Abstract
Metallo-β-lactamases (MBLs) can confer resistance to most β-lactams, including carbapenems. Their emergence in gram-negative pathogens is a matter of major concern. Italy was the first European country to report the presence of acquired MBLs in gram-negative pathogens and is one of the countries where MBL producers have been detected repeatedly. Here, we present the results of the first Italian nationwide survey of acquired MBLs in gram-negative pathogens. Of 14,812 consecutive nonreplicate clinical isolates (12,245 Enterobacteriaceae isolates and 2,567 gram-negative nonfermenters) screened for reduced carbapenem susceptibility during a 4-month period (September to December 2004), 30 isolates (28 Pseudomonas aeruginosa isolates, 1 Pseudomonas putida isolate, and 1 Enterobacter cloacae isolate) carried acquired MBL determinants. MBL producers were detected in 10 of 12 cities, with a predominance of VIM-type enzymes over IMP-type enzymes (4:1). Although having an overall low prevalence (1.3%) and significant geographical differences, MBL-producing P. aeruginosa strains appeared to be widespread in Italy, with a notable diversity of clones, enzymes, and integrons carrying MBL gene cassettes.
β-Lactamase production is the major mechanism of resistance to β-lactam antibiotics in gram-negative pathogens. Carbapenemases, including the metallo-β-lactamases (MBLs) and the class A and class D serine carbapenemases, are emerging β-lactamases of major importance due to their ability to confer resistance to carbapenems in major gram-negative pathogens (27).
The acquired MBLs, in particular, are resistance determinants of remarkable clinical concern due to their very broad substrate spectra (including expanded-spectrum cephalosporins and carbapenems) and nonsusceptibility to therapeutic serine-β-lactamase inhibitors (5, 27, 30). Acquired MBL genes are usually clustered with other resistance determinants on mobile DNA elements, and their presence is a virtually constant marker for multidrug resistance (27, 30). The first identified acquired MBL, the IMP-1 enzyme, was discovered in the late 1980s in Japan, in multidrug-resistant (MDR) isolates of Pseudomonas aeruginosa (39). Thenceforth, enzymes of the IMP type or of five additional types of acquired MBLs (VIM, SPM, GIM, SIM, and AIM) have been detected worldwide in clinical isolates of gram-negative nonfermenters (GNNFs) showing complex MDR phenotypes and sometimes causing large nosocomial disease outbreaks (27, 30, 40). The IMP- and VIM-type MBLs are currently the most widespread and have also been detected in several enterobacterial species (30).
Although the number of reports on acquired MBLs has steadily increased during the past decade, most reports deal only with sporadic isolates or small outbreaks in individual institutions, while data from larger-scale epidemiological surveys are considerably more limited (5). Italy was the first European country (and the second country after Japan) to report the emergence of acquired MBLs (7, 20), as well as one of the countries where MBL-producing strains have repeatedly been detected (6, 8, 21-23, 33, 36) and even caused relatively large outbreaks of infection (18, 25).
In this work, we report the results of the first Italian nationwide survey of acquired MBLs among GNNFs and Enterobacteriaceae.
MATERIALS AND METHODS
Study design.
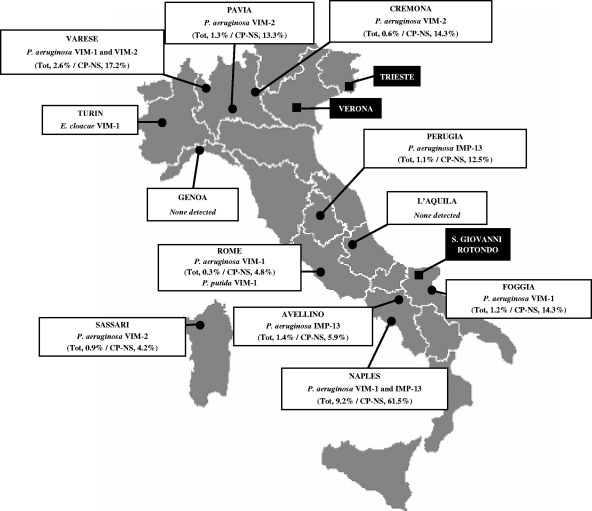
Fourteen clinical microbiology laboratories from 12 Italian cities located across the national territory participated in the study (Fig. 1). In each laboratory, from September to December 2004, all consecutive nonreplicate isolates of Enterobacteriaceae and GNNFs were screened for reduced susceptibility to imipenem. Since the impact of MBL production on carbapenem susceptibility in different species can be variable and no specific guidelines on carbapenem MIC breakpoints for the screening of MBL producers are available (5), we adopted the following imipenem MIC breakpoints to indicate MBL production (based on data in the literature and on personal experience): P. aeruginosa, Proteus spp., Morganella morganii, and Providencia spp., >4 μg/ml; Acinetobacter spp. and other GNNFs other than P. aeruginosa, >2 μg/ml; and Enterobacteriaceae other than Proteus spp., M. morganii, and Providencia spp., >1 μg/ml. All isolates fitting these criteria were collected and subjected to confirmatory imipenem MIC testing. If an imipenem MIC higher than the screening breakpoint was confirmed, the isolate was subjected to a phenotypic analysis of MBL production and to the molecular characterization of MBL determinants. Confirmed MBL-producing isolates were further investigated for susceptibility to anti-gram-negative pathogen agents, genotypic relatedness, and the integron context of the MBL determinant. Due to their intrinsic resistance to carbapenems mediated by resident MBL production, isolates of Stenotrophomonas maltophilia, Elizabethkingia meningoseptica, and Chryseobacterium indologenes were not considered eligible for inclusion in the study.
FIG. 1.
Locations of the clinical microbiology laboratories participating in the survey (3 laboratories, whose data were pooled, were in Turin, while the remaining 11 were in the other cities) and distribution of the MBL-producing isolates. For each city, the MBL-producing species and enzyme types are indicated. For P. aeruginosa, the percentages of MBL producers among the total number of isolates (tot) and the total number of carbapenem-nonsusceptible isolates (CP-NS) are also shown in parentheses. The locations of Verona, where the VIM-1-producing P. aeruginosa index strain VR-143/97 was first detected (20), and Trieste and San Giovanni Rotondo, where two major outbreaks of infection with MBL-producing P. aeruginosa have been reported previously (18, 25), are also shown.
Bacterial identification and antibiotic susceptibility testing.
In all laboratories collecting isolates, identification to the species level and antimicrobial susceptibility testing were carried out with the Phoenix automated microbiology system (Becton Dickinson Diagnostic Systems, Sparks, MD). Confirmatory imipenem MIC testing of collected isolates and susceptibility testing of MBL producers were carried out by Etest (AB Biodisk, Solna, Sweden). Results of susceptibility testing were interpreted as recommended by the CLSI (4).
Phenotypic analysis of MBL production.
Reference spectrophotometric assays for the detection of MBL activity were carried out as described previously (20). Positivity was demonstrated when an imipenem-hydrolyzing specific activity of ≥5 nmol/min/mg of protein was detectable in crude cell extracts and the activity was inhibited by at least 80% after the incubation of the extracts for 20 min in the presence of 5 mM EDTA at 20°C. All GNNF isolates were also tested for MBL production with the MBL Etest (AB Biodisk) using Mueller-Hinton agar plates (Oxoid) as recommended by the manufacturer (AB Biodisk). All P. aeruginosa isolates were also tested for MBL production with the EPI microdilution test as described previously (24). The test measures imipenem MICs in the presence or absence of a mixture of EDTA plus 1,10-phenanthroline. Isolates are considered positive for MBL production when the imipenem MICs are decreased by at least fourfold in the presence of the chelator mixture (24). P. aeruginosa ATCC 27853 and P. aeruginosa 32SM (an imipenem-resistant clinical isolate from our collection that lacks OprD and constitutively produces the AmpC enzyme but does not produce any MBL enzyme) were used as negative controls. P. aeruginosa 101/1477 (producing IMP-1) (19) and P. aeruginosa VR-143/97 (producing VIM-1) (20) were used as positive controls.
Molecular analysis of MBL determinants and integrons.
The presence of blaIMP-like and blaVIM-like MBL genes was investigated by a colony blot assay using random-primed 32P-labeled DNA probes as described previously (23). Probes consisted of mixtures (1:1 molar ratio) of PCR-generated amplicons containing either the blaVIM-1 and blaVIM-2 or the blaIMP-2 and blaIMP-12 genes for the detection of blaVIM or blaIMP alleles, respectively. The nature of MBL genes was preliminarily determined by the direct sequencing of PCR amplification products obtained using primers VIM-DIA/f (5′-CAGATTGCCGATGGTGTTTGG) and VIM-DIA/r (5′-AGGTGGGCCATTCAGCCAGA) or IMP-DIA/f (5′-GGAATAGAGTGGCTTAATTCTC) and IMP-DIA/r (5′-GTGATGCGTCYCCAAYTTCACT), designed based on conserved regions of blaVIM and blaIMP genes as described previously (8). Nucleotide sequences of complete MBL genes were determined from both strands of PCR products generated with primers corresponding to regions outside the coding sequences. The structures of the variable regions of class 1 integrons containing MBL gene cassettes were determined by a PCR mapping and sequencing strategy as described previously (32).
Analysis of genomic relatedness of P. aeruginosa isolates.
P. aeruginosa isolates producing MBLs were investigated for genomic relatedness by macrorestriction profiling of genomic DNA after the digestion of DNA with SpeI and fragment separation by pulsed-field gel electrophoresis as described previously (11). Scanned gel pictures were interpreted using the Fingerprinting II version 3.0 software (Bio-Rad Laboratories, Segrate, Italy) and the Dice coefficient analysis. The unweighted-pair group method with arithmetic averages was applied, and the bandwidth tolerance was set at 1.2%. Clonal relatedness was inferred as recommended by Tenover et al. (35). P. aeruginosa VR-143/97 (20), P. aeruginosa TS-832347 (17), and P. aeruginosa AV65 (25) were included for comparison as representatives of MBL-producing P. aeruginosa clones involved in previously reported disease outbreaks in Italy.
Nucleotide sequence accession numbers.
The nucleotide sequences of the variable regions of In75, In88, and In89, determined in this work, have been submitted to the GenBank/EMBL database and assigned accession numbers FJ172675 (In75), FJ172674 (In88), and FJ172676 (In89).
RESULTS
Prevalence and sources of MBL-producing gram-negative bacteria.
During the study period (September to December 2004), a total of 14,812 nonreplicate isolates of eligible gram-negative bacteria (12,245 Enterobacteriaceae isolates and 2,567 GNNFs) were handled by the participating laboratories. Overall, imipenem MICs for 13 of 12,245 enterobacteria (0.1%) and 247 of 2,567 GNNFs (9.6%) were higher than the screening breakpoints indicating MBL production. The 13 enterobacterial isolates included 9 Proteus mirabilis and 2 Enterobacter cloacae isolates, 1 Enterobacter aerogenes isolate, and 1 Klebsiella pneumoniae isolate. The 247 GNNFs included 222 P. aeruginosa, 21 Acinetobacter baumannii, 2 Burkholderia cepacia, and 2 Pseudomonas putida isolates (Table 1).
TABLE 1.
MBL production in nonreplicate clinical isolates of gram-negative isolates included in the survey
| Species of isolate surveyed (n = 14,812) | No. (%) of isolates collecteda | Range of imipenem MICs (μg/ml)b | No. of collected isolates positive for MBL production by:
|
||
|---|---|---|---|---|---|
| MBL production reference assayc | MBL Etestd | EPI teste | |||
| Enterobacteriaceae | |||||
| P. mirabilis (n = 990) | 9 (0.9) | 8 | 0 | NA | NA |
| E. cloacae (n = 614) | 2 (0.3) | 4-8 | 1 | NA | NA |
| E. aerogenes (n = 248) | 1 (0.4) | 16 | 0 | NA | NA |
| K. pneumoniae (n = 1,214) | 1 (0.1) | 2 | 0 | NA | NA |
| Other species (n = 9,179) | 0 | NAf | NA | NA | NA |
| Total (n = 12,245) | 13 (0.1) | NA | 1 | NA | NA |
| GNNFs | |||||
| P. aeruginosa (n = 2,103) | 222 (10.6) | 8->256 | 28 | 29g | 28h |
| P. putida (n = 60) | 2 (3.3) | 16-32 | 1 | 1i | NA |
| A. baumannii (n = 237) | 21 (8.9) | 4-64 | 0 | 4 | NA |
| B. cepacia complex (n = 50) | 2 (4.0) | 8-32 | 0 | 0 | NA |
| Other species (n = 117) | 0 | NA | NA | NA | NA |
| Total (n = 2,567) | 247 (9.6) | NA | 29 | 34 | NA |
Isolates were collected from among the surveyed isolates on the basis of imipenem MICs higher than the tentative breakpoints set for the various species.
Imipenem MICs for the collected isolates.
In positive isolates, imipenem-hydrolyzing activity in crude extracts was ≥40 nmol/min/mg of protein.
Carried out on the collected GNNFs.
Carried out on the collected P. aeruginosa isolates.
NA, not applicable.
Including 27 isolates positive in the reference assay.
Corresponding to the 28 isolates positive in the reference assay.
Corresponding to the isolate positive in the reference assay.
The reference spectrophotometric assay detected the production of MBL activity in 30 (11.5%) of the 260 isolates collected, including 28 P. aeruginosa isolates, 1 P. putida isolate, and 1 E. cloacae isolate. The production of EDTA-resistant carbapenemase activity was never detected. The MBL Etest was positive for 29 P. aeruginosa isolates (including 27 of those positive in the spectrophotometric assay), 4 A. baumannii isolates, and 1 P. putida isolate (corresponding to that found to be positive in the spectrophotometric assay). The EPI test was positive for the 28 P. aeruginosa isolates determined to be positive by the spectrophotometric assay (Table 1).
MBL-producing isolates were detected in 10 of the 12 cities. MBL-producing P. aeruginosa isolates were detected in nine cities, with remarkably variable prevalences (either overall or among carbapenem-resistant isolates) (Fig. 1).
Isolates carrying MBL determinants were most frequently obtained from urine (47%). The remainder of these isolates were obtained from blood samples (17%), lower respiratory tract specimens (13%), pus samples (10%), surgical wound specimens (10%), and vascular catheter specimens (3%). Most patients (47%) providing the samples were hospitalized in intensive care units (Table 2).
TABLE 2.
Epidemiological data, MBL determinants, clonal lineages, and resistance phenotypes for the MBL-producing isolates
| Isolatea | Specimen | Wardh | Enzyme | Clone | Integronb | MICc (μg/ml) of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | CAZ | FEP | ATM | IMP | MEM | COL | AMK | GEN | CIP | LEV | ||||||
| TO-16X-51 | Urine | ICU | VIM-1 | NAd | NA | >128 | >128 | >128 | 128 | 8 | 4 | 2 | 0.25 | 24 | 2 | >32 | >32 |
| RM-11X-11 | Urine | ICU | VIM-1 | NA | NA | >128 | >128 | >128 | >128 | 8 | >32 | >32 | 0.25 | 1 | 4 | 0.06 | 0.25 |
| FG-17X-14 | Sputum | ICU | VIM-1 | A1 | In70e | >128 | >128 | >128 | >128 | 6 | >32 | >32 | 1 | 8 | >128 | >32 | >32 |
| NA-14X-21 | Urine | ICU | VIM-1 | A2 | In70 | >128 | >128 | >128 | >128 | 8 | >32 | >32 | 2 | 24 | >128 | >32 | >32 |
| NA-14X-26 | Urine | ICU | VIM-1 | A2 | NA | >128 | >128 | >128 | >128 | 8 | >32 | >32 | 2 | 32 | >128 | >32 | >32 |
| NA-14X-32 | Urine | ICU | VIM-1 | A2 | NA | >128 | >128 | >128 | >128 | 6 | >32 | >32 | 2 | 24 | >128 | >32 | >32 |
| NA-14X-34 | Urine | ICU | VIM-1 | A3 | In70 | >128 | >128 | >128 | >128 | 6 | >32 | >32 | 2 | 24 | >128 | >32 | >32 |
| FG-17X-08 | Sputum | ICU | VIM-1 | A3 | NA | >128 | >128 | >128 | >128 | 64 | >32 | >32 | 0.5 | 16 | >128 | >32 | >32 |
| VA-06X-69 | Urine | Nephrology | VIM-1 | D | In110e | >128 | >128 | >128 | >128 | 4 | >32 | >32 | 1 | 8 | >128 | >32 | >32 |
| VA-06X-63 | Blood | Nephrology | VIM-1 | D1 | NA | >128 | >128 | >128 | >128 | 4 | >32 | >32 | 1 | 8 | >128 | >32 | >32 |
| VA-06X-16 | Wound specimen | Geriatrics | VIM-1 | D2 | In110 | >128 | >128 | >128 | >128 | 4 | >32 | >32 | 1 | 8 | >128 | >32 | >32 |
| VA-06X-17 | Wound specimen | Pneumology | VIM-1 | D2 | In110 | >128 | >128 | >128 | >128 | 6 | >32 | >32 | 1 | 8 | >128 | >32 | >32 |
| VA-06X-65 | Sputum | Pneumology | VIM-1 | D3 | In110 | >128 | >128 | >128 | >128 | 0.25 | >32 | >32 | 0.5 | 8 | >128 | >32 | >32 |
| VA-06X-47 | Urine | Nephrology | VIM-1 | D4 | In110 | >128 | >128 | >128 | >128 | 4 | >32 | >32 | 1 | 8 | >128 | >32 | >32 |
| VA-06X-29 | Pus | Otology | VIM-1 | E | In80e | >128 | >128 | >128 | >128 | 8 | >32 | >32 | 2 | >128 | >128 | >32 | >32 |
| RM-11X-09 | Wound specimen | ICU | VIM-1 | F | In75f | >128 | >128 | >128 | >128 | 16 | >32 | >32 | 1 | 8 | >128 | >32 | >32 |
| NA-14X-13 | Blood | ICU | VIM-1 | G | In105e | >128 | >128 | >128 | >128 | 8 | >32 | >32 | 2 | 128 | >128 | >32 | >32 |
| SS-09X-45 | Urine | Nephrology | VIM-2 | B1 | NAg | >128 | >128 | >128 | >128 | 6 | >32 | >32 | 2 | 128 | >128 | >32 | >32 |
| VA-06X-20 | Urine | Orthopedics | VIM-2 | B2 | NA | 64 | 48 | 64 | 64 | 8 | >32 | >32 | 1 | >128 | 8 | >32 | >32 |
| VA-06X-28 | Blood | Geriatrics | VIM-2 | B2 | NA | 64 | 48 | 64 | 64 | 8 | >32 | >32 | 1 | >128 | 8 | >32 | >32 |
| VA-06X-38 | Wound specimen | Geriatrics | VIM-2 | B2 | In59e | 32 | 32 | 48 | 48 | 4 | >32 | >32 | 1 | >128 | 8 | >32 | >32 |
| CR-04X-14 | Blood | ICU | VIM-2 | B3 | NA | >128 | >128 | 64 | 64 | 8 | >32 | >32 | 2 | >128 | 8 | >32 | >32 |
| PV-05X-02 | Wound specimen | Medicine | VIM-2 | B3 | In59 | 48 | 32 | 32 | 32 | 8 | >32 | 8 | 1 | >128 | 8 | >32 | >32 |
| PV-05X-07 | Vascular catheter specimen | Pneumology | VIM-2 | B3 | NA | 48 | 32 | 32 | 48 | 8 | >32 | 8 | 1 | >128 | 8 | >32 | >32 |
| AV-13X-28 | Urine | ICU | IMP-13 | C1 | In88f | 24 | 16 | >128 | >128 | 8 | 16 | 8 | 2 | 8 | >128 | >32 | >32 |
| NA-14X-02 | Urine | Urology | IMP-13 | C2 | In89f | 24 | 16 | >128 | >128 | 16 | >32 | >32 | 0.5 | 16 | >128 | >32 | >32 |
| NA-14X-10 | Blood | ICU | IMP-13 | C2 | NA | 16 | 16 | >128 | >128 | 16 | >32 | >32 | 0.5 | 16 | >128 | >32 | >32 |
| NA-14X-11 | Urine | Urology | IMP-13 | C2 | NA | 16 | 16 | >128 | >128 | 16 | >32 | >32 | 0.5 | 16 | >128 | >32 | >32 |
| NA-14X-12 | Urine | Urology | IMP-13 | C2 | NA | 16 | 16 | >128 | >128 | 16 | >32 | >32 | 0.5 | 16 | >128 | >32 | >32 |
| PG-12X-03 | Sputum | ICU | IMP-13 | C3 | InPSGe | 16 | 16 | >128 | >128 | 16 | >32 | >32 | 0.5 | >128 | >128 | >32 | >32 |
The first two letters of the name indicate the city of origin (Fig. 1), as follows: TO, Turin; RM, Rome; FG, Foggia; NA, Naples; VA, Varese; SS, Sassari; CR, Cremona; PV, Pavia; AV, Avellino; and PG, Perugia. The first two isolates listed, TO-16X-51 and RM-11X-11, were E. cloacae and P. putida, respectively; all other isolates listed were P. aeruginosa.
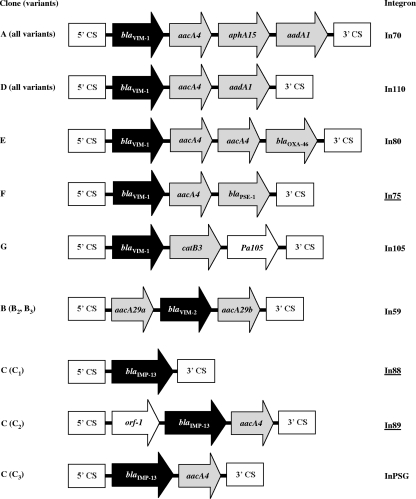
The structures of the variable regions of class 1 integrons carrying MBL gene cassettes in P. aeruginosa isolates representative of different clonal lineages and variants thereof were determined.
Abbreviations for antimicrobial agents: PIP, piperacillin; TZP, piperacillin-tazobactam; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; IPM, imipenem; MEM, meropenem; COL, colistin; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; and LEV, levofloxacin.
NA, not applicable.
New integron detected in this work.
Amplification of the blaVIM-2-containing integron was unsuccessful with this isolate.
Ward in which the patient providing the sample was hospitalized. ICU, intensive care unit.
Nature and distribution of the acquired MBL determinants.
Colony blot hybridization confirmed the presence of known MBL determinants in each of the 30 isolates positive in the spectrophotometric assay, while no blaIMP or blaVIM genes were detected in any of the remaining 230 isolates.
The sequencing of the MBL determinants revealed the presence of blaIMP-13 in 6 P. aeruginosa isolates from three centers, that of blaVIM-2 in 7 P. aeruginosa isolates from four centers, and that of blaVIM-1 in 15 P. aeruginosa isolates from four centers, as well as in the single P. putida and E. cloacae isolates (Fig. 1; Table 2).
Clonal diversity of MBL-producing P. aeruginosa isolates.
The clonal diversity of the 28 MBL-producing P. aeruginosa isolates was investigated by pulsed-field gel electrophoresis macrorestriction profiling after the digestion of DNA with SpeI. Overall, clonal relatedness among isolates producing different MBL types or variants was not apparent (Table 2).
The VIM-1-producing isolates appeared to belong in five clonal lineages, of which one (clone A) included six isolates from two different cities, while the others included isolates detected in single centers (Table 2). Interestingly, the isolates of clone A were clonally related to the serotype O11 ST227 VIM-1-producing index strain originally detected in Verona and subsequently involved in a large disease outbreak occurring in Trieste (11, 17, 20) (data not shown).
All isolates producing VIM-2 appeared to be related to one another (clone B), and they were also clonally related to the VIM-2-producing P. aeruginosa strain involved in the outbreak in Trieste (17) (data not shown).
Finally, all isolates producing IMP-13 appeared to be related to one another (clone C), and they were also clonally related to the IMP-13-producing P. aeruginosa strain previously involved in a large nosocomial disease outbreak that occurred in San Giovanni Rotondo (25) (data not shown).
Integron context of the MBL-encoding gene cassettes.
The structures of the variable regions of class 1 integrons carrying the blaVIM or blaIMP gene cassettes in several P. aeruginosa isolates representative of different clonal lineages and variants were determined.
Different integron structures were found in different clones and also in different variants of the IMP-13-producing clone. On the other hand, a conserved integron structure was shared by variants of each VIM-1-producing clone and by variants of the VIM-2-producing clone (Table 2; Fig. 2).
FIG. 2.
Structures of the variable regions of class 1 integrons carrying MBL gene cassettes in different clones and variants of MBL-producing P. aeruginosa. The gene cassettes are indicated by arrows (those representing cassettes carrying MBL genes are black; those representing cassettes carrying other known resistance genes are gray). 5′ conserved segments (5′CS) and 3′ conserved segments (3′CS) flanking the cassette arrays are also indicated. Underlined integrons are those first detected in this study.
Several of these integrons (In59, In70, In80, In110, In105, and InPSG) have already been identified in MBL-producing isolates from Italy or France and have been described previously (11, 12, 21, 25, 28, 31), while the structures of three of them were original (Fig. 2).
Susceptibility of MBL-positive isolates to clinically relevant drugs.
MBL-positive P. aeruginosa isolates were always characterized by an MDR phenotype including resistance to carbapenems, cephalosporins, fluoroquinolones, and gentamicin. Some remained susceptible to amikacin (50%), aztreonam (75%), and piperacillin-tazobactam (39%), while all were susceptible to colistin. In contrast, the MBL-producing P. putida isolate showed a resistance phenotype limited to β-lactams (except aztreonam), whereas aminoglycosides and fluoroquinolones retained activity against this strain. Finally, the VIM-1-producing E. cloacae isolate showed high-level resistance to penicillins, cephalosporins, and fluoroquinolones, while it showed intermediate susceptibility to amikacin and was susceptible to aztreonam and gentamicin. Carbapenem MICs for this isolate also remained in the range indicating susceptibility, although they were close to the breakpoint (Table 2).
DISCUSSION
The emergence of acquired MBLs among gram-negative pathogens including GNNFs and enterobacteria is an increasing problem worldwide (27, 30). However, the epidemiology of these resistance determinants remains largely unknown (5). This study reports the results of the first countrywide survey of the prevalence of acquired MBLs among gram-negative pathogens in Italy, one of the first countries where the problem has been detected (7, 20). Results revealed the widespread presence of these resistance determinants in P. aeruginosa, while they appeared to be very uncommon in enterobacteria and virtually absent in Acinetobacter spp. The overall MBL prevalence in P. aeruginosa was relatively low (1.3%), being similar to those reported in earlier large-scale surveys in Japan (1.3 to 1.9%) (15, 16) but somewhat higher than that reported previously in Greece (0.8%) (10) and much higher than that reported in a contemporary survey from Spain (0.1%) (13). Although in most cities, the prevalence was close to the average national value, in a single center (Naples), it was approximately eightfold higher (Fig. 1). Similar high prevalences of MBL producers in the Calgary Health Region of Canada (26) and also in the University Hospital of Thessaly in Greece (29) have recently been reported, suggesting that in some settings the epidemiological impact of MBLs can be consistently greater than that reported in general.
Concerning the very low prevalence of acquired MBLs in Enterobacteriaceae observed in this survey, it seems very unlikely to have resulted from a low sensitivity of the criteria adopted for screening. In fact, the screening breakpoints were overall consistent with the most recent recommendations issued by the CLSI in order to suspect carbapenemase production in Enterobacteriaceae (4).
Molecular analysis revealed three types of MBL genes circulating in Italy, namely, blaVIM-1, blaVIM-2, and blaIMP-13. These correspond to the ones most frequently reported in Italy (1, 3, 6, 20, 22, 33) and also those reported to be involved in large outbreaks (17, 25). blaVIM-type determinants were overall more prevalent and widespread than blaIMP-type ones. For MBL-producing P. aeruginosa, a notable diversity was also observed at the clonal and integron levels, suggesting that both clonal expansion and horizontal gene transfer phenomena are playing a role in the dissemination of MBL-producing P. aeruginosa in our setting. The multiclonal diffusion of the VIM-1 MBL determinant in P. aeruginosa and the presence of this determinant in other species also may reflect an overall higher propensity of the VIM-1 MBL determinant for horizontal spread.
The MBL Etest, which is broadly used for the screening of MBL producers, was able to correctly categorize most MBL-producing P. aeruginosa isolates and the single MBL-producing P. putida isolate. However, it yielded false-positive results for two P. aeruginosa and four A. baumannii isolates. It also yielded two false-negative results, those for two IMP-13-producing P. aeruginosa isolates, as reported previously (25).
In conclusion, despite an overall low prevalence of acquired MBLs, MBL production was found to be a widespread resistance mechanism in P. aeruginosa strains circulating in Italy, involving multiple clones and enzyme types. Although at the time of the survey, acquired MBLs were found to be exceedingly uncommon in Enterobacteriaceae, they have recently shown a remarkable dissemination in different enterobacterial species and Acinetobacter in Greece (9, 37, 38), and outbreaks of infection with MBL-producing strains of K. pneumoniae and other Enterobacteriaceae have also started to be reported in Spain (34), France (2, 14), and Italy (1, 3). Altogether, these findings underscore that resistance mediated by MBL production can rapidly evolve and may become a major problem in the near future, and they emphasize the need for the continuous surveillance of these emerging resistance determinants.
Acknowledgments
The excellent contribution of many colleagues is gratefully acknowledged: L. Andrini (Ospedale Mauriziano, Turin); G. Bruno (Ospedale di L'Aquila, L'Aquila); P. Buonopane (A. O. San Giuseppe Moscati, Avellino); M. Casini Lemmi (Ospedale Galliera, Genoa); G. Cherchi (Ospedale Santissima Annunziata, Sassari); A. Di Taranto (Ospedali Riuniti, Foggia); L. Ferrari (Ospedale di Cremona, Cremona); G. Fucale (Centro Traumatologico Ortopedico, Turin); E. Giacobone (IRCCS San Matteo, Pavia); A. Lavitola (Università Federico II, Naples); A. Mencacci (Università di Perugia, Perugia); B. Pini (Ospedale di Circolo, Varese); M. Tronci, (Ospedale San Camillo, Rome); and G. Viberti (Ospedale San Luigi Gonzaga, Orbassano).
This work was partially supported by a research grant from Becton Dickinson Europe and by a grant from the FP6 European research framework (LSHM-CT2005-018705) to G.M.R.
Footnotes
Published ahead of print on 22 September 2008.
REFERENCES
- 1.Aschbacher, R., M. Doumith, D. M. Livermore, C. Larcher, and N. Woodford. 2008. Linkage of acquired quinolone resistance (qnrS1) and metallo-β-lactamase (blaVIM-1) genes in multiple species of Enterobacteriaceae from Bolzano, Italy. J. Antimicrob. Chemother. 61:515-523. [DOI] [PubMed] [Google Scholar]
- 2.Biendo, M., B. Canarelli, D. Thomas, F. Rousseau, F. Hamdad, C. Adjide, G. Laurans, and F. Eb. 2008. Successive emergence of extended-spectrum β-lactamase-producing and carbapenemase-producing Enterobacter aerogenes isolates in a university hospital. J. Clin. Microbiol. 46:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cagnacci, S., L. Gualco, S. Roveta, S. Mannelli, L. Borgianni, J. D. Docquier, F. Dodi, M. Centanaro, E. Debbia, A. Marchese, and G. M. Rossolini. 2008. Bloodstream infections caused by multidrug-resistant Klebsiella pneumoniae producing the carbapenem-hydrolysing VIM-1 metallo-β-lactamase: first Italian outbreak. J. Antimicrob. Chemother. 61:296-300. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Cornaglia, G., M. Akova, G. Amicosante, R. Canton, R. Cauda, J. D. Docquier, M. Edelstein, J. M. Frere, M. Fuzi, M. Galleni, H. Giamarellou, M. Gniadkowski, R. Koncan, B. Libisch, F. Luzzaro, V. Miriagou, F. Navarro, P. Nordmann, L. Pagani, L. Peixe, L. Poirel, M. Souli, E. Tacconelli, A. Vatopoulos, and G. M. Rossolini. 2007. Metallo-β-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380-388. [DOI] [PubMed] [Google Scholar]
- 6.Cornaglia, G., A. Mazzariol, L. Lauretti, G. M. Rossolini, and R. Fontana. 2000. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin. Infect. Dis. 31:1119-1125. [DOI] [PubMed] [Google Scholar]
- 7.Cornaglia, G., M. L. Riccio, A. Mazzariol, L. Lauretti, R. Fontana, and G. M. Rossolini. 1999. Appearance of IMP-1 metallo-β-lactamase in Europe. Lancet 353:899-900. [DOI] [PubMed] [Google Scholar]
- 8.Docquier, J. D., M. L. Riccio, C. Mugnaioli, F. Luzzaro, A. Endimiani, A. Toniolo, G. Amicosante, and G. M. Rossolini. 2003. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47:1522-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galani, I., M. Souli, E. Koratzanis, G. Koratzanis, Z. Chryssouli, and H. Giamarellou. 2007. Emerging bacterial pathogens: Escherichia coli, Enterobacter aerogenes and Proteus mirabilis clinical isolates harbouring the same transferable plasmid coding for metallo-β-lactamase VIM-1 in Greece. J. Antimicrob. Chemother. 59:578-579. [DOI] [PubMed] [Google Scholar]
- 10.Giakkoupi, P., G. Petrikkos, L. S. Tzouvelekis, S. Tsonas, N. J. Legakis, and A. C. Vatopoulos. 2003. Spread of integron-associated VIM-type metallo-β-lactamase genes among imipenem-nonsusceptible Pseudomonas aeruginosa strains in Greek hospitals. J. Clin. Microbiol. 41:822-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giske, C. G., B. Libisch, C. Colinon, E. Scoulica, L. Pagani, M. Fuzi, G. Kronvall, and G. M. Rossolini. 2006. Establishing clonal relationships between VIM-1-like metallo-β-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J. Clin. Microbiol. 44:4309-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giuliani, F., J. D. Docquier, M. L. Riccio, L. Pagani, and G. M. Rossolini. 2005. OXA-46, a new class D β-lactamase of narrow substrate specificity encoded by a blaVIM-1-containing integron from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 49:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez, O., C. Juan, E. Cercenado, F. Navarro, E. Bouza, P. Coll, J. L. Perez, and A. Oliver. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob. Agents Chemother. 51:4329-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kassis-Chikhani, N., D. Decre, V. Gautier, B. Burghoffer, F. Saliba, D. Mathieu, D. Samuel, D. Castaing, J. C. Petit, E. Dussaix, and G. Arlet. 2006. First outbreak of multidrug-resistant Klebsiella pneumoniae carrying blaVIM-1 and blaSHV-5 in a French university hospital. J. Antimicrob. Chemother. 57:142-145. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, S., J. Alba, K. Shiroto, R. Sano, Y. Niki, S. Maesaki, K. Akizawa, M. Kaku, Y. Watanuki, Y. Ishii, and K. Yamaguchi. 2005. Clonal diversity of metallo-β-lactamase-possessing Pseudomonas aeruginosa in geographically diverse regions of Japan. J. Clin. Microbiol. 43:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurokawa, H., T. Yagi, N. Shibata, K. Shibayama, and Y. Arakawa. 1999. Worldwide proliferation of carbapenem-resistant gram-negative bacteria. Lancet 354:955. [DOI] [PubMed] [Google Scholar]
- 17.Lagatolla, C., E. Edalucci, L. Dolzani, M. L. Riccio, F. De Luca, E. Medessi, G. M. Rossolini, and E. A. Tonin. 2006. Molecular evolution of metallo-β-lactamase-producing Pseudomonas aeruginosa in a nosocomial setting of high-level endemicity. J. Clin. Microbiol. 44:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagatolla, C., E. A. Tonin, C. Monti-Bragadin, L. Dolzani, F. Gombac, C. Bearzi, E. Edalucci, F. Gionechetti, and G. M. Rossolini. 2004. Endemic carbapenem-resistant Pseudomonas aeruginosa with acquired metallo-β-lactamase determinants in European hospital. Emerg. Infect. Dis. 10:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J. M. Frere, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lombardi, G., F. Luzzaro, J. D. Docquier, M. L. Riccio, M. Perilli, A. Coli, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2002. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-β-lactamase. J. Clin. Microbiol. 40:4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luzzaro, F., J. D. Docquier, C. Colinon, A. Endimiani, G. Lombardi, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2004. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-β-lactamase encoded by a conjugative plasmid. Antimicrob. Agents Chemother. 48:648-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luzzaro, F., A. Endimiani, J. D. Docquier, C. Mugnaioli, M. Bonsignori, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2004. Prevalence and characterization of metallo-β-lactamases in clinical isolates of Pseudomonas aeruginosa. Diagn. Microbiol. Infect. Dis. 48:131-135. [DOI] [PubMed] [Google Scholar]
- 24.Migliavacca, R., J. D. Docquier, C. Mugnaioli, G. Amicosante, R. Daturi, K. Lee, G. M. Rossolini, and L. Pagani. 2002. Simple microdilution test for detection of metallo-β-lactamase production in Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4388-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagani, L., C. Colinon, R. Migliavacca, M. Labonia, J. D. Docquier, E. Nucleo, M. Spalla, B. M. Li, and G. M. Rossolini. 2005. Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-β-lactamase. J. Clin. Microbiol. 43:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitout, J. D., B. L. Chow, D. B. Gregson, K. B. Laupland, S. Elsayed, and D. L. Church. 2007. Molecular epidemiology of metallo-β-lactamase-producing Pseudomonas aeruginosa in the Calgary Health Region: emergence of VIM-2-producing isolates. J. Clin. Microbiol. 45:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel, L., J. D. Pitout, and P. Nordmann. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2:501-512. [DOI] [PubMed] [Google Scholar]
- 28.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pournaras, S., M. Maniati, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 51:1409-1414. [DOI] [PubMed] [Google Scholar]
- 30.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossolini, G. M., M. L. Riccio, G. Cornaglia, L. Pagani, C. Lagatolla, L. Selan, and R. Fontana. 2000. Carbapenem-resistant Pseudomonas aeruginosa with acquired blaVIM metallo-β-lactamase determinants, Italy. Emerg. Infect. Dis. 6:312-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tato, M., T. M. Coque, P. Ruiz-Garbajosa, V. Pintado, J. Cobo, H. S. Sader, R. N. Jones, F. Baquero, and R. Canton. 2007. Complex clonal and plasmid epidemiology in the first outbreak of Enterobacteriaceae infection involving VIM-1 metallo-β-lactamase in Spain: toward endemicity? Clin. Infect. Dis. 45:1171-1178. [DOI] [PubMed] [Google Scholar]
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toleman, M. A., D. Biedenbach, D. M. Bennett, R. N. Jones, and T. R. Walsh. 2005. Italian metallo-β-lactamases: a national problem? Report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 55:61-70. [DOI] [PubMed] [Google Scholar]
- 37.Tsakris, A., A. Ikonomidis, S. Pournaras, L. S. Tzouvelekis, D. Sofianou, N. J. Legakis, and A. N. Maniatis. 2006. VIM-1 metallo-β-lactamase in Acinetobacter baumannii. Emerg. Infect. Dis. 12:981-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vatopoulos, A. 24 January 2008, posting date. High rates of metallo-β-lactamase-producing Klebsiella pneumoniae in Greece—a review of the current evidence. Euro Surveill. 13:pii=8023. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8023. [PubMed]
- 39.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yong, D., J. M. Bell, B. Ritchie, R. Pratt, M. A. Toleman, and T. R. Walsh. 2007. A novel sub-group metallo-β-lactamase (MBL), AIM-1, emerges in Pseudomonas aeruginosa (PSA) from Australia. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. abstr. C1-593.