GFP-3XASP-myosin-expressing cells failed to divide in suspension, as had been reported for 3XASP-myosinexpressing cells (Egelhoff et al., 1993). Conversely, GFP-3XALA-myosin-expressing cells did divide in suspension, as had been previously observed for 3XALA-myosin-expressing cells (Egelhoff et al., 1993).
On an adhesive surface, both the GFP-3XALA-myosin- and GFP-3XASP-myosin-expressing cells underwent cell division, and we were able to image the dynamics of the myosin localization in these cells. GFP-3XASP-myosin was found almost exclusively in the cytoplasm with very little evidence of cortical localization (Figure 2, 3XASP). During cell division, GFP-3XASP-myosin remained diffusely located throughout the cytoplasm and no accumulations were noted.
GFP-3XALA-myosin was found both in the cytoplasm and in the cortex (Figure 2, 3XALA). Punctate accumulations of GFP-3XALA-myosin were seen throughout the cytoplasm at all stages of the cell cycle. During cell division, as GFP-3XALA-myosin localized to the cleavage furrow, it was simultaneously lost from the polar regions of the cell. This was in contrast to GFP-myosin which did not show appreciable distal cytoplasmic loss as it accumulated in the furrow (Figure 2, compare WT and 3XALA). The timing of accumulation of GFP-3XALA-myosin in the furrow was similar to that of GFP-myosin (Figure 2), although the amount of accumulation was dramatically increased.
The amount of myosin accumulation was quantified by comparing the average pixel intensity of all pixels within certain regions of each cell (Figure 3). These regions, shown by the white outlines in Figure 3, A and B, were drawn on the cell images based on the cell shape. The ragged nature of these outlines reflects the boundaries of the pixels themselves. The pixel intensities within these regions were averaged, and that number was used as a measure of the amount of GFP-labeled myosin in that region.
There was variability in the expression level of GFP-labeled proteins from cell to cell. However, by measuring the ratio of fluorescence intensity of different regions of the same cell, we could pool data from many cells in the population. Figure 3, A and B, shows a cell expressing GFP-myosin in early mitosis (Figure 3A) and during furrowing (Figure 3B). Two ratio measurements were used. The first ratio was between the average intensity in the furrow and the intensity in the adjacent cytoplasm during furrowing (Figure 3B). This ratio reflects the enrichment of GFP-myosin in the furrow compared with the cytoplasm at one time. The second ratio was between the average intensity in the furrow (regions in Figure 3B) and the cortex of the same cell at an earlier time point just as mitosis was occurring (Figure 3A). This ratio reflects the enrichment of GFP-myosin in the furrow compared with the cortex of the same cell earlier in time.
We were unable to make any statements about the absolute increases in concentration in GFP-myosin in the furrow because the fluorescence intensity reflects both the concentration of the fluorophore and the cellular volume under that pixel. However, given that cells expressing any of the three GFP constructs have similar shapes, we could make accurate relative measurements.
Hence, the ratio accumulations in the furrow of GFP-3XASP-myosin and GFP-3XALA-myosin was normalized to the ratio accumulations in the furrow of GFP-myosin. The normalized value of 1 represents the ratio accumulations of GFP-myosin in the furrow compared with the cytoplasm (Figure 3C, solid bars) and compared with the cortex of the same cell in early mitosis (Figure 3C, striped bars).
When the ratio of myosin accumulation in the furrow versus the cortex of the same cell in early mitosis was quantified, we found that the increase in GFP-3XASP-myosin fluorescence intensity was only 5% of that seen with GFP-myosin (Figure 3C, striped bars). Conversely, GFP-3XALA-myosin had an increase in accumulation 3.5-fold greater than that seen with GFP-myosin.
We also compared the ratio of myosin accumulation in the furrow versus an area of adjacent perinuclear cytoplasm at the same time (Figure 3, B and C, solid bars). GFP-3XASP-myosin amounts were apparently less in the furrow than in the cytoplasm, and this is reflected in the negative value for GFP-3XASP-myosin (Figure 3C, solid bars). GFP-3XALA-myosin had an increase in accumulation 2.2-fold greater than that seen with GFP-myosin (Figure 3C, solid bars).
Rates of Cleavage Furrow Constriction and Cell Migration Are Constant
The fluorescent signal from GFP-myosin allows one to accurately image the cell edges and thus determine three distances over time: the width of the cleavage furrow, the distance from one edge of the cell to the other along the long axis of the dividing cell, and the distance between the nuclei (Figure 4A). All cells were analyzed in this way, and a representative analysis of a GFP-myosin cell is shown in Figure 4B. As mentioned above, the nuclei could be recognized by exclusion of the GFP-myosin protein (see Figures 1C and 2).
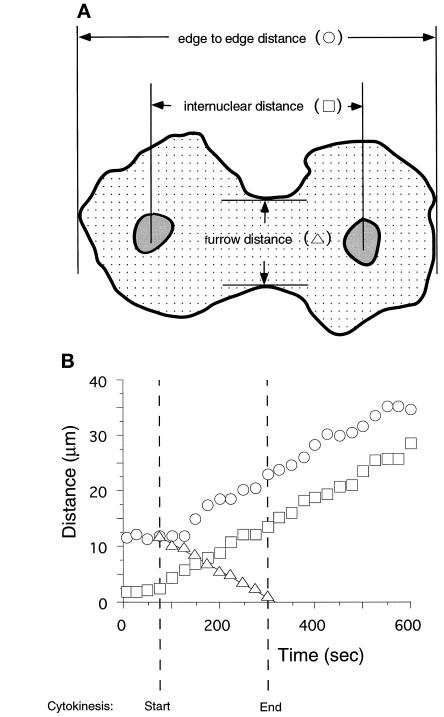
Figure 4.
(A) Determination of the rates of cleavage, nuclear movement, and cell migration. The distances indicated were measured at various times before, during, and after cytokinesis. (B) All three distances changed over time in a strictly linear manner. Note that the rates of cell migration (circles) and nuclear movement (squares) were established during cytokinesis (between the hatched time lines) and maintained after the cells separated.
The width of the cleavage furrow decreased with time in a strictly linear manner (Figure 4, triangles). This was seen not only in GFP-myosin cells but also with GFP-3XASP-myosin and GFP-3XALA-myosin cells. The average R2 value of the regression of these data onto a single line was 0.98 (SD 0.01, n = 57) for GFP-myosin cells, 0.98 (SD 0.01, n = 13) for GFP-3XASP-myosin cells, and 0.99 (SD 0.01, n = 21) for GFP-3XALA-myosin cells. The rates of furrow constriction and of cell migration and nuclear movement for GFP-myosin- and for GFP-3XASP-myosin-expressing cells are shown in Table 1. We could not determine a rate of nuclear movement or of cell migration for GFP-3XALA-myosin-expressing cells because the fluorescent signal was lost from all but the central region of the cell, making it difficult to measure these distances (Figure 2, 3XALA). However, all of the other rates for cells expressing either of the mutated myosins are statistically identical with each other and with those cells expressing wild-type myosin.
Rates of Cell Migration Are Established during Cytokinesis
The rates of nuclear movement and cell migration that were measured for GFP-myosin cells during cytokinesis remained unchanged after the daughter cells separated completely (Figure 4B). Note that the nuclei began moving apart just as the cleavage furrow formed, as shown by the boxes (Figure 4) at the start of cytokinesis. This rate of nuclear movement was maintained throughout cytokinesis and was similar to the rate of cell migration (Figure 4, boxes).
GFP-myosin cells did not elongate until the cleavage furrow was formed (Figure 4B, circles). As the cleavage furrow formed (Figure 4B, triangles), the cell began elongating, and this rate of migration during cytokinesis was maintained after cell separation for the duration of the imaging session (Figure 4B, circles). This pattern was seen in all 22 cells for which we have the data. Interestingly, this period of linear cell migration was accompanied in 21 of 22 cells by a localization of myosin in the posterior end of the daughter cell as shown in Figure 2, WT.
DISCUSSION
Myosin Localizes to the Central Region of the Cell before Cleavage Furrow Formation
It has been suggested that cytokinesis occurs in two sequential steps, each with its own separate mechanism (Dan, 1988). According to this model, the first step uses an astral mechanism, and the second step uses a contractile ring mechanism. The first step is active very early to create the early furrow. It uses astral microtubule arrays radiating from opposite poles to generate an anisotropic deformation of the spherical surface of the cell. This would then allow the recruitment to the central cortex of the molecules involved in the formation of the contractile ring. As these molecules accumulate in the furrow, the second step would allow the furrow to constrict using a contractile ring mechanism. One crucial prediction of this two step model is that the accumulation of the components of the contractile ring, including myosin, in the central region of the cell would not occur until after the early furrow had formed (Inoue, 1990).
We addressed this specific point by examining the timing of myosin accumulation in relation to the shape changes of the cell and found that myosin localizes to the central region of the cell before the morphological changes that define the furrow. Our results are consistent with other reports that myosin accumulates in the central region of the cell before a morphological furrow is apparent (Fujiwara and Pollard, 1976; Mittal et al., 1987; Kitanishi-Yumura and Fukui, 1989; Mabuchi, 1994). Hence, it is likely that the shape changes that occur both early and later in cell division can be explained by contraction of an actomyosin contractile ring in the cleavage furrow. Our observations do not support Dan’s two-step model for cytokinesis but rather are consistent with Schroeder’s contractile ring hypothesis whereby all the morphological changes that occur in going from a spherical cell to a furrowed one to two daughter cells can be explained by the contraction of a myosin containing contractile ring (Schroeder, 1968). Further studies on the intimate relationship between the motor function of myosin and the creation and function of the cleavage furrow are being carried out with various myosin mutants in Dictyostelium (see, for example, Zang et al., 1997).
Heavy Chain Phosphorylation Regulates Myosin Localization in the Dividing Cell
Myosin II from either muscle or nonmuscle cells can form bipolar thick filaments by antiparallel association of the heavy chain tail domain (Clarke and Spudich, 1974; Huxley, 1963). In Dictyostelium, this filament formation is regulated by heavy chain phosphorylation of three-tail-domain threonine residues at positions 1823, 1833, and 2029 (Vaillancourt et al., 1988; Luck-Vielmetter et al., 1990; Egelhoff et al., 1993). It is thought that myosin must be in the form of bipolar thick filaments to carry out its role in cytokinesis. This is supported by genetic evidence that Dictyostelium cells expressing only mutant myosins that cannot form bipolar thick filaments, such as 3XASP-myosin, fail to divide when grown in suspension (Egelhoff et al., 1993).
Although GFP-3XASP-myosin cells cannot divide when grown in suspension, they can divide on an adhesive surface. These divisions are carried out with very little, if any, myosin accumulating in the cleavage furrow. Cells with no myosin heavy chain can also undergo an adhesive surface-dependent cell division that morphologically resembles normal cytokinesis (Fukui et al., 1990; Neujahr et al., 1997; Zang et al. 1997). We call this myosin-independent form of cell division cytokinesis B (see Zang et al., 1997).
3XALA-myosin overassembles into a Triton-insoluble cytoskeletal fraction (Egelhoff et al., 1993). Unlike cells expressing 3XASP-myosin, those expressing 3XALA-myosin are partially competent to divide in suspension. GFP-3XALA-myosin, which mimics the dephosphorylated myosin state, accumulates to a greater degree in the cleavage furrow than does GFP-myosin. Indeed, GFP-3XALA-myosin is depleted from the polar regions of the cell during division and almost all of the myosin in the cell is found in the furrow at this time.
These experiments suggest that heavy chain dephosphorylation is required to localize myosin to the cleavage furrow. Whether this dephosphorylation requirement reflects the need to be in the form of bipolar thick filaments to be transported or to be stabilized in the furrow is unknown. It is possible that the binding of myosin to other molecules is phosphorylation state dependent and that this binding is required for normal localization. How myosin moves to the furrow or is stabilized there is unknown. This interesting issue is presently being addressed by examining the localization of various myosin mutants.
The Pattern of Myosin Localization Persists after Cell Division
The karyokinetic axis is established by the spindle in dividing cells and is thought partly to reflect the interaction of astral microtubules with the polar regions of the cell. It is likely that this polarity allows the cell to elongate in shape along this axis as the cleavage furrow constricts. We found that daughter cells continue to migrate along the karyokinetic axis for more than 10 min after cell division with their leading edges moving at the same rate established during cytokinesis. During this migration, myosin is localized to the posterior region of the daughter cell. This suggests that this regulation of cell motility begins in anaphase. Does this polarized migration depend on the asymmetrical localization of myosin? Apparently not, given that polarized daughter cell migration occurs in the GFP-3XASP-myosin-expressing cells where asymmetrical myosin localization is not seen. Furthermore, Dictyostelium cells lacking myosin, which fail to undergo cytokinesis in suspension, do show polar pseudopod formation at the appropriate time in their cell cycle (Zang et al., 1997). This suggests that something other than myosin localization is setting up the migration axis parallel to the karyokinetic axis.
The maintenance of these migration rates in the daughter cells has yet another important consequence for the cell. It suggests that the random walk migration of the interphase cell is not reestablished until well after cell division. This implies that whatever biochemical signal triggers the transition from cytokinesis to interphase, it is not active until the end of this directed migratory period where the true interphase random walk migration is established. It is not known what signal triggers the exit from cytokinesis. It may involve the timed destruction of a ubiquitinated species, as has been postulated for exit from mitosis (King et al., 1994). However, given that Dictyostelium has a very short G1 phase (Weeks and Weijer, 1994), it may also reflect the onset of activation of the G1 cyclins.
These studies establish that GFP-myosin can accurately reflect the dynamic localization of myosin in live cells, and a number of novel features of myosin localization during cytokinesis have been revealed that were not apparent from previous studies of fixed cells. These features include the accumulation of myosin in the future furrow region during anaphase, the regulation of this accumulation by the phosphorylation of the myosin heavy chain, and the identification of a post-cell division state characterized by directed cell migration at rates established during cell division. Further studies using this technique with specific, well characterized Dictyostelium myosin mutants will allow the elucidation of the molecular mechanism not only of the regulation of this dramatic event but also of the interactions necessary for generating the force to divide the cell.
ACKNOWLEDGMENTS
We thank Hans Warrick for doing the immunofluorescence experiment; Janet Smith, Scot Munroe, and Hans Warrick for critical comments on the manuscript; and the members of the Spudich laboratory for many hours of intense conversation on the wonderful world of myosin. This work was supported by grant GM40509 from the National Institutes of Health to J.A.S.; J.-H.Z. was supported by a Howard Hughes Medical Institute predoctoral award; and J.H.S. was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (DRG-073).
Footnotes
GFP, green-fluorescent protein; GFP-myosin, green-fluorescent protein fused to myosin; GFP-3XALA-myosin, green-fluorescent protein fused to 3XALA-myosin; GFP-3XASP-myosin, green-fluorescent protein fused to 3XASP-myosin.
REFERENCES
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burns CG, Larochelle DA, Erickson H, Reedy M, De Lozanne A. Single-headed myosin II acts as a dominant negative mutation in Dictyostelium. Proc Natl Acad Sci USA. 1995;92:8244–8248. doi: 10.1073/pnas.92.18.8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Spudich JA. Biochemical and structural studies of actomyosin-like proteins from non-muscle cells. J Mol Biol. 1974;86:209–222. doi: 10.1016/0022-2836(74)90013-8. [DOI] [PubMed] [Google Scholar]
- Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of Aequorea green-fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- Dan K. Mechanism of equal cleavage of sea urchin egg: transposition from astral mechanism to constricting mechanism. Zool Sci. 1988;5:507–517. [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- Egelhoff TT, Manstein DJ, Spudich JA. Complementation of myosin null mutants in Dictyostelium discoideum by direct functional selection. Dev Biol. 1990;137:359–367. doi: 10.1016/0012-1606(90)90260-p. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Pollard TD. Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J Cell Biol. 1976;71:848–875. doi: 10.1083/jcb.71.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- Inoue S. Dynamics of mitosis and cleavage. Ann NY Acad Sci. 1990;582:1–14. doi: 10.1111/j.1749-6632.1990.tb21662.x. [DOI] [PubMed] [Google Scholar]
- Karess RE, Chang X-j, Edwards KA, Kulkarni S, Aguilera I, Kiehart DP. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Mabuchi I, Inoue S. Evidence that myosin does not contribute force production in chromosome movement. J Cell Biol. 1982;94:165–178. doi: 10.1083/jcb.94.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Jackson PK, Kirschner MW. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Kitanishi-Yumura T, Fukui Y. Actomyosin organization during cytokinesis: reversible translocation and differential redistribution in Dictyostelium. Cell Motil Cytoskeleton. 1989;12:78–89. doi: 10.1002/cm.970120203. [DOI] [PubMed] [Google Scholar]
- Knecht DA, Loomis WF. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science. 1987;236:1081–1085. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- Kuczmarski ER, Spudich JA. Regulation of myosin self-assembly: phosphorylation of Dictyostelium heavy chain inhibits formation of thick filaments. Proc Natl Acad Sci USA. 1980;77:7292–7296. doi: 10.1073/pnas.77.12.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczmarski ER, Tafuri SR, Parysek LM. Effect of heavy chain phosphorylation of the polymerization and structure of Dictyostelium myosin filaments. J Cell Biol. 1987;105:2989–2997. doi: 10.1083/jcb.105.6.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck-Vielmetter D, Schleicher M, Grabatin B, Wippler J, Gerisch G. Replacement of threonine residues by serine and alanine in a phosphorylatable heavy chain fragment of Dictyostelium myosin II. FEBS Lett. 1990;269:239–243. doi: 10.1016/0014-5793(90)81163-i. [DOI] [PubMed] [Google Scholar]
- Mabuchi I. Cleavage furrow: timing of emergence of contractile ring actin filaments and establishment of the contractile ring by filament bundling in sea urchin eggs. J Cell Sci. 1994;107:1853–1862. doi: 10.1242/jcs.107.7.1853. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan RK, Pardee JD. Assembly mechanism of Dictyostelium myosin II: regulation by K+, Mg2+, and actin filaments. Biochemistry. 1996;35:15504–15514. doi: 10.1021/bi9618981. [DOI] [PubMed] [Google Scholar]
- Manstein DJ, Titus MA, De Lozanne A, Spudich JA. Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P, Pollard TD. Arrangement of actin filaments and myosin-like filaments in the contractile ring and of actin-like filaments in the mitotic spindle of dividing HeLa cells. J Ultrastruct Mol Struct Res. 1986;94:92–103. doi: 10.1016/0889-1605(86)90055-8. [DOI] [PubMed] [Google Scholar]
- Mittal B, Sanger JM, Sanger JW. Visualization of myosin in living cells. J Cell Biol. 1987;195:1753–1760. doi: 10.1083/jcb.105.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB. Spindle and kinetochore morphology of Dictyostelium discoideum. J Cell Biol. 1976;68:113–122. doi: 10.1083/jcb.68.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores SL, Sabry JH, Spudich JA. Myosin dynamics in live Dictyostelium cells. Proc Natl Acad Sci USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr R, Heizer C, Gerish G. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system, and formation of the cleavage furrow. J Cell Sci. 1997;110:123–127. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- Pasternak C, Flicker P, Ravid S, Spudich JA. Intermolecular versus intramolecular interactions of Dictyostelium myosin: possible regulation by heavy chain phosphorylation. J Cell Biol. 1989;109:203–210. doi: 10.1083/jcb.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid S, Spudich JA. Myosin heavy chain kinase from developed Dictyostelium cells. J Biol Chem. 1989;264:15144–15150. [PubMed] [Google Scholar]
- Ruppel KM, Uyeda TQ, Spudich JA. Role of highly conserved lysine 130 of myosin motor domain. In vivo and in vitro characterization of site specifically mutated myosin. J Biol Chem. 1994;269:18773–18780. [PubMed] [Google Scholar]
- Satterwhite LL, Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- Schroeder TM. Cytokinesis: filaments in the cleavage furrow. Exp Cell Res. 1968;53:272–318. doi: 10.1016/0014-4827(68)90373-x. [DOI] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Vaillancourt JP, Lyons C, Cote GP. Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J Biol Chem. 1988;263:10082–10087. [PubMed] [Google Scholar]
- Weeks G, Weijer CJ. The Dictyostelium cell cycle and its relationship to differentiation. FEMS Microbiol Lett. 1994;124:123–130. doi: 10.1016/0378-1097(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Wolpert L. The mechanics and mechanism of cleavage. Int Rev Cytol. 1960;10:163–216. [Google Scholar]
- Yumura S, Fukui Y. Reversible cyclic AMP-dependent change in distribution of myosin thick filaments in Dictyostelium. Nature. 1985;314:194–196. doi: 10.1038/314194a0. [DOI] [PubMed] [Google Scholar]
- Zang, J., Caret, G., Sabry, J.H., Wagner, P., Moores, S.L. and Spudich, J.A., On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and non-adhesive conditions. Mol. Biol. Cell. 8, 2617–2629. [DOI] [PMC free article] [PubMed]