Abstract
Pyrosequencing was compared to Sanger dideoxy sequencing to detect mutations in FKS1 responsible for reduced echinocandin susceptibility in Candida albicans. These methods were in complete agreement for 10 of 12 clinical isolates with elevated echinocandin MICs, supporting the potential feasibility of pyrosequencing to detect mutations within diploid fungi.
Each clinically available member of the echinocandin class of antifungals (anidulafungin, caspofungin, and micafungin) has been shown to be effective in the treatment of invasive candidiasis, with an excellent safety and drug-drug interaction profile (10, 13, 17, 21). Several case reports have detailed clinical failures, with members of this class associated with reduced susceptibility (6-9, 12, 14, 18, 19). In many of these cases, this reduced echinocandin susceptibility has been associated with mutations in the FKS1 gene that resulted in amino acid changes in the β-1,3-d-glucan synthase. In C. albicans, these point mutations are dominant, resulting in phenotypic resistance as either homozygous or heterozygous genotypes (18). Thus, detection of these mutations in diploid organisms may be problematic and requires the analysis of both cloned alleles when using traditional sequencing methods (6, 8). An allele-specific real-time PCR assay to overcome this obstacle has previously been described (2). Pyrosequencing is a bioluminometric, nonelectrophoretic technique that employs a cascade of coupled enzymatic reactions to monitor DNA synthesis that has been used to detect point mutations in Saccharomyces cerevisiae and Aspergillus fumigatus (1, 4, 22) and can rapidly screen a large collection of samples for known polymorphisms (11). We evaluated the utility of pyrosequencing technology to detect point mutations in regions of the FKS1 gene known to confer reduced echinocandin susceptibility in a collection of C. albicans clinical isolates.
Twelve C. albicans isolates were acquired from the University of Texas Health Science Center at San Antonio Fungus Testing Laboratories and were subcultured twice prior to susceptibility testing and sequence analysis. C. albicans SC5314 served as the reference isolate for susceptibility testing and sequencing. Echinocandin stock solutions were prepared by dissolving drug powders in dimethyl sulfoxide (anidulafungin; Pfizer, Inc., New York, NY) or water (caspofungin [Merck & Co., Inc., Whitehouse Station, NJ], micafungin [Astellas Pharmaceuticals, Deerfield, IL]) and further dilution in RPMI medium buffered to pH 7.0 with 0.165 M 4-morpholinepropanesulfonic acid (MOPS). Microdilution broth susceptibility testing was performed as described for CLSI M27-A3 methodology in the presence and absence of 50% human serum (Sigma, St. Louis, MO) (3). Trays were read at 24 h, with the MIC defined as the lowest concentration of drug causing a significant decrease in turbidity compared to growth control results.
Genomic DNA was extracted from cells grown overnight in yeast extract-peptone-dextrose broth by use of a MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI). Prior to Sanger dideoxy sequencing and pyrosequencing, PCR amplification was performed with primers specific for FKS1 (GenBank accession no. XM_716336). Dideoxy sequencing was performed in duplicate with a CEQ dye terminator cycle sequencing Quick Start kit (Beckman Coulter, Fullerton, CA), and sequencing analyses were performed with CEQ 8000 genetic analysis system software (Beckman Coulter). Pyrosequencing was conducted as previously described (5). Briefly, a solution of binding buffer and streptavidin-coated Sepharose beads (Amersham Biosciences, Piscataway, NJ) was prepared and added to each sample of the PCR product. Single-stranded DNA was obtained by immobilizing samples in 70% ethanol followed by denaturation solution and washing buffer. Single-stranded DNA was then hybridized to a sequencing primer followed by the addition of substrates, enzymes, and deoxynucleoside triphosphates from an SNP reagent kit (Biotage, Uppsala, Sweden). Samples were analyzed using a Pyrosequencing 96MA instrument (Biotage) in triplicate experiments.
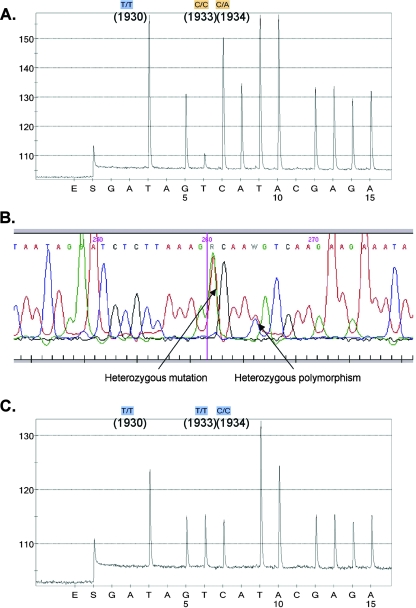
Sequence data identifying point mutations in the FKS1 gene were in complete agreement between the dideoxy sequencing and pyrosequencing results for 10 of 12 isolates (Table 1). T1922C homozygosity, corresponding to an amino acid change from phenylalanine to serine at codon 641 (F641S), was identified in 8 of 12 isolates, and T1933C-homozygous mutations were found in 3 other isolates. For one of the two isolates with results not in full agreement (isolate 5415), dideoxy sequencing identified a T1933C mutation whereas pyrosequencing identified this nucleic acid change plus a heterozygous T1934A mutation (Fig. 1A). For the second isolate, dideoxy sequencing identified a T1933Y mutation that was not found by pyrosequencing (Fig. 1B and C). When segments of the FKS1 openreading frame containing this hot spot were PCR amplified, cloned, and sequenced from these two isolates (18), the sequences obtained from each clone were in agreement with those obtained by dideoxy sequencing. A previous study that evaluated pyrosequencing and dideoxy sequencing using 4,747 synthetic DNA fragments inserted into yeast gene deletion strains reported these methods to be in full agreement for 80.8% of samples, with similar rates of failure (4.8% and 4.17%, respectively) for identifying the correct putative sequence (4). One limitation of pyrosequencing is the short read length (<50 bp) (1, 11). Although this limitation hampers the usefulness of this technique in detecting unknown resistance mechanisms, this is overcome when determining known point mutations as described here. Thus, these results support the feasibility of pyrosequencing for rapid screening of point mutations known to confer reduced antifungal activity. However, as our data suggest, further work is needed to optimize this assay to detect heterozygous mutations in C. albicans.
TABLE 1.
Sanger dideoxy and pyrosequencing results
| Strain | Dideoxy sequencing result | Pyrosequencing result | Protein sequenceb |
|---|---|---|---|
| SC5314 | WTa | WT | FLTLSLRDP |
| 4715 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 2762 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 32746 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 4254 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 41301 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 41509 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 43001 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 42996 | T1922C | T1922C homozygous | SLTLSLRDP (F641S) |
| 42379 | T1933C | T1933C homozygous | FLTLPLRDP (S645P) |
| 53264 | T1933C | T1933C homozygous | FLTLPLRDP (S645P) |
| 5415 | T1933C | T1933C homozygous, C1934A heterozygous | FLTL-P/H-LRDP (S645P/H) |
| 42286 | T1933Y | No mutation detected | FLTL-S/P-LRDP (S645P/S) |
WT, wild type.
Boldface and italicized letters refer to amino acid changes within protein sequences that occurred as a result of the nucleic acid point mutation within FKS1.
FIG. 1.
FKS1 sequence results for C. albicans for the two isolates not in full agreement, 5415 (A) and 42286 (B and C), as determined by pyrosequencing (A and C) and by Sanger dideoxy sequencing (C).
Although drug MICs for each of the 12 clinical isolates were consistently elevated for caspofungin (range, 2 to 8 μg/ml), anidulafungin (MIC range, 0.125 to 1 μg/ml) and micafungin (MIC range, 0.5 to 4 μg/ml) appeared to maintain potency against some of these isolates. However, this enhanced potency was negated with the addition of serum to the growth medium (Table 2). These results are consistent with previous findings (15), including those of studies that reported similar levels of in vivo efficacy among members of this class despite greater in vitro potency for anidulafungin and micafungin in the absence of serum (16, 23). Interestingly, in these studies the in vivo activity correlated better with the in vitro potency when tested in the presence of serum. This raises concerns about switching to another echinocandin when resistance is noted, either phenotypically or genotypically, for another member of this class, as others have previously reported (20).
TABLE 2.
MICs for anidulafungin, caspofungin, and micafungin after 24 h of incubation in the presence and absence of 50% human serum
| Isolate | Anidulafungin MIC (μg/ml)
|
Caspofungin MIC (μg/ml)
|
Micafungin MIC (μg/ml)
|
|||
|---|---|---|---|---|---|---|
| No HSa | 50% HS | No HS | 50% HS | No HS | 50% HS | |
| SC5314 | 0.03 | 0.5 | 0.125 | 0.25 | 0.06 | 1 |
| 4715 | 0.25 | 8 | 2 | 8 | 1 | 16 |
| 2762 | 0.5 | 4 | 4 | 8 | 1 | 16 |
| 32746 | 0.25 | 8 | 4 | 4 | 1 | 16 |
| 4254 | 0.125 | 2 | 2 | 4 | 0.5 | 8 |
| 41301 | 0.25 | 8 | 4 | 8 | 0.5 | 16 |
| 41509 | 0.5 | 8 | 4 | 8 | 0.5 | 32 |
| 43001 | 0.5 | 4 | 4 | 8 | 2 | 16 |
| 42996 | 0.125 | 4 | 4 | 8 | 1 | 32 |
| 42379 | 0.5 | 8 | 8 | >32 | 4 | >32 |
| 53264 | 1 | 32 | 4 | >32 | 4 | >32 |
| 5415 | 1 | 16 | 8 | 32 | 1 | 32 |
| 42286 | 0.125 | 1 | 2 | 4 | 1 | 8 |
HS, human serum.
In conclusion, these results demonstrate the utility of pyrosequencing for the detection of known point mutations conferring echinocandin resistance. Further studies are warranted to optimize this assay and assess pyrosequencing in screening for other mechanisms of antifungal resistance.
(Presented in part at the 47th Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], Chicago, IL, 2007 [10a].)
Acknowledgments
The study was funded in part by a grant from the Society of Infectious Diseases Pharmacists (to N.P.W.) and by NIH grant AI069397 (to D.S.P.) and a Pfizer grant (to D.S.P.).
N.P.W. has received research support from Pfizer and Schering-Plough. D.S.P. has received research support from Pfizer and Merck and serves on advisory panels for Merck, Pfizer, and Astellas.
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Ahmadian, A., M. Ehn, and S. Hober. 2006. Pyrosequencing: history, biochemistry and future. Clin. Chim. Acta 363:83-94. [DOI] [PubMed] [Google Scholar]
- 2.Balashov, S. V., S. Park, and D. S. Perlin. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50:2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard, 3rd ed., M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Gharizadeh, B., Z. S. Herman, R. G. Eason, O. Jejelowo, and N. Pourmand. 2006. Large-scale pyrosequencing of synthetic DNA: a comparison with results from Sanger dideoxy sequencing. Electrophoresis 27:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabinski, J. L., L. S. Smith, G. B. Chisholm, R. Drengler, G. I. Rodriguez, A. S. Lang, S. P. Kalter, A. M. Garner, L. M. Fichtel, J. Hollsten, B. H. Pollock, and J. G. Kuhn. 2006. Genotypic and allelic frequencies of SULT1A1 polymorphisms in women receiving adjuvant tamoxifen therapy. Breast Cancer Res. Treat. 95:13-16. [DOI] [PubMed] [Google Scholar]
- 6.Hakki, M., J. F. Staab, and K. A. Marr. 2006. Emergence of a Candida krusei isolate with reduced susceptibility to caspofungin during therapy. Antimicrob. Agents Chemother. 50:2522-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez, S., J. L. Lopez-Ribot, L. K. Najvar, D. I. McCarthy, R. Bocanegra, and J. R. Graybill. 2004. Caspofungin resistance in Candida albicans: correlating clinical outcome with laboratory susceptibility testing of three isogenic isolates serially obtained from a patient with progressive Candida esophagitis. Antimicrob. Agents Chemother. 48:1382-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahn, J. N., G. Garcia-Effron, M. J. Hsu, S. Park, K. A. Marr, and D. S. Perlin. 2007. Acquired echinocandin resistance in a Candida krusei isolate due to modification of glucan synthase. Antimicrob. Agents Chemother. 51:1876-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krogh-Madsen, M., M. C. Arendrup, L. Heslet, and J. D. Knudsen. 2006. Amphotericin B and caspofungin resistance in Candida glabrata isolates recovered from a critically ill patient. Clin. Infect. Dis. 42:938-944. [DOI] [PubMed] [Google Scholar]
- 10.Kuse, E. R., P. Chetchotisakd, C. A. da Cunha, M. Ruhnke, C. Barrios, D. Raghunadharao, J. S. Sekhon, A. Freire, V. Ramasubramanian, I. Demeyer, M. Nucci, A. Leelarasamee, F. Jacobs, J. Decruyenaere, D. Pittet, A. J. Ullmann, L. Ostrosky-Zeichner, O. Lortholary, S. Koblinger, H. Diekmann-Berndt, and O. A. Cornely. 2007. Micafungin versus liposomal amphotericin B for candidaemia and invasive candidosis: a phase III randomised double-blind trial. Lancet 369:1519-1527. [DOI] [PubMed] [Google Scholar]
- 10a.Lee, S. A., J. L. Grabinski, G. Garcia-Effron, D. S. Perlin, and N. P. Wiederhold. 2007. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother., abstr. M-531.
- 11.Lindström, A., J. Odeberg, and J. Albert. 2004. Pyrosequencing for detection of lamivudine-resistant hepatitis B virus. J. Clin. Microbiol. 42:4788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, C. D., B. W. Lomaestro, S. Park, and D. S. Perlin. 2006. Progressive esophagitis caused by Candida albicans with reduced susceptibility to caspofungin. Pharmacotherapy 26:877-880. [DOI] [PubMed] [Google Scholar]
- 13.Mora-Duarte, J., R. Betts, C. Rotstein, A. L. Colombo, L. Thompson-Moya, J. Smietana, R. Lupinacci, C. Sable, N. Kartsonis, and J. Perfect. 2002. Comparison of caspofungin and amphotericin B for invasive candidiasis. N. Engl. J. Med. 347:2020-2029. [DOI] [PubMed] [Google Scholar]
- 14.Moudgal, V., T. Little, D. Boikov, and J. A. Vazquez. 2005. Multiechinocandin- and multiazole-resistant Candida parapsilosis isolates serially obtained during therapy for prosthetic valve endocarditis. Antimicrob. Agents Chemother. 49:767-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odabasi, Z., V. Paetznick, J. H. Rex, and L. Ostrosky-Zeichner. 2007. Effects of serum on in vitro susceptibility testing of echinocandins. Antimicrob. Agents Chemother. 51:4214-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paderu, P., G. Garcia-Effron, S. Balashov, G. Delmas, S. Park, and D. S. Perlin. 2007. Serum differentially alters the antifungal properties of echinocandin drugs. Antimicrob. Agents Chemother. 51:2253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pappas, P. G., C. M. Rotstein, R. F. Betts, M. Nucci, D. Talwar, J. J. De Waele, J. A. Vazquez, B. F. Dupont, D. L. Horn, L. Ostrosky-Zeichner, A. C. Reboli, B. Suh, R. Digumarti, C. Wu, L. L. Kovanda, L. J. Arnold, and D. N. Buell. 2007. Micafungin versus caspofungin for treatment of candidemia and other forms of invasive candidiasis. Clin. Infect. Dis. 45:883-893. [DOI] [PubMed] [Google Scholar]
- 18.Park, S., R. Kelly, J. N. Kahn, J. Robles, M. J. Hsu, E. Register, W. Li, V. Vyas, H. Fan, G. Abruzzo, A. Flattery, C. Gill, G. Chrebet, S. A. Parent, M. Kurtz, H. Teppler, C. M. Douglas, and D. S. Perlin. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier, R., I. Alarie, R. Lagace, and T. J. Walsh. 2005. Emergence of disseminated candidiasis caused by Candida krusei during treatment with caspofungin: case report and review of literature. Med. Mycol. 43:559-564. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller, M. A., D. J. Diekema, L. Ostrosky-Zeichner, J. H. Rex, B. D. Alexander, D. Andes, S. D. Brown, V. Chaturvedi, M. A. Ghannoum, C. C. Knapp, D. J. Sheehan, and T. J. Walsh. 2008. Correlation of MIC with outcome for Candida species tested against caspofungin, anidulafungin, and micafungin: analysis and proposal for interpretive MIC breakpoints. J. Clin. Microbiol. 46:2620-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reboli, A. C., C. Rotstein, P. G. Pappas, S. W. Chapman, D. H. Kett, D. Kumar, R. Betts, M. Wible, B. P. Goldstein, J. Schranz, D. S. Krause, and T. J. Walsh. 2007. Anidulafungin versus fluconazole for invasive candidiasis. N. Engl. J. Med. 356:2472-2482. [DOI] [PubMed] [Google Scholar]
- 22.Trama, J. P., E. Mordechai, and M. E. Adelson. 2005. Detection of Aspergillus fumigatus and a mutation that confers reduced susceptibility to itraconazole and posaconazole by real-time PCR and pyrosequencing. J. Clin. Microbiol. 43:906-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiederhold, N. P., L. K. Najvar, R. Bocanegra, D. Molina, M. Olivo, and J. R. Graybill. 2007. In vivo efficacy of anidulafungin and caspofungin against Candida glabrata and association with in vitro potency in the presence of sera. Antimicrob. Agents Chemother. 51:1616-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]