Abstract
The bacterium Clostridium saccharolyticum K10, isolated from a fecal sample obtained from a healthy donor who had received long-term tetracycline therapy, was found to carry three tetracycline resistance genes: tet(W) and the mosaic tet(O/32/O), both conferring ribosome protection-type resistance, and a novel, closely linked efflux-type resistance gene designated tet(40). tet(40) encodes a predicted membrane-associated protein with 42% amino acid identity to tetA(P). Tetracycline did not accumulate in Escherichia coli cells expressing the Tet(40) efflux protein, and resistance to tetracycline was reduced when cells were incubated with an efflux pump inhibitor. E. coli cells carrying tet(40) had a 50% inhibitory concentration of tetracycline of 60 μg/ml. Analysis of a transconjugant from a mating between donor strain C. saccharolyticum K10 and the recipient human gut commensal bacterium Roseburia inulinivorans suggested that tet(O/32/O) and tet(40) were cotransferred on a mobile element. Sequence analysis of a 37-kb insert identified on the basis of tetracycline resistance from a metagenomic fosmid library again revealed a tandem arrangement of tet(O/32/O) and tet(40), flanked by regions with homology to parts of the VanG operon previously identified in Enterococcus faecalis. At least 10 of the metagenomic inserts that carried tet(O/32/O) also carried tet(40), suggesting that tet(40), although previously undetected, may be an abundant efflux gene.
Tetracycline has been used extensively for more than 50 years, both therapeutically and prophylactically, to combat bacterial infections in humans and animals (34). The estimated use of antimicrobial agents in animal husbandry and agriculture far outweighs the total use in humans (19), and there is scientific evidence linking the use of antibiotics in agriculture and the emergence of bacterial antimicrobial resistance in humans (16, 36) and in the environment (5). Tetracycline resistance (Tcr) is one of the most common bacterial antibiotic resistances.
Tcr genes that are less than 80% identical fall into different classes (15), and to date 39 distinct Tcr genes have been described, conferring resistance by four different mechanisms: ribosomal protection (RP) mechanism, tetracycline efflux, enzymatic inactivation of tetracycline, and modification of the ribosomal target (28). Mosaic derivatives of RP-type genes, in which part of the gene is recombined between two or more different classes of RP-type Tcr genes, have been described recently (26, 38, 39, 40). This is thought to be a recent event in the evolution of Tcr genes, driven by intense selection pressure and the presence of multiple resistance genes in the same bacterium (39). Multiple RP-type Tcr genes have been identified in several bacterial species, including Butyrivibrio fibrisolvens [tet(O) and tet(W) (2)] and Streptococcus pyogenes [tet(O) and tet(M) (10)], while both RP- and efflux-type Tcr genes have been identified in Enterococcus and Streptococcus spp. [tet(M) and tet(L) (12, 29)] and Clostridium perfringens [tetA408(P) linked to tet(M) (18) and tetA(P) with tetB(P) (37)].
Many Tcr genes are found on mobile genetic elements, such as plasmids and conjugative transposons (CTn) (28), contributing to their widespread distribution. The type of mobile element with which a specific Tcr determinant is associated influences the ability to spread horizontally to new bacterial genera. CTn are found in both gram-positive and gram-negative bacteria (24, 31). The RP-type genes tet(M), tet(W), and tet(Q) are the most widespread Tcr genes (27) and are often associated with CTn.
The human gut contains a dense microbial population of more than 500 bacterial species and may represent an ideal situation for horizontal gene transfer. Commensal gut bacteria carry a variety of plasmids and mobile genetic elements that can be transferred by conjugation (35). Furthermore, bacterial adaptation to the presence of antibiotic resistance genes can largely abolish the selective disadvantages incurred by possessing resistance genes in the absence of antibiotic selection (30). Isolation of antibiotic-resistant bacteria from healthy individuals (4, 25, 33, 42) proves that the human gut microbiota is an important reservoir of antimicrobial resistance, and this can explain the resurgence of antimicrobial resistance after administration of antibiotics (16, 21).
Clostridium saccharolyticum K10 was previously reported to carry two RP-type resistance genes, tet(W) and the mosaic tet(O/32/O) (21, 26, 40). We report here the discovery of a new Tcr gene, herein named tet(40), that is located in tandem with the tet(O/32/O) gene in C. saccharolyticum K10. Metagenomic libraries provided a powerful approach for analyzing Tcr genes in human gut bacteria and indicated that these two genes commonly occur in tandem and are likely to be associated with mobile genetic elements.
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
Anaerobic strains were routinely grown at 37°C in anaerobic M2GSC broth (23) containing tetracycline (10 μg/ml). Strains were cultured in Bellco tubes under 100% CO2 or in an anaerobic cabinet in an atmosphere of 10% CO2, 10% H2, 80% N2. Escherichia coli TransforMax EPI300 (Epicentre, Madison, WI) and OneShot Top10 (DH10B, a derivative of DH101; Invitrogen, Paisley, United Kingdom) were grown in LB medium (32) at 37°C. E. coli containing fosmid clones was screened for Tcr by positive selection on LB agar plates supplemented with 5 or 10 μg/ml tetracycline. All chemicals, unless otherwise stated, were purchased from Sigma-Aldrich.
Analysis of IC of tetracycline.
The gene encoding tet(40) was cloned into the T vector (pGEM-T Easy vector; Promega, Southampton, United Kingdom) and electroporated into TransforMax EPI300 E. coli cells (Epicentre). The inhibitory concentration (IC) of tetracycline was determined by inoculating 0.1 ml of an overnight culture into 5 ml fresh LB medium containing serial dilutions of tetracycline (0 to 140 μg/ml), in triplicate. Tubes were incubated at 37°C for 16 h, and the optical density at 650 nm (OD650) was subsequently read (LKB Novaspec II; Pharmacia). The lowest concentration of tetracycline reducing the growth of the bacterial cells by 50% (illustrated by a 50% reduction in the OD650 compared to that of a control culture grown in the absence of tetracycline) was defined as the 50% inhibitory concentration (IC50). The effect of the efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on the growth of E. coli cells containing either tet(40) or tet(W) in the presence of tetracycline was established. Triplicate tubes containing three concentrations of tetracycline (40, 60, and 80 μg/ml) were incubated in the presence or absence of 2.5 μg/ml CCCP, and the OD650 was measured after 18 h of growth.
Detection of tetracycline in samples.
Triplicate cultures were set up by inoculating 5 ml LB containing 5 μg/ml tetracycline with clones containing pGEM-T Easy (negative control), tet(40), tet(W), tet(X), or tetA(P). After overnight growth, cultures were spun for 10 min at 5,400 × g. The cleared supernatant was removed and 100 μl mixed with 900 μl acetonitrile (3) in a 1.5-ml Eppendorf tube. The remaining pellets were resuspended in 300 μl of water and sonicated for 10 to 15 s (amplitude, 9 μm). The resulting cell lysates were centrifuged at 4°C for 15 min at maximum speed to remove the cell debris, and 100 μl of the supernatant was mixed with 900 μl acetonitrile. Samples were then mixed vigorously on a Whirlimixer for 15 s, incubated for 10 min at room temperature, and spun down for 10 min at maximum speed. Final supernatants were transferred to fresh 1.5-ml Eppendorf tubes. The acetonitrile contained the internal standard demeclocycline hydrochloride at a final concentration of 500 pg/μl to allow quantification of the tetracycline.
The concentration of tetracycline in each fraction was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS-MS) at room temperature, basically as described previously (3), using an Agilent 1100 high-performance liquid chromatography system (Agilent Technologies, Wokingham, United Kingdom) with a Jupiter 5-μm, C18 column (Phenomenex, Macclesfield, United Kingdom) and an organic mobile phase. Mobile-phase solvents were a mixture of solutions A and B, where solution A was water containing 0.1% formic acid and solution B was acetonitrile containing 0.1% formic acid. The gradient program started at 95% of solution A held for 5 min, followed by 3% of solution A held for 5 min and then 95% of solution A held for 4 min in preparation for the next injection. The flow rate was 300 μl/min, and the injection volume was 5 μl. The LC eluent was directed into, without splitting, a Q-Trap triple quadrupole mass spectrometer (Applied Biosystems, Warrington, United Kingdom) with a Turbo ion spray source fitted in positive ion mode for the detection and quantitation of the antibiotics. Tetracycline and demeclocycline hydrochloride were detected using multiple reaction monitoring transitions, which were calculated by infusing standards directly into the mass spectrometer, via a syringe pump, at a concentration of 5 ng/μl. Data were normalized according to the detection of antibiotics in the pGEM-T control and tabulated as the means of six replicates from two independent growth experiments.
Genome walking and PCR.
DNA preparation and genome walking were carried out as previously described (13). PCR amplification was conducted using forward primer AP1 and nested primer AP2 (provided in a Universal GenomeWalker kit; Clontech) in combination with reverse primer Tet32-3′ or nested primer TetExt (Table 1) to amplify regions downstream of the gene.
TABLE 1.
Oligonucleotide primers used in this work
| Procedure | Primer name | Primer sequence | Reference or source |
|---|---|---|---|
| Genome walking | Tet32-3′ | TCATTCTGAAAGGAGAAATCCCTGCTAG | This work |
| TetExt | ACCGCAGGAATATCTCTCACGGGCGTA | This work | |
| Tcr gene amplification | TetOFF2 | TTGTTTTGGGGCTATTGGAG | 26 |
| TetOFR3 | TATATGACTTTTGCAAGCTG | 26 | |
| TetWarray-for | GGAGGAAAATACCGACATA | 27 | |
| TetWarray-rev | AATCTTACAGTCCGTTACG | 27 | |
| Tet32(2)array-rev | CTCTTTCATAGCCAGGCC | 27 | |
| TetQarray-for | CAAGATGTCCTGTTTATGC | 27 | |
| TetQarray-rev | GAATCCCTTCAAAAACGC | 27 | |
| Sequencing | 519r | GWATTACCGCGGCKGCTG | |
| Fosmid subcloning | adapt-I | AATTCGGCACGAGG | This work |
| adapt-IIa | O3P-CCTCGTGCCG | This work |
This primer is phosphorylated at the 5′ end.
Extraction of high-molecular-weight DNA (for metagenomic library).
A stool sample from donor Ab1 was resuspended in an equal volume (wt/vol) of buffer A (100 mM Tris-HCl, pH 8.0, 50 mM EDTA, and 500 mM NaCl), and 2-ml fractions were frozen in liquid nitrogen and crushed using a mortar and pestle precooled in liquid nitrogen. The crushed sample was then pulverized using a motorized hammer mill (Spex 6700 freezer/mill; Glen Creston Ltd., Middlesex, United Kingdom) for 2 min at full speed. Proteinase K (100 μg/ml) was added and the mixture incubated for 1 h at 50°C. Sodium dodecyl sulfate was added to the sample (1% final concentration), and incubation was continued for 1 h at 55°C. The sample was briefly pelleted (2,000 rpm for 2 min) and the supernatant collected in a 50-ml Falcon tube. Equal volumes of molten (70°C) 2% agarose type VI-A dissolved in buffer A were mixed with the sample, and the mix was poured into 5-ml petri dishes. Agarose slabs approximately 0.5 cm thick were transferred into 50-ml Falcon tubes and washed six times in TE50 buffer (20 mM Tris-HCl, 50 mM EDTA, pH 8.0) at 4°C, with changes of buffer approximately every 12 h. Finally, the slabs were washed overnight in storage buffer (50% glycerol, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and stored at −20°C until required.
Fosmid metagenomic library construction.
DNA embedded in agarose was size fractionated using pulsed-field gel electrophoresis. Slabs were placed in a 1% pulse field-certified agarose gel (SeaKem gold; Cambrex, United Kingdom) dissolved in 0.5× TAE buffer (32) and subjected to contour-clamped homogeneous electric-field gel electrophoresis (CHEF-DR II; Bio-Rad) at 6 V/cm for 1 h at 14°C with a current switch of 10 to 1 s, using 0.5× TAE as running buffer. The size range of electrophoresed DNA was estimated by comparison with DNA Hyperladder VI (Bioline). Agarose containing low-molecular-mass DNA (<10 kb) was excised and discarded, and electrophoresis continued for 20 h, changing the pulse field current switch to 0.4 to 1.5 s. The DNA fraction of 30 to 50 kb was excised from the gel, and the DNA was electroeluted from the agarose into dialysis tubing (32). The electroeluted DNA was dialyzed against 1× Tris-EDTA buffer for 2 h prior to ethanol precipitation and resuspension in 40 μl of 10 mM Tris-HCl buffer, pH 8.0. The DNA was end repaired by incubation for 2 h at 37°C with T4 DNA polymerase, T4 polynucleotide kinase, and Klenow enzyme following the manufacturer's guidelines (Roche). Enzymes were heat inactivated at 70°C for 15 min, and the DNA was purified by gel exclusion chromatography using Chromaspin 1000-TE columns (Clontech) following the manufacturer's guidance.
Finally, 250 ng of concentrated, size-fractionated, end-repaired DNA was ligated with 500 ng of blunt-ended fosmid vector (pcc1FOS; Epicentre) at 16°C for 16 h and the ligation mix was packaged using MaxPack packaging extracts (Epicentre) and transformed into E. coli TransforMax EPI300 cells (Epicentre) following the manufacturer's guidelines. Colonies were selected using a BioRobotics BioPick colony picker (Genomic Solutions, Ann Arbor, MI) and arrayed into 384-well microtiter plates containing 70 μl of freezing mix (2× LB medium supplemented with 10% glycerol), grown overnight at 37°C, and stored at −70°C.
Fosmid DNA extraction and PCR screening of Tcr fosmid clones.
The copy number of fosmids contained within Tcr colonies was increased using 1× copy number induction solution (Epicentre) following the manufacturer's protocol. Fosmid DNA was purified from these cultures using a QIAprep miniprep kit (Qiagen) following the manufacturer's instructions, with appropriate modifications for recovery of large plasmids from larger culture volumes. DNA was finally eluted using 50 μl of 5 mM prewarmed (70°C) Tris-HCl, pH 8.0, and concentrated to ∼10 μl in a vacuum concentrator before being stored at −20°C until further use.
DNA purified from Tcr fosmids was screened by PCR amplification for known Tcr genes, using conditions described previously (26, 27). Primers used were specific for tet(O), tet(W), tet(O/32/O), and tet(Q) (Table 1).
Bacterial 16S rRNA gene analysis.
Approximately 100 ng of metagenomic DNA, purified as described above, was used as a template for PCR amplification of 16S rRNA with eubacterial universal primers fD1 and rP2 (43). Touchdown PCR amplification was done under standard conditions with a Bio-Rad MyCycler thermal cycler, using an initial annealing temperature 5°C higher than optimal. Cycling conditions were an initial cycle of 94°C for 3 min followed by 10 cycles of 94°C for 45 s, 62°C for 45 s (with a decrement of 0.5°C for each subsequent cycle), and 72°C for 2 min. A further 10 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 2 min were followed by a final cycle step at 72°C for 10 min. This method ensured amplification of specific products by combining stringent conditions for primer hybridization and simultaneously allowing underrepresented bacterial species to be amplified during successive cycles and minimizing the PCR bias associated with a higher number of cycles. Resulting PCR products were ligated into the pGEM-T Easy vector (Promega). A total of 96 transformed colonies were selected and arrayed in a 96-well microtiter plate. PCR products amplified from these colonies using vector-specific primers M13 Forward and pGEM-R were sequenced using the eubacterial universal primer 519r, and the partial sequence was analyzed by a BLAST search at the Ribosomal Database Project (http://rdp.cme.msu.edu/index.jsp). Sequences of single representative clones in the array were extended using fD1 and rP2 primers and assembled using the program CAP integrated to work under the program BioEdit (11). Phylogenetic analysis was completed using ClustalX (41), and the resulting phylogenetic tree was edited using the program Molecular Evolutionary Genetic Analysis, version 3.1 (MEGA 3.1, http://www.megasoftware.net/).
Sequencing of fosmid clones.
Two different approaches were used to sequence the selected fosmid clone T45, which carries multiple Tcr genes. First, in vitro random insertion transposon mutagenesis using transposon EZ-Tn5<kan> (Epicentre) was carried out following the manufacturer's guidelines. Fosmid DNA was then extracted from 384 clones selected on kanamycin LB agar plates and sequenced using transposon EZ-Tn5-specific primers. Second, a shotgun plasmid library of fosmid clone T45 was prepared using DNA purified from 1 liter LB medium by CsCl gradient centrifugation (32) to eliminate E. coli chromosomal DNA. Purified DNA, resuspended in 750 μl of shearing buffer (Tris-EDTA supplemented with 10% glycerol), was randomly sheared using a nebulizer (Invitrogen, Paisley, United Kingdom) for 1 min at a pressure of ∼100 kPa g−1 in a single-head diaphragm pump (Laboport; KNF Neuberger, Denmark). Sheared DNA was ethanol precipitated, resuspended in 40 μl sterile water, and end repaired. DNA was then ligated with 2.0 μM of specific adaptor primers (adapt-I and adapt-II [Table 1]) and finally size fractionated in a size exclusion column (chromaspin 1000-TE; Clontech). This DNA was used as a template for standard PCR amplification with the adapt-I primer in a 50-μl reaction mix using a 35-cycle protocol with an annealing temperature of 47°C. The PCR products were ligated into the pGEM-T Easy vector and colonies in the shotgun library arrayed in six 96-well microtiter plates containing 150 μl LB medium. Following overnight growth, colony PCR amplification using the M13 Forward and pGEM-R primer set was carried out, and the PCR products were sequenced using nested primers T7 promoter and M13 Reverse.
Sequences at each end of the fosmid clones carrying Tcr genes were determined using vector-specific primers, following the manufacturer's guidance (Epicentre). Sequence assembly was performed using the package Phred/Phrap/Consed (http://www.phrap.org/phredphrapconsed.html) in the Rowett Research Institute's in-house computer facility (openMosix Beowulf cluster), and genome analysis and annotation were carried out using release 8 of the Artemis program (http://www.sanger.ac.uk/Software/Artemis/). Identification of open reading frames (ORFs) was conducted using the heuristic-model option for gene prediction of GeneMarK (http://exon.biology.gatech.edu/GeneMark/heuristic_hmm2.cgi). Computer-assisted analysis of ORFs was carried out using the SIP BLAST network service hosted by the Swiss Institute of Bioinformatics BlastP (http://us.expasy.org/tools/blast/). Conserved regions were further analyzed using Pfam, Prosite, Interpro, and Print databases.
Nucleotide sequence accession numbers.
The partial DNA sequence of fosmid clone T45, including the tet(40) gene, has been deposited in the EMBL database under the accession number AM419751. The accession number for the 16S rRNA sequence of C. saccharolyticum K10 is EU305624, and that for the tandem tet(O/32/O) and tet(40) genes in this bacterial strain is AJ295238.
RESULTS
Identification of a new Tcr gene, tet(40), in C. saccharolyticum K10.
The mosaic tet(O/32/O) gene was identified previously in C. saccharolyticum K10 isolated from a healthy human fecal sample received from an individual (Ab1) undergoing long-term tetracycline therapy (21). The full 16S rRNA sequence of C. saccharolyticum K10 was assembled and had 99% sequence identity to the butyrate-producing human gut isolates M62/1 and SM4/1, which were themselves most closely related to C. saccharolyticum (17).
The sequence of the tet(O/32/O) gene (20, 39) was extended by genome walking. The 150-nucleotide (nt) sequence upstream of the start codon had 100% identity to a similar region in tet(O). This included the regulatory regions for tet(O), which differ from those of tet(W) and tet(M) in lacking a leader peptide sequence (22). Immediately downstream of the tet(O/32/O) gene there was an ORF of 1,220 nt potentially encoding a 406-amino-acid protein. The gene, which has been designated tet(40) (15), was located 50 nt downstream of the tet(O/32/O) stop codon and had its own ribosome binding site (AGGAG) as well as the canonical Pribnow-Gilbert box (−10 TATAA and −35 TTAACA). The DNA percent G+C content of the coding region for tet(O/32/O) was 41%, compared to 56.5% for tet(40), indicating that the two genes probably originated from different donor microorganisms.
The protein encoded by tet(40) had 42% amino acid sequence identity to the TetA(P) Tcr efflux protein from C. perfringens (37) and 43% identity to TetA408(P) (18). Alignment of the novel protein Tet(40) with TetA(P) and TetA408(P) showed that the efflux protein motif E60xP62xxxxxD68xxxR72R73 was strongly conserved and very similar in sequence to the consensus motif ExPxxxxxDxxxRK (Fig. 1). Other amino acids of putative functional importance were also conserved among the proteins P62, T63, A119, A122, G137, E233, D236, and S361, including three glutamic acid residues shown to be functionally important in TetA(P) (1, 14), specifically, E53, E60, and E90 (Fig. 1) in Tet(40). Secondary-structure analysis of Tet(40) revealed that there were 12 transmembrane segments, indicative of membrane localization.
FIG. 1.
Multiple alignment of the new Tcr protein Tet(40) against efflux proteins from Clostridium species: TetA408(P) from C. perfringens (BAB71965.1), TetA(P) from C. septicum (BAB71966.1), and a multiple drug resistance type (MDR-typ) from C. acetobutylicum (AAK79415.1) (accession numbers are from the EMBL database). The alignment was carried out using the program ClustalW. Functional glutamate residues E53, E60, and E90 are indicated with an asterisk, and residues comprising the conserved efflux protein motif are indicated with a circle.
Functional analysis of the Tet(40) protein.
The full ORF of the tet(40) gene together with the upstream regions containing predicted ribosomal binding site and promoter sequences were specifically amplified by PCR and cloned into the pGEM-T Easy vector. Expression of the gene in E. coli indicated that the presence of tet(40) itself, not in tandem with tet(O/32/O), was sufficient for expression of the Tcr phenotype. The IC50 of tetracycline for sensitive E. coli EPI300 cells (IC50 of ∼2 μg/ml) containing the tet(40) gene cloned in the pGEM-T vector was 60 μg/ml.
The cloned tet(40) gene was also assessed for activity as an efflux pump by determination of the relative amounts of tetracycline in the supernatant and cell lysate following overnight bacterial incubation (3). The LC-MS-MS results obtained for the tet(40) clones were compared with those obtained for clones containing genes conferring Tcr by alternative mechanisms: tet(W), ribosome protection; tet(X), tetracycline inactivation; and tetA(P), an efflux pump. Clones containing tet(40) or tetA(P) accumulated less tetracycline than those containing the RP gene tet(W) (Fig. 2), confirming that tet(40) confers Tcr by actively pumping tetracycline out of the bacterial cell. Tetracycline was virtually undetectable in fractions of samples containing tet(X), due to modification of the tetracycline molecule into an undetectable form.
FIG. 2.
LC-MS-MS analysis of efflux pump activity compared to other Tcr mechanisms. The control culture was a broth of E. coli EPI300 cells transformed with the native pGEM-T vector. The Tcr genes were all full-length PCR amplicons cloned into the pGEM-T vector and transformed into competent E. coli host cells. The amount of tetracycline detected in the control sample is assumed to represent 100% recovery of the introduced tetracycline, and all other values are normalized against this. Slightly more tetracycline was recovered in the supernatants from all of the test cultures than from the control sample, resulting in values of >1 for the cleared supernatants. No tetracycline was detected in the final cellular pellet fraction of any of the samples tested. The results show the average normalized data from two biological replicates, each carried out in triplicate; error bars represent standard deviations.
The efflux pump inhibitors phenyl-arginine-β-naphthylamide and CCCP both reduced the growth of E. coli cells containing tet(40) in the presence of tetracycline but had no effect on cells containing tet(W). In the presence of 40 to 60 μg/ml tetracycline and CCCP, the growth of cells containing tet(40) was reduced by ∼50% compared to the growth of cells containing tetracycline only, while the effect on cells containing tet(W) was negligible (<1% reduction in growth) (Table 2). A growth reduction of 32% was observed for tetA(P) under the same conditions (data not shown).
TABLE 2.
Effects of tetracycline with or without CCCP on growth of E. coli cells containing tet(40) or tet(W)
| Gene | Tetracycline concn (μg/ml) | Avg (±SD) OD650a
|
% Growthb | |
|---|---|---|---|---|
| Plus tetracycline | Plus tetracycline and CCCP | |||
| tet(40) | 40 | 0.47 ± 0.04 | 0.26 ± 0.03 | 56.5 |
| 60 | 0.33 ± 0.07 | 0.17 ± 0.03 | 53.4 | |
| 80 | 0.20 ± 0.005 | 0.03 ± 0.004 | 16.5 | |
| tet(W) | 40 | 0.91 ± 0.04 | 0.87 ± 0.02 | 96.4 |
| 60 | 0.80 ± 0.09 | 0.79 ± 0.09 | 99.2 | |
| 80 | 0.73 ± 0.06 | 0.55 ± 0.03 | 75.4 | |
The OD650 measurements given are the averages (±standard deviations) from triplicate tubes for a single experiment, except those quoted in the presence of 60 μg/ml tetracycline, which are the averages from triplicate tubes for three separate experiments.
Percentage of growth in the presence of CCCP and tetracycline compared to that in the presence of tetracycline alone.
Human colon metagenomic library.
At the time of isolation of C. saccharolyticum K10, almost 100% of the cultivable anaerobes in feces from individual Ab1 were tetracycline resistant. More than 90% of anaerobes remained resistant 3 years later, although there had been no tetracycline therapy in the intervening period. DNA extracted from the second Ab1 fecal sample was screened for the presence of multiple Tcr genes by use of a macroarray (27). This confirmed the presence of four of the most prevalent RP-type Tcr genes [tet(W), tet(O), tet(Q), and tet(O/32/O)], but known efflux genes, including tetA(P), were not detected (data not shown). This DNA was used to construct a metagenomic library in the E. coli fosmid vector pcc1Fos. The average insert size was estimated to be 35 kb, following restriction analysis (data not shown). Eighty out of approximately 4,000 fosmid clones grew on 10 μg/ml tetracycline, and of these, 33 randomly selected Tcr clones were screened by PCR amplification using specific primers targeting the four RP Tcr genes known to be present in the Ab1 sample. Two clones were found to contain tet(O) and tet(W), 17 carried tet(O/32/O) and tet(W), and one tet(O/32/O) and tet(Q). In nine further clones, tet(O/32/O) was the only resistance gene detected, and one clone contained only tet(W). The three remaining clones failed to amplify with tet(O)-, tet(W)-, or tet(Q)-specific primer pairs.
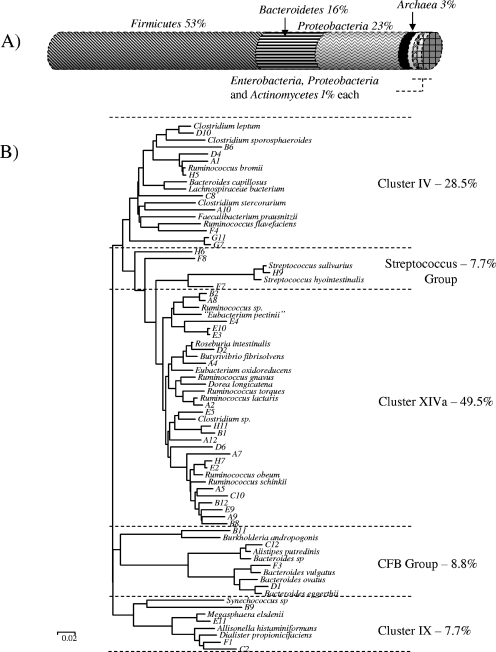
Bacterial diversity of the Ab1 DNA sample was examined by direct amplification and sequencing of 16S rRNA genes. Phylogenetic analysis indicated a distribution of bacterial species in sample Ab1 that is fairly typical of that for human fecal samples, with approximately 90% of sequences related to Firmicutes (including 50% in clostridial cluster XIVa and 29% in clostridial cluster IV) and 9% to Bacteroidetes (Fig. 3). Sequencing the ends of the 80 fosmid inserts conferring Tcr suggested that these metagenomic inserts were also derived from diverse bacterial groupings, with the largest number (approximately 50%) related to sequences from Firmicutes, based on BlastX searches (Fig. 3).
FIG. 3.
(A) Diagram depicting the results of sequencing each end of some of the Tcr fosmid clones isolated from the Ab1 metagenomic fosmid library by use of vector-specific primers. Sequences were analyzed using the program BlastX at the network service hosted by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Results were distributed within one of the bacterial phyla shown and expressed as percentages. For comparison, bacteria included in phylogroups IV, IX, and XIVa and the streptococcus group (below) all belong to the phylum Firmicutes. (B) Phylogenetic analysis of 96 cloned 16S rRNA sequences amplified using eubacterial universal primers from DNA extracted from the Ab1 fecal sample. The phylogenetic tree (rooted phylogram) was created using the ClustalX neighbor-joining method and edited using MEGA 3.1. The number of bootstrap trials was set to 1,000. The proportion of bacterial species falling into each of the five phylogroups detected is shown. CFB, Cytophaga-Flavobacterium-Bacteroidetes group, corresponds to Bacteroidetes in panel A.
Sequencing of fosmid clones carrying tandem Tcr genes.
The sequences of 10 selected clones harboring tet(O/32/O) were extended downstream and in all cases revealed the presence of the tet(40) gene. The sequences of both the tet(40) and tet(O/32/O) genes were >99.9% identical to those identified in C. saccharolyticum K10. The T45 clone contains an insert of ∼37 kb, and sequences of ∼21 kb and ∼8 kb were assembled from each end by creating and sequencing a small-insert shotgun library. Despite repeated efforts using different methods, we were unable to sequence across the central gap, estimated to be 7 to 9 kb, and orf20 and orf21, flanking the gap, were truncated (did not include start/stop codons, respectively). Sequences upstream of the tandem Tcr genes tet(O/32/O) and tet(40) were highly homologous (with between 31 and 83% amino acid identity) to the region of the VanG genetic element between orfG11 and orfG20 (Fig. 4; also see Table S1 in the supplemental material). The VanG element identified in Enterococcus faecalis is a mobile element carrying a set of genes conferring vancomycin resistance (the VanG operon) downstream of orfG22 (6, 19). In clone T45, the tandem Tcr genes are located after orf10, which has 41% identity to orfG22 in the VanG element in E. faecalis (7, 20) (Fig. 4). Downstream of the tandem Tcr genes, clone T45 orf18 encodes a protein with 52% identity to a RecA DNA repair protein from Clostridium thermocellum (see Table S1 in the supplemental material). The VanG element of E. faecalis does not contain xis or int genes that are typical of the Tn916-like CTn, and a RecA protein is postulated to perform this function (7). The DNA percent G+C contents of ORFs after the sequence gap in T45 range from 46 to 58%, whereas those of the ORFs in the first 21-kb sequence range from 35 to 46%, with the exception of tet(40), which has a higher G+C content (∼56%). The closest relationships of the ORFs in T45 are summarized in Table S1 in the supplemental material.
FIG. 4.
Diagram showing genome structure of DNA insert in fosmid clone T45. Sequence analysis and editing were carried out using the program Artemis (http://www.sanger.ac.uk/Software/Artemis/), and the locations of genes were predicted using the heuristic approach for prokaryotic-gene predictions under the program GeneMark (http://exon.biology.gatech.edu/GeneMark/). Numbers inside arrows represent orfs in T45. Amino acid sequence identities to equivalent ORFs in the VanG operon in E. faecalis (shown in gray) are shown underneath the arrows. Tandem Tcr genes are highlighted in black. The locations of primer pairs used for assessing the presence of the transposon-like element and tandem Tcr genes are shown. BlastP results for all ORFs are summarized in Table S1 in the supplemental material. The gap in the sequence between orfs 20 and 21 is indicated (//).
PCR screening to identify the transposon-like element carrying tandem Tcr genes.
A PCR-based screen was conducted, using primers designed from the sequence of fosmid clone T45, on DNA from the other metagenomic clones and C. saccharolyticum K10 (detailed in Fig. 4). Fourteen primer sets were specifically designed, each amplifying a region of 2 kb, contiguously in both parts of the T45 sequence. Amplicons of the tet(O/32/O) and tet(40) genes were designed to be approximately 2.5 kb for ease of identification (Fig. 5).
FIG. 5.
PCR screening of new mobile element carrying the tandem Tcr genes in other clones in the metagenomic library (T2 to T21), C. saccharolyticum K10, and a representative R. inulinivorans-K10 transconjugant (Tc) genomic DNA. Primers were designed from the sequence of T45 in such a way that the PCR products were contiguous along the length of the clone, as shown in Fig. 4. The sizes of the PCR products were predicted to be 2 kb in all cases, except for tet(O/32/O) and tet(40), which were approximately 2.5 kb each (marked by asterisk). The gap in the sequence of fosmid clone T45 between primer pairs 10 and 11 is indicated (//).
In addition to T45, six fosmid clones containing the tet(O/32/O) gene carried the linked tet(40) gene and many of the same flanking ORFs. Certain flanking ORFs were not amplified in three inserts, indicating variability in the organization of the putative transposable element in one case (Fig. 5, T14) and variation in the cloned sequences in clones T10 and T18. C. saccharolyticum K10 and a transconjugant derived from a mating between C. saccharolyticum K10 and Roseburia inulinivorans (K. Scott, unpublished data) were also screened. Products were obtained for all amplicons upstream of tet(O/32/O) and tet(40) and for those based on the first 6 kb downstream of tet(40) in fosmid clone T45 in these bacteria (Fig. 5), indicating that the sequences diverge sometime after this point. This suggests strongly that the tandem Tcr genes are transferred via a transposon-like mobile genetic element which finishes more than 6 kb after the end of tet(40), corresponding to the gap in the T45 sequence. Clone T45 and related clones T2, T9, and T21 do not appear to originate from C. saccharolyticum K10, since they possess different sequences downstream of orf20.
DISCUSSION
The new tet(40) gene described here encodes an efflux pump with similarities to tetA(P) isolated in some Clostridium species. Tetracycline efflux proteins are membrane-associated efflux pumps belonging to the major facilitator superfamily (MFS) that actively export tetracycline from bacterial cells against the concentration gradient (6). The efflux pump activity of Tet(40) was inhibited by CCCP. This compound acts as a proton uncoupler and inhibits MFS efflux pumps that use proton-motive force antiport systems to pump tetracycline out of bacterial cells. There are six groups of Tet efflux proteins based on amino acid sequence identity. TetA(P) and presumably also Tet(40) belong to group 4, characterized by the presence of 12 transmembrane α-helices and their prevalence in gram-positive Clostridium species (6). Key functional residues and domains were conserved between the amino acid sequences of Tet(40) and TetA(P) (1). tetA(P) is part of the Tet P determinant consisting of two genes overlapping by 17 bp. tetA(P) encodes a transmembrane protein that mediates active efflux of tetracycline, whereas tetB(P) encodes an RP type of tetracycline resistance. Although the RP tet(O/32/O) and the efflux tet(40) genes present in C. saccharolyticum K10 do not overlap and have distinct ribosome binding sites, they were cotransferred on the same transferable element, TnK10. Presumably the tandem presence of RP and efflux genes confers a greater level of tetracycline resistance on the host cell. All of the metagenomic clones analyzed in detail in this study contained the tandem arrangement of tet(O/32/O) and tet(40). Since tet(40) has not been described previously, its distribution has not been assessed, and we do not know whether either gene exists independently.
Overall, 2% of clones in the metagenomic library constructed from the Ab1 sample conferred resistance to 10 μg/ml of tetracycline. If we assume an average bacterial genome size of 5 Mb and an average insert size of 35 kb, then the 4,000 clones screened correspond to approximately 28 bacterial genomes. If each bacterial genome harbored one Tcr gene, we would expect 28 resistant clones out of 4,000 screened. The recovery of 80 resistant clones therefore implies the recovery of more than one chromosomal region conferring resistance from each genome. Furthermore, individual positive clones contained up to three Tcr genes. Thus, there was an extraordinarily high incidence of Tcr genes in the metagenomic library. The fecal sample used to prepare the library was obtained from donor Ab1, who had received repeated therapeutic doses of tetracycline for many years and for whom more than 90% of fecal bacteria were tetracycline resistant (21). The most abundant Tcr gene in the metagenomic library was tet(O/32/O), detected in 27 out of 33 inserts expressing resistance to 10 μg tetracycline/ml. tet(W) was present in 19 inserts, tet(O) in 2 inserts, and tet(Q) in 1 insert, while 17 inserts contained both tet(O/32/O) and tet(W). More clones contained the tet(O/32/O) and tet(W) combination than single genes, and these genes were both also present in C. saccharolyticum K10. Whereas tet(O/32/O) and the novel tet(40) gene were cotransmissible in matings from C. saccharolyticum K10, tet(W) was not cotransferred. Thus, despite the fact that tet(W) and tet(O/32/O) are close enough to be recovered in the same inserts in several clones in the metagenomic library, they did not reside on the same transmissible element.
Novel mobile elements carrying the tandem Tcr genes were detected in C. saccharolyticum K10 (21) and also by sequencing in several clones from the metagenomic library. Recent findings indicate that tet(O/32/O) is one of the most abundant genes in fecal samples from pigs and humans (26), and it seems likely that the new transposon-like element TnK10 has been responsible for at least some of the spread of this Tcr gene. It is also probable that the closely linked tet(40) gene may prove to be as abundant as tet(O/32/O). Efflux genes have previously been found mainly in gram-negative bacteria (28). The sequence of the putative transposon in clone T45 contained regions with strong identity to the VanG transposon (7), which itself is homologous to Tn1549, another CTn conferring vancomycin resistance (9). The mobile conjugative element identified previously in the related cluster XIVa anaerobe B. fibrisolvens 1.230, TnB1230, is also similar to Tn1549 (22). Thus, it appears that both of these transposons containing transferable Tcr genes are related to enterococcal transposons conferring vancomycin resistance and that the Tcr genes replace the vancomycin resistance gene cassette. The two transposons from the commensal anaerobes are, however, more similar to their respective enterococcal homologues than to each other.
Based on the information available from sequencing and PCR amplification, it appears that at least part of the CTn present in C. saccharolyticum K10 (TnK10) and clone T45 are very similar. The lack of amplification of any of the ORFs downstream of the gap in clone T45 indicates that the 3′ end of the TnK10 transposon occurs within this sequence gap. The successful amplification of T45 orf1 (PCR 1) in all of the fosmid clones, C. saccharolyticum K10, and the R. inulinivorans transconjugant implies that the 5′ end of the transposon is not contained in clone T45. It is possible that the element in clone T45 is a composite transposon with the central part homologous to TnK10. Thus, the recA gene (orf18) could be instrumental in the conjugative transfer of TnK10, whereas the transposase and integrase encoded on orf21 and orf22 could be part of a larger composite transposon present in fosmid clones T45, T2, T9, and T21. The differences in DNA %G+C strongly indicate different origins for the two parts of the sequence.
In conclusion, the tet(40) gene reported here represents a new efflux-type resistance determinant, found to be present in tandem with an RP-type resistance gene. The potential abundance of this new gene among gut bacteria, at least in individuals with a history of oral tetracycline therapy, is suggested by its recovery in many inserts conferring Tcr from a human fecal metagenomic library. This is the first report of the use of a metagenomic approach for the analysis of tetracycline resistance in bacteria associated with the human colon, although a new Tcr gene encoding a novel NADPH-dependent oxidoreductase that enzymatically inactivates tetracycline, tet(37), was identified in a human oral metagenomic library (8). Metagenomic approaches are therefore potentially valuable for investigating the occurrence of antibiotic resistance genes and for the recovery of novel genes, especially from microorganisms that cannot easily be cultivated under laboratory conditions and that may represent an important reservoir of antibiotic resistance in the environment.
Supplementary Material
Acknowledgments
We thank Donna Henderson for automated DNA sequencing and Gill Campbell for automatic colony picking. Many thanks are also given to Anthony Travis for helpful assistance in sequence assembly using the RRI computer facility and to Gary Duncan for the LC-MS-MS analysis.
K.A.K. is currently supported by the Department for Environment, Food and Rural Affairs (DEFRA), and M.T.R. was supported by European Union grant GEMINI (QLRT-2001-02056). The Rowett Research Institute is supported by SG-RERAD (Scottish Government Rural and Environment Research and Analysis Directorate).
Footnotes
Published ahead of print on 8 September 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Bannam, T. L., and J. I. Rood. 1999. Identification of structural and functional domains of the tetracycline efflux protein TetA(P) from Clostridium perfringens. Microbiology 145:2947-2955. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, T. M., K. P. Scott, K. Forbes, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 3.Beaudry, F., and J. R. E. del Castillo. 2005. Determination of chlortetracycline in swine plasma by LC-ESI/MS/MS. Biomed. Chromatogr. 19:523-528. [DOI] [PubMed] [Google Scholar]
- 4.Calva, J. J., J. Sifuentes-Osornio, and C. Ceron. 1996. Antimicrobial resistance in fecal flora: longitudinal community-based surveillance of children from urban Mexico. Antimicrob. Agents Chemother. 40:1699-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Depardieu, F., M. G. Bonora, P. E. Reynolds, and P. Courvalin. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931-948. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Torres, M. L., R. McNab, D. A. Spratt, A. Villedieu, N. Hunt, M. Wilson, and P. Mullany. 2003. Novel tetracycline resistance determinant from the oral metagenome. Antimicrob. Agents Chemother. 47:1430-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 10.Giovanetti, E., A. Brenciani, R. Lupidi, M. C. Roberts, and P. E. Varaldo. 2003. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob. Agents Chemother. 47:2844-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Huys, G., K. D'Haene, J. M. Collard, and J. Swings. 2004. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 70:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazimierczak, K. A., H. J. Flint, and K. P. Scott. 2006. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob. Agents Chemother. 50:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennan, R. M., L. M. McMurry, S. B. Levy, and J. I. Rood. 1997. Glutamate residues located within putative transmembrane helices are essential for TetA(P)-mediated tetracycline efflux. J. Bacteriol. 179:7011-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 17.Louis, P., S. H. Duncan, S. I. McCrae, J. Millar, M. S. Jackson, and H. J. Flint. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyras, D., and J. I. Rood. 1996. Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob. Agents Chemother. 40:2500-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwen, S. A., and P. J. Fedorka-Cray. 2002. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3):S93-S106. [DOI] [PubMed] [Google Scholar]
- 20.McKessar, S. J., A. M. Berry, J. M. Bell, J. D. Turnidge, and J. C. Paton. 2000. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob. Agents Chemother. 44:3224-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melville, C. M., K. P. Scott, D. K. Mercer, and H. J. Flint. 2001. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob. Agents Chemother. 45:3246-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melville, C. M., R. Brunel, H. J. Flint, and K. P. Scott. 2004. The Butyrivibrio fibrisolvens tet(W) gene is carried on the novel conjugative transposon TnB1230, which contains duplicated nitroreductase coding sequences. J. Bacteriol. 186:3656-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazaki, K., J. C. Martin, R. Marinsek-Logar, and H. J. Flint. 1997. Degradation of xylans by the rumen anaerobe Prevotella bryantii (formerly Prevotella ruminicola subsp. brevis) B14. Anaerobe 3:373-381. [DOI] [PubMed] [Google Scholar]
- 24.Nikolich, M. P., G. Hong, N. B. Shoemaker, and A. A. Salyers. 1994. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl. Environ. Microbiol. 60:3255-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osterblad, M., A. Hakanen, R. Manninen, T. Leistevuo, R. Peltonen, O. Meurman, P. Huovinen, and P. Kotilainen. 2000. A between-species comparison of antimicrobial resistance in enterobacteria in fecal flora. Antimicrob. Agents Chemother. 44:1479-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson, A., M. T. Rincon, H. J. Flint, and K. P. Scott. 2007. Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob. Agents Chemother. 51:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson, A. J., R. Colangeli, P. Spigaglia, and K. P. Scott. 2007. Distribution of specific tetracycline and erythromycin resistance genes in environmental samples assessed by macroarray detection. Environ. Microbiol. 9:703-715. [DOI] [PubMed] [Google Scholar]
- 28.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Avial, I., C. Rodriguez-Avial, E. Culebras, and J. J. Picazo. 2003. Distribution of tetracycline resistance genes tet(M), tet(O), tet(L) and tet(K) in blood isolates of viridans group streptococci harbouring erm(B) and mef(A) genes. Susceptibility to quinupristin/dalfopristin and linezolid. Int. J. Antimicrob. Agents 21:536-541. [DOI] [PubMed] [Google Scholar]
- 30.Salyers, A. A., and C. F. Amabile-Cuevas. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shlaes, D. M. 2006. An update on tetracyclines. Curr. Opin. Investig. Drugs 7:167-171. [PubMed] [Google Scholar]
- 35.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silbergeld, E. K., J. Graham, and L. B. Price. 2008. Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 29:151-169. [DOI] [PubMed] [Google Scholar]
- 37.Sloan, J., L. M. McMurry, D. Lyras, S. B. Levy, and J. I. Rood. 1994. The Clostridium perfringens Tet P determinant comprises two overlapping genes: tetA(P), which mediates active tetracycline efflux, and tetB(P), which is related to the ribosomal protection family of tetracycline-resistance determinants. Mol. Microbiol. 11:403-415. [DOI] [PubMed] [Google Scholar]
- 38.Stanton, T. B., and S. B. Humphrey. 2003. Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl. Environ. Microbiol. 69:3874-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanton, T. B., J. S. McDowall, and M. A. Rasmussen. 2004. Diverse tetracycline resistance genotypes of Megasphaera elsdenii strains selectively cultured from swine feces. Appl. Environ. Microbiol. 70:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanton, T. B., S. B. Humphrey, K. P. Scott, and H. J. Flint. 2005. Hybrid tet genes and tet gene nomenclature: request for opinion. Antimicrob. Agents Chemother. 49:1265-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villedieu, A., M. L. Diaz-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.