Abstract
The hydroxyanilide fenhexamid, one of the latest antibotrytis fungicides, active especially against leotiomycete plant-pathogenic fungi, inhibits 3-ketoreductase of the C-4-demethylation enzyme complex during ergosterol biosynthesis. We isolated Botrytis cinerea strains resistant to various levels of fenhexamid from French and German vineyards. The sequence of the gene encoding 3-ketoreductase, erg27, varied according to levels of resistance. Highly resistant isolates, termed HydR3+, all presented a modification of the phenylalanine at the C terminus of the putative transmembrane domain at position 412, either to serine (85% of the isolates), to isoleucine (11.5% of the isolates), or to valine (3.5% of the isolates). The introduction of the 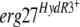 allele into a fenhexamid-sensitive strain by means of a replicative plasmid conferred fenhexamid resistance on the resulting transformants, showing that the mutations at position 412 are responsible for fenhexamid resistance. Weakly to moderately resistant isolates, termed HydR3−, showed different point mutations between the strains in the sequenced regions of the erg27 gene, corresponding to amino acid changes between positions 195 and 400 of the protein. The
allele into a fenhexamid-sensitive strain by means of a replicative plasmid conferred fenhexamid resistance on the resulting transformants, showing that the mutations at position 412 are responsible for fenhexamid resistance. Weakly to moderately resistant isolates, termed HydR3−, showed different point mutations between the strains in the sequenced regions of the erg27 gene, corresponding to amino acid changes between positions 195 and 400 of the protein. The 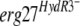 alleles on the replicative vector introduced into a sensitive strain did not confer resistance to fenhexamid. Genetic crosses between HydR3− and sensitive strains showed strict correlation between the sequenced mutation in the erg27 gene and the resistance phenotypes, suggesting that these mutations are linked to fenhexamid resistance. The HydR3 mutations possibly modify the affinity of the 3-ketoreductase enzyme for its specific inhibitor, fenhexamid.
alleles on the replicative vector introduced into a sensitive strain did not confer resistance to fenhexamid. Genetic crosses between HydR3− and sensitive strains showed strict correlation between the sequenced mutation in the erg27 gene and the resistance phenotypes, suggesting that these mutations are linked to fenhexamid resistance. The HydR3 mutations possibly modify the affinity of the 3-ketoreductase enzyme for its specific inhibitor, fenhexamid.
The ergosterol biosynthesis pathway is a target for many antifungals in the medical and the agricultural sector. The available sterol biosynthesis inhibitors include inhibitors of (i) squalene epoxidase, (ii) 14α-demethylase, (iii) Δ14-reductase and/or Δ8-Δ7-isomerase, and (iv) 3-ketoreductase involved in C-4-demethylation (18, 20). The principal antifungals used in medicine and agriculture are 14α-demethylation inhibitors (DMIs), represented principally by triazole derivatives such as epoxiconazole, tebuconazole, or fluconazole (1, 18, 20). Among the C-4-demethylation inhibitors, the sole fungicide used in agriculture is fenhexamid, which is active against the gray mold agent Botrytis cinerea and related species (Sclerotinia spp. and Monilinia spp.) (28). The target site of this hydroxyanilide is the 3-ketoreductase of the C-4-demethylation enzyme complex (7). B. cinerea strains resistant to fenhexamid have been isolated and described previously. They have been classified into three categories, HydR1, HydR2, and HydR3 (19). Strains of the HydR1 category have been easily detected in field populations of B. cinerea before the introduction of fenhexamid on the market, but apparently their resistance does not affect fenhexamid efficacy in the field. This feature may be due to the fact that HydR1 strains exhibit resistance to fenhexamid only during mycelial growth, not during germ tube formation. In fact, they belong to another species, Botrytis pseudocinerea, which is naturally resistant to fenhexamid and part of the B. cinerea species complex (14, 15); In axenic cultures, B. pseudocinerea strains are more susceptible than B. cinerea sensu stricto strains to various fungicides, including DMIs and inhibitors of sterol Δ14-reductase (e.g., fenpropimorph and fenpropidin) (19). Sequence polymorphism of the genes erg27 and CYP51, encoding 3-ketoreductase and eburicol 14α-demethylase, respectively, could explain the fenhexamid-resistant and DMI-hypersensitive phenotypes (2, 3). Moreover, B. pseudocinerea strains metabolize fenhexamid more rapidly than do B. cinerea strains (19, 30).
Strains belonging to the HydR2 and HydR3 categories are B. cinerea (sensu stricto) strains resistant to fenhexamid, isolated in Germany and Japan prior to the registration of this botryticide. They exhibited moderate (HydR2) to high (HydR3) resistance levels toward fenhexamid in tests performed on mycelia, but only HydR3 isolates presented fenhexamid resistance during germ tube elongation (22). Sequence analysis of the erg27 gene putatively encoding 3-ketoreductase revealed two mutations in the Erg27 protein of both HydR3 isolates (F412I and R496T), whereas no mutations were detected in the erg27 alleles of both analyzed HydR2 isolates (2).
Fenhexamid was registered in France in 2000 with the limitation of one application per season. HydR3 isolates were first detected in Champagne vineyards in 2004 and, from 2005 on, also in other French vineyards. Frequencies of HydR3 strains in B. cinerea populations vary from less than 30% in 10 to 20% of the tested Champagne vineyards (three treatments per season including fenhexamid) to more than 50% in the Loire region with one sole fenhexamid treatment per season since 2000. However, HydR2 strains have never been detected in France (23). It should be noted that fenhexamid treatments remain efficient despite high frequencies of B. cinerea strains highly resistant to fenhexamid, suggesting reduced fitness of these strains (P. Leroux et al., unpublished data). In this study we analyzed the sequences of erg27 alleles of B. cinerea strains isolated from French and some German vineyards showing moderate to high fenhexamid resistance. Using a transformation protocol based on a replicative plasmid, as well as crosses, we are able to assess for the first time the functional relationship between erg27 mutations and fenhexamid resistance.
MATERIALS AND METHODS
Fungal strains, media, and culture condition.
Botrytis cinerea natural isolates are listed in Table 1. B05.10 (6) was the recipient strain for transformation. B. cinerea strains were grown in the synthetic complete medium MY (2 g liter−1 of malt extract, 2 g liter−1 of yeast extract, 12.5 g liter−1 of agar) at 21°C under continuous light for conidiation. Liquid cultures were made in YSS medium [2 g liter−1 of yeast extract, 10 g liter−1 of glucose, 2 g liter−1 of KH2PO4, 1.5 g liter−1 of K2HPO4, 1 g liter−1 of (NH4)2SO4, 0.5 g liter−1 of MgSO4·7H2O] at 23°C and shaken at 150 rpm. For DNA isolation, 107 spores were used to inoculate 100 ml of liquid medium and grown for 16 h. Growth tests were performed on YSS plates supplemented with 50 μg ml−1 hygromycin B (Sigma-Aldrich) in the case of transformants and with fenhexamid (technical product kindly provided by Bayer CropScience, Lyon, France) at concentrations indicated in the figure legends. Plates were inoculated with nonsporulating mycelium plugs from 4-day-old cultures on MY medium and were then incubated at 21°C for 4 to 8 days without light. Germination assays were carried out on the same media as those used in the growth tests in 24-well microtiter plates. A 2.5-ml amount of the medium was inoculated with 104 spores and incubated at 21°C without light for 7 days.
TABLE 1.
Strains used in this studya
| Resistance class and strain | Yr of isolation | Reference or origin | EC50 (μg/ml)
|
|
|---|---|---|---|---|
| GTb | Mycc | |||
| HydS | ||||
| SAS405 | 1988 | 13 | <0.1 | <0.1 |
| SAS56 | 1989 | 13 | <0.1 | <0.1 |
| B05.10 | 1994 | 6 | <0.1 | <0.03 |
| HydR3+ | ||||
| 1837 | 1998 | 2 | >10 | >10 |
| 05-190 | 2005 | Champagne | >10 | >10 |
| 05-221 | 2005 | Gers | >10 | 7 |
| 05-AVB | 2005 | Champagne | >10 | 5 |
| 05-PV Reims | 2005 | Champagne | >10 | >10 |
| 118 | 2006 | Champagne | >10 | >10 |
| 179 | 2006 | Champagne | >10 | >10 |
| 214 | 2006 | Champagne | 10 | >10 |
| 223a | 2006 | Champagne | >10 | >10 |
| 223b | 2006 | Champagne | >10 | >10 |
| 264 | 2006 | Champagne | >10 | >10 |
| 312 | 2006 | Bordeaux | >10 | >10 |
| 379a | 2006 | Champagne | >10 | >10 |
| 379b | 2006 | Champagne | >10 | >10 |
| 440a | 2006 | Loire | >10 | >10 |
| 440b | 2006 | Loire | >10 | >10 |
| 506a | 2006 | Bordeaux | >10 | >10 |
| 506b | 2006 | Bordeaux | >10 | >10 |
| 520a | 2006 | Bordeaux | 10 | 3 |
| 520b | 2006 | Bordeaux | 8 | 3 |
| 533 | 2006 | Bordeaux | >10 | 10 |
| MK3-1 | 2006 | Wachenheim (DE) | >5 | NA |
| MK3-31 | 2006 | Wachenheim (DE) | >5 | NA |
| MK3-36 | 2006 | Wachenheim (DE) | >5 | NA |
| MK3-9 | 2006 | Wachenheim (DE) | >5 | NA |
| MK5-14 | 2006 | Bad Duerkheim (DE) | >5 | NA |
| MK5-2 | 2006 | Bad Duerkheim (DE) | >5 | NA |
| HydR3− | ||||
| 05-1.27 | 2005 | Alsace | 2.5 | 0.2 |
| 05-ABA | 2005 | Champagne | 0.8 | 0.2 |
| 57 | 2006 | Champagne | 2 | <0.3 |
| 202 | 2006 | Champagne | 2 | <0.3 |
| 221 | 2006 | Champagne | 0.5 | 0.2 |
| 286 | 2006 | Champagne | 4 | 1.5 |
| 365 | 2006 | Champagne | 2 | 0.3 |
| 452 | 2006 | Bordeaux | 2.5 | 1.5 |
| 453a | 2006 | Bordeaux | 3 | 2 |
| 453b | 2006 | Bordeaux | 2 | 1.5 |
| 454 | 2006 | Bordeaux | 0.5 | <0.3 |
If not otherwise indicated, the original regions are located in France. The fenhexamid resistance phenotypes are classified into HydS (sensitive), HydR3+ (highly resistant), and HydR3− (moderately to slightly resistant). For further details see the text. NA, not analyzed; DE, Germany.
Fifty percent inhibition of germ tube elongation after 24 h.
Fifty percent inhibition of mycelium growth measured over 5 days.
In order to establish the effective concentrations for 50% growth inhibition (EC50s), the effect of fenhexamid on the germination rate and mycelial growth was measured as described by Albertini and Leroux (2) and Leroux et al. (21).
Osmotically stabilized MMVS medium (4 g liter−1 NaNO3, 400 g liter−1 saccharose, 2 g liter−1 KH2PO4, 1 g liter−1 MgSO4·7H2O, 1 g liter−1 KCl, 0.2 g liter−1 FeSO4·7H2O, 15 g liter−1 agar) was used for protoplast regeneration after transformation (see below).
Crosses.
B. cinerea strains 05-ABA, 453b, SAS405, and SAS56 were cultured as described in reference 12 for sclerotium and microconidium production. Crosses between the HydR3− and tester strains (13) were performed by placing the mature sclerotia in a 12-well sterile microtiter dish in 3 ml of a microconidium-water solution of the corresponding mating partner. After being sealed, the microtiter plates were incubated under a 12-h light/dark period at 10°C for 2 to 4 months. Controls for autofertilization were performed in parallel with sclerotia and microconidia of the same parental strain. The mature apothecia were detached and dissected in a water droplet using a sterile surgical blade. The released ascospores were then resuspended in 1.5 ml sterile water and filtered through a 25-μm cloth filter. One hundred fifty microliters of each solution was spread on MY medium plates. Germinating ascospores were picked after 24 h at 20°C on fresh MY plates and incubated at 21°C under white light for further analyses.
DNA manipulations.
Genomic DNA was extracted from mycelium using a sarcosyl-based protocol (9). Gel electrophoresis, restriction enzyme digestions, and Southern blot experiments were performed using standard protocols (29). The oligonucleotides used in this study are listed in Table 2. For erg27 allele sequencing, the genomic loci surrounding the erg27 gene were amplified from the genomic DNA of the strains listed in Table 1 using the primer pair erg27LP2-Xho/erg27RP-Bam or erg27Beg/erg27End (Table 2). The purified PCR products (Macherey and Nagel, Dueren, Germany) were sequenced on both strands with the primers indicated in Table 2. Sequence editing and alignment were performed using the CodonCode Aligner software (CodonCode Corporation, Dedham, MA) including the phred-phrap-consed package (10, 11).
TABLE 2.
Oligonucleotides used in this studya
| Name | 5′ end | Strand | Sequence (5′-3′) |
|---|---|---|---|
| erg27LP2-Xho | 490 | D | AGTCTCGAGATGGAGCGGCAGCGGGTAAT |
| erg27Beg | 1261 | D | TGGGATTACCACCATGGGAGACAAGTG |
| erg1800down | 1764 | D | CCGCCACTTATTCCGCAGATGTT |
| erg2600down | 2554 | D | TGTAAGATGGATGGGAAGCCAATG |
| erg1900up | 1906 | R | CCCTGCATTCAAGACTACAACATCCAG |
| erg2000up | 2067 | R | TCGGAGGGTTTGGCTTGTTTTG |
| erg2600up | 2670 | R | GAAGCTGCCCCGTCCATGTTATC |
| erg3200up | 3295 | R | GCTTGGGCTACTTTAGATGTGA |
| erg27End | 2789 | R | CAATGGTTCCGCATTTCTTTGCCTCCC |
| erg27RP-Bam | 3408 | R | AGGATCCGCACGAGGCGTGCCTAACTCA |
B. cinerea specific sequences are in bold; adapter sequences are in lightface. The positions of the 5′ ends are indicated relative to the erg27 gene presented in Fig. 1. D, direct; R, reverse.
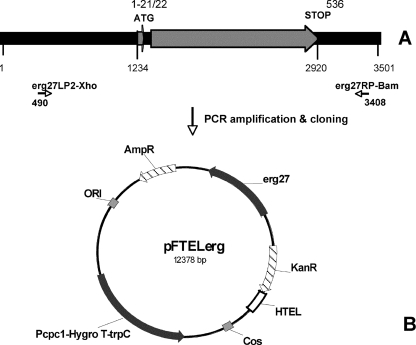
For cloning, the different erg27 alleles were amplified with a high-fidelity thermostable DNA polymerase (Phusion; Finnzymes) between positions 490 and 3408 using the oligonucleotides erg27LP2-Xho and erg27RP-Bam, respectively. After BamHI and XhoI digestion, the column-purified PCR fragments were ligated to the 8.5-kb BglII-XhoI fragment of pFAC1 (4), resulting in the pFTELerg plasmids. Plasmid constructs were verified by PCR and restriction analysis.
B. cinerea transformation.
Protocols for protoplast formation and transformation were described previously by Levis et al. (24) and adapted according to the work of Proctor et al. (27) for protoplast freezing. Transformation was carried out using 2 μg of each pFTEL-erg plasmid. Transformed protoplasts were plated on MMVS medium, containing 50 μg ml−1 of hygromycin B (Sigma-Aldrich), and cultivated at 21°C under constant white light until conidiation. Conidia were then picked onto fresh MY plates containing hygromycin B and incubated at 21°C under white light.
RESULTS
Phenotypic analysis of fungicide resistance.
B. cinerea strains isolated in 2005 and 2006 from French vineyards were subjected to our current fungicide resistance tests (21) in order to establish the resistance categories. For fenhexamid-resistant isolates, we performed dose-response measurements in order to evaluate the fenhexamid concentration (EC50) causing a 50% reduction in germ tube length or in mycelium growth rate, indicated as GT and Myc, respectively, in Table 1. The results showed very diverse fenhexamid resistance phenotypes, ranging from high EC50s (more than 10 μg ml−1 in the case of germ tube length and mycelial growth rate) to quite low EC50s (below 1 μg ml−1) with intermediate sensitivities. Generally the EC50 (GT) values were higher than the EC50 (Myc) values. All strains were tested on fenpropimorph in order to distinguish fenhexamid-resistant B. cinerea isolates from the naturally resistant B. pseudocinerea HydR1 strain (2, 14), which is hypersensitive to this sterol biosynthesis inhibitor (22). Roughly 1 to 2% of the analyzed isolates belonged to the HydR1 category described above (data not shown).
All strains corresponding to the HydR3 category of fenhexamid-resistant isolates according to our previous classification (2) (i.e., resistant to fenhexamid at concentrations higher than 0.1 μg ml−1 during spore germination and mycelium growth) are listed in Table 1. However, in a global comparison of the EC50s, one can differentiate at least two HydR3 classes. One shows EC50 (GT) values above 5 μg ml−1 and EC50 (Myc) values higher than 2 μg ml−1, designated HydR3+. The other class, with EC50s below these thresholds, was designated HydR3−. No isolate matching the HydR2 (22) criteria was found among all tested strains.
Sequence analysis of the erg27 alleles.
Albertini and Leroux (2) previously identified two amino acid changes in the Erg27HydR3 proteins, F412I and R496T. Given the differences observed in fenhexamid resistance, we analyzed the erg27 coding sequences of all new HydR3 isolates (see the supplemental material). Starting with the 1998 and 2005 isolates, we sequenced between positions 1299 and 2750 (3016 in the case of strain 1837) of the erg27 gene locus presented in Fig. 1, using the oligonucleotides listed in Table 2. In these isolates, namely, 1837, 05-PV Reims, 05-AVB, 05-221, 05-190, 05-ABA, and 05-1.27, we identified changes in only two amino acid residues that might be linked to the fenhexamid resistance phenotypes. At position 412, phenylalanine was replaced by isoleucine, valine, or serine in the highly resistant HydR3+ isolates. In the HydR3− strains, at position 400 a leucine-to-serine mutation was found (Table 3) in 05-ABA or leucine was replaced by phenylalanine in 05-1.27. The arginine-to-threonine (R496T) mutation was not detected in any of the isolates sequenced until codon 496. However, three positions were subject to changes regardless of the resistance phenotypes, namely, the silent changes at positions 2461 and 2518 as well as a proline-to-serine transition at residue 238 (nucleotide position 2024) (reference 2 and data not shown).
FIG. 1.
Schematic representation of the erg27 genomic locus (A) and pFTELerg plasmid (B). (A) The sequence positions indicated correspond to the genomic sequence extracted from the B. cinerea B05.10 genome (http://www.broad.mit.edu/annotation/genome/botrytis_cinerea/), supercontig 1.2, from positions 695500 to 699000, covering the complete erg27 coding region with 1,000-bp 5′ untranslated region and 600-bp 3′ untranslated region. Locations of the coding sequences with their respective positions on the supercontig fragment are indicated under the boxes; numbers of the amino acid residues are indicated above. The coding sequence is interrupted by an intron (positions 1299 to 1377). (B) The pFTELerg plasmid is a derivative of pFAC1 (4). Bacterial selection markers are represented by the hatched boxes; fungal genes are represented by the dark gray boxes. Pcpc1-Hygro T-trpC, hygromycin resistance marker under the control of the cpc1 promoter and the trpC terminator; HTEL, human telomeric sequence; ORI, origin of replication in Escherichia coli.
TABLE 3.
Resistance phenotypes and associated mutations in the Erg27 proteina
| Resistance type and strain(s) | Substitution for residue in Erg27:
|
||||||
|---|---|---|---|---|---|---|---|
| L195 | V309 | A314 | S336 | N369 | L400 | F412 | |
| HydR3+ | |||||||
| 05-190 | S | ||||||
| 05-221 | S | ||||||
| 05-AVB | S | ||||||
| 118 | S | ||||||
| 179 | S | ||||||
| 214 | S | ||||||
| 223a and 223b | S | ||||||
| 264 | S | ||||||
| 379a and 379b | S | ||||||
| 440b | S | ||||||
| 506a and 506b | S | ||||||
| 520a and 520b | S | ||||||
| 533 | S | ||||||
| MK3-1 to MK5-2 | S | ||||||
| 312 | D | I | |||||
| 1837 | I | ||||||
| 440a | I | ||||||
| 05-PV Reims | V | ||||||
| HydR3− | |||||||
| 221 | F | ||||||
| 57 | M | ||||||
| 365 | M | ||||||
| 454 | V | ||||||
| 452 | C | D | |||||
| 286 | C | ||||||
| 453a and 453b | D | ||||||
| 202 | F | ||||||
| 05-1.27 | F | ||||||
| 05-ABA | S | ||||||
Sequences of isolates are available in the supplemental material.
From our 2006 isolates, we analyzed the erg27 region comprised between positions 1810 and 2640. The mutation of the phenylalanine at position 412 was revealed to be predominant in all strains studied. In 85% of the cases, the phenylalanine was replaced by a serine residue (codon UCC in most cases, UCU in one case), in 11.5% it was replaced by an isoleucine residue, and the remaining isolate (05-PV Reims) had a valine at the same position (Table 3). It is interesting that the mutation at position 412 was observed in those isolates that we classified in the previous section as HydR3+ (highly resistant). Six other amino acids were subject to modifications (Table 3) in the strains showing reduced resistance levels, the HydR3− isolates. They include L195F, V309M, A314V, S336C, N369D, and L400F or L400S. Two of the strains accumulated two mutations: the HydR3+ strain 312 (N369D and F412I) and the HydR3− strain 452 (S336C and N369D). The same polymorphic changes at positions 2024, 2461, and 2518 (see above) were present in 71% of the analyzed isolates. In the case of the HydR3− isolates, all showed the same sequence at these positions (data not shown).
Transformation of a HydS strain with erg27HydR3 alleles.
In order to determine whether the identified mutations are responsible for the fenhexamid-resistant phenotypes, we introduced different erg27HydR3 alleles into the fenhexamid-sensitive B05.10 strain and analyzed fenhexamid sensitivities of the resulting merodiploid transformants. We amplified 3-kb fragments comprising the putative promoter and coding regions, as well as the 400-bp sequence downstream from the stop codon (for details, see Materials and Methods). This fragment was cloned into a BglII-XhoI restriction fragment of the fungal replicative vector pFAC1 (4), resulting in pFTELerg plasmids (Fig. 1). The pFTEL plasmids harbor a bacterial plasmid backbone, the hygromycin resistance marker for fungal transformation, and one human telomeric sequence cassette, in addition to the cloned erg27 allele. As a negative control, we used the empty pFAC1 vector after EcoRV digestion and ligation, resulting in plasmid pFACR5. B05.10 transformants were selected and propagated on medium containing hygromycin B.
The pFAC1 plasmid and its derivatives harboring a telomeric sequence are nonintegrative plasmids, mitotically unstable unless the transformants are grown under selective pressure (4; C. Lanen and S. Fillinger, unpublished data). In B. cinerea they are present at approximately one copy per genome under these conditions.
Mutations of phenylalanine at position 412 confer resistance to fenhexamid.
We tested two of the 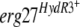 alleles showing a modification at position 412 in the Erg27 protein. The transformants TELergF412I and TELergF412S harbor the
alleles showing a modification at position 412 in the Erg27 protein. The transformants TELergF412I and TELergF412S harbor the 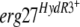 alleles of strains 1837 (F412I) and 223b (F412S), respectively. They were grown on rich medium containing hygromycin B until conidiation. The collected spores were spread on synthetic complete medium, complemented with hygromycin B and variable fenhexamid concentrations (1 to 20 μg ml−1 [Fig. 2B]). As control strains, we tested B05.10 transformants with the empty vector (TR5.10) or the pFTELergWT plasmid with the erg27WT allele of the sensitive B05.10 strain (TELergWT, Fig. 2B). In parallel, we tested the parental strains on the same medium without hygromycin B (Fig. 2A).
alleles of strains 1837 (F412I) and 223b (F412S), respectively. They were grown on rich medium containing hygromycin B until conidiation. The collected spores were spread on synthetic complete medium, complemented with hygromycin B and variable fenhexamid concentrations (1 to 20 μg ml−1 [Fig. 2B]). As control strains, we tested B05.10 transformants with the empty vector (TR5.10) or the pFTELergWT plasmid with the erg27WT allele of the sensitive B05.10 strain (TELergWT, Fig. 2B). In parallel, we tested the parental strains on the same medium without hygromycin B (Fig. 2A).
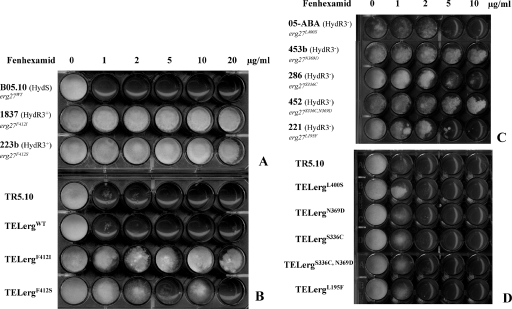
FIG. 2.
Growth tests of HydS and HydR3 parental strains (A and C) and transformants (B and D) on fenhexamid. (A) HydR3+ strains compared to the B05.10 reference strain. (B) B05.10 transformants harboring the indicated 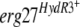 allele on the pFTELerg plasmid. (C) HydR3− strains. (D) B05.10 transformants harboring the indicated
allele on the pFTELerg plasmid. (C) HydR3− strains. (D) B05.10 transformants harboring the indicated 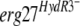 allele on the pFTELerg plasmid. TR5.10, transformants carrying the empty pFACR5 vector (see Materials and Methods). Growth was monitored on YSS medium with the indicated fenhexamid concentrations after 7 days at 21°C. The growth medium was supplemented with hygromycin B at 50 μg ml−1 for testing growth of the transformants (B and D). All strains and transformants are in bold; the erg27 alleles are in italics.
allele on the pFTELerg plasmid. TR5.10, transformants carrying the empty pFACR5 vector (see Materials and Methods). Growth was monitored on YSS medium with the indicated fenhexamid concentrations after 7 days at 21°C. The growth medium was supplemented with hygromycin B at 50 μg ml−1 for testing growth of the transformants (B and D). All strains and transformants are in bold; the erg27 alleles are in italics.
The recipient strain B05.10 developed only on medium without fenhexamid, even after 7 days of incubation, whereas both HydR3+ strains, 1837 and 223b, developed dense mycelium on fenhexamid concentrations up to 20 μg ml−1.
On the other hand, both types of control transformants, TR5.10 and TELergWT, showed a slight increase in fenhexamid resistance in comparison to the recipient B05.10 strain. Indeed, the spores germinated and formed mycelium on medium complemented with fenhexamid at 2 μg ml−1, suggesting that the incorporation of the replicative plasmids pFACR5 and pFTELergWT results in increased resistance to fenhexamid. Comparable results were obtained on other fungicides, e.g., dicarboximides and anilinopyrimidines, for yet-unknown reasons (data not shown). However, much better growth could be observed for both transformants harboring an 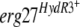 allele: according to microscopic observations, approximately 10% of the transformant spores developed mycelium on fenhexamid concentrations up to 20 μg ml−1 in the case of TELergF412I, comparable to the corresponding HydR3+ strain 1837. The remaining spores behaved like the untransformed parental strain. TELergF412S also showed growth at high fenhexamid concentrations, although to a lesser extent than did the corresponding 223b strain.
allele: according to microscopic observations, approximately 10% of the transformant spores developed mycelium on fenhexamid concentrations up to 20 μg ml−1 in the case of TELergF412I, comparable to the corresponding HydR3+ strain 1837. The remaining spores behaved like the untransformed parental strain. TELergF412S also showed growth at high fenhexamid concentrations, although to a lesser extent than did the corresponding 223b strain.
These results show that the Erg27 mutation of F412 in the HydR3+ strains is responsible for fenhexamid resistance in B. cinerea.
Other erg27 mutations cannot directly be linked to fenhexamid resistance.
We then tested the 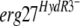 alleles by the same approach. We cloned the erg27 alleles of the following isolates: 221, 286, 453b, 452, and 05-ABA, harboring the mutations L195F, S336C, N369D, S336C plus N369C, or L400S, respectively. The resulting pFTELerg plasmids were used to transform the B05.10 HydS strain. Spores of hygromycin-resistant transformants were inoculated as described above on medium containing fenhexamid and hygromycin B. After 7 days of incubation, the transformants (TELerg as indicated in Fig. 2D) did not show significantly higher fenhexamid resistance than did the control transformants TELergWT and TR5.10 (Fig. 2B and D). We also analyzed the fenhexamid resistance of the transformants during mycelial growth by using mycelial plugs on media containing different fenhexamid concentrations. Also under these conditions the
alleles by the same approach. We cloned the erg27 alleles of the following isolates: 221, 286, 453b, 452, and 05-ABA, harboring the mutations L195F, S336C, N369D, S336C plus N369C, or L400S, respectively. The resulting pFTELerg plasmids were used to transform the B05.10 HydS strain. Spores of hygromycin-resistant transformants were inoculated as described above on medium containing fenhexamid and hygromycin B. After 7 days of incubation, the transformants (TELerg as indicated in Fig. 2D) did not show significantly higher fenhexamid resistance than did the control transformants TELergWT and TR5.10 (Fig. 2B and D). We also analyzed the fenhexamid resistance of the transformants during mycelial growth by using mycelial plugs on media containing different fenhexamid concentrations. Also under these conditions the 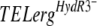 transformants behaved like the control transformants (data not shown). One possible explanation is that the
transformants behaved like the control transformants (data not shown). One possible explanation is that the 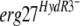 allele is recessive toward the wild-type allele in the merodiploid transformants.
allele is recessive toward the wild-type allele in the merodiploid transformants.
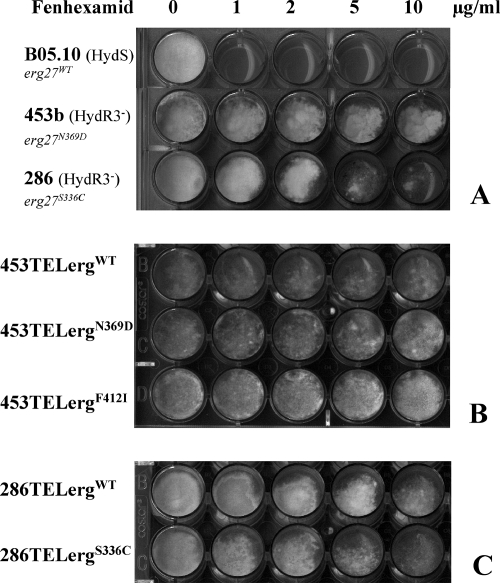
To test this hypothesis, we used two of the HydR3− strains, 453b and 286, as recipients for transformation with different erg27 alleles. Fenhexamid resistance analysis using the spore germination assay showed that all transformants presented the same resistance profile as did the recipient strains (Fig. 3), regardless of the erg27 allele integrated. These results indicate that the 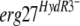 allele is not recessive per se in an
allele is not recessive per se in an 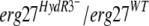 merodiploid. Another possible explanation is that in the HydR3− isolates, fenhexamid resistance is not linked to the erg27 alleles.
merodiploid. Another possible explanation is that in the HydR3− isolates, fenhexamid resistance is not linked to the erg27 alleles.
FIG. 3.
Dominance test of HydR3− alleles. Different erg27 alleles were introduced into the HydR3− strains 453b and 286. Growth of the parental strains (A), transformants of strain 453b (B), and transformants of strain 286 (C) on YSS medium with different fenhexamid concentrations after 7 days at 21°C. The growth medium was supplemented with hygromycin B at 50 μg ml−1 for testing growth of the transformants (B and C). All strains and transformants are in bold; the erg27 alleles are in italics.
HydR3− progeny analysis.
We crossed the HydR3− 453b strain with the fenhexamid-sensitive SAS405 (Table 1). After batch ascospore isolation, we analyzed 30 progeny ascospores on increasing fenhexamid concentrations. We found a ratio of roughly 1:1 between sensitive and resistant progeny (data not shown), suggesting that the mutation of the HydR3− fenhexamid resistance phenotype is monogenic in strain 453b. We sequenced the erg27 genes of six resistant and six sensitive strains derived from the cross. The results indicate a clear link between the identified mutation in the erg27 gene and the resistance phenotypes: all of the resistant but none of the sensitive strains harbored the mutation of the parental HydR3− mutant (N369D). The cross of a second HydR3− strain, 05-ABA, with SAS405 gave rise to comparable results (data not shown).
DISCUSSION
One of the most recent botryticides is the hydroxyanilide fenhexamid. Along with the naturally resistant species B. pseudocinerea, fenhexamid-resistant B. cinerea isolates have rapidly emerged (22), some of which carry mutations in the erg27 gene, potentially encoding the fenhexamid target enzyme, 3-ketoreductase (2). In the present study we analyzed recent fenhexamid-resistant isolates from French and German vineyards.
Fungicide sensitivity measurements revealed two fenhexamid-resistant isolate categories, named HydR3+ and HydR3−, highly and moderately resistant, respectively. HydR3+ strains present resistance factors higher than 50 or even 100, whereas HydR3− phenotypes are below this threshold with resistance factors ranging between 2 and 40. The development of two categories of resistance to fenhexamid has also been observed by others in laboratory isolates (8, 31). Whether they correspond to the naturally isolated HydR3+ and HydR3− categories remains to be established.
Sequence analysis of the erg27 alleles in both resistance categories showed a clear relationship between the phenotypes and the protein sequence. In the case of HydR3+ isolates the same residue was mutated in all isolates. The phenylalanine at position 412 was changed in 85% of the cases to serine, in 11.5% to isoleucine, and in one case to valine. Despite identical sequences, the EC50s of HydR3+ isolates varied. Examples include isolates 520a and 520b, which have the F412S mutation as in other HydR3+ strains but lower EC50s, suggesting that natural variations among these B. cinerea strains may account to some extent for different fenhexamid susceptibilities.
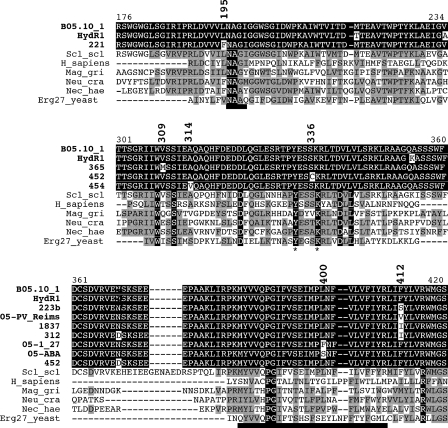
In the case of the HydR3− isolates, at least six mutations could be identified from the sequence comparisons reported in Table 3 and Fig. 4. The mutation of L400 is localized in the putative transmembrane domain (2), as is the HydR3+ mutation of F412. The mutations of L195 and S336 are in the vicinity of the conserved enzymatic domains, the NAGI domain with unknown function and the active site, respectively. Also the mutations of V309, and to a lesser extent of A314, are close to highly conserved residues, W308 and S311, respectively, although their function remains unknown. Only the mutation of N369 is located in a domain that is not conserved among the different Erg27 proteins (Fig. 4). It is noticeable that none of the identified mutations corresponds to a modification found in the Erg27 protein of HydR1 strains (2). Only the codon 314 (GCA) corresponds to a silent modification (GCG) in the HydR1 isolates (data not shown). Whether all these modifications of the Erg27 protein interfere with its affinity for fenhexamid or modify its enzymatic properties remains to be shown by site-directed mutagenesis.
FIG. 4.
Erg27 protein sequence comparison. The three regions covering the identified HydR3 mutations were plotted. The upper parts present the alignment of the mutated Erg27 protein sequences from the tested B. cinerea isolates as indicated by the bold letters, compared to those of the HydS and HydR1 strains. In the lower sections, the B. cinerea Erg27 protein sequence (GenBank accession number AY220532) was aligned with the homologous fungal proteins and the human β-17-HSD-7 protein. Similarities are shown as black on gray, identical amino acids are shown as white on black, and differences are shown as black on white. The thick underlines indicate the NAGI domain of the human β-17-HSD-7 protein and the putative transmembrane domain of the B. cinerea protein, respectively; the conserved amino acids Y and K of the catalytic site are indicated by asterisks. Numbering is according to the B. cinerea Erg27 protein. Scl_scl, Sclerotinia sclerotiorum XP_001598240; H_sapiens, Homo sapiens HSD17B7, P56937; Neu_cra, Neurospora crassa XP_958799.1; Mag_gri, Magnaporthe grisea XP_363377.1; Nec_hae, Nectria haematococca jgi|Necha2|91272| (http://genome.jgi-psf.org/Necha2/Necha2.home.html); Erg27_yeast, Saccharomyces cerevisiae Q12452.
When we introduced an 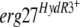 allele carried on a replicative plasmid derived from pFAC1 (4) into a B. cinerea sensitive recipient strain, the resulting transformants acquired fenhexamid resistance, demonstrating the involvement of the F412S and F412I mutations in the HydR3+ phenotype. On the other hand, the functional validation of
allele carried on a replicative plasmid derived from pFAC1 (4) into a B. cinerea sensitive recipient strain, the resulting transformants acquired fenhexamid resistance, demonstrating the involvement of the F412S and F412I mutations in the HydR3+ phenotype. On the other hand, the functional validation of 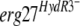 alleles using the same approach did not result in fenhexamid-resistant transformants. The possibility that these alleles could be recessive in an
alleles using the same approach did not result in fenhexamid-resistant transformants. The possibility that these alleles could be recessive in an 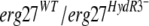 merodiploid was excluded using the complementary experiment: introducing the erg27WT allele into a HydR3− strain. The resulting transformants kept the HydR3− phenotype. One possible explanation for the lack of fenhexamid resistance in the
merodiploid was excluded using the complementary experiment: introducing the erg27WT allele into a HydR3− strain. The resulting transformants kept the HydR3− phenotype. One possible explanation for the lack of fenhexamid resistance in the 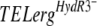 transformants might be the weak expression of the plasmid-borne erg27 alleles. This is corroborated by the low percentage of TELergF412S or TELergF412I transformant spores that present fenhexamid resistance.
transformants might be the weak expression of the plasmid-borne erg27 alleles. This is corroborated by the low percentage of TELergF412S or TELergF412I transformant spores that present fenhexamid resistance.
Finally, we could show the physical link between the HydR3− mutations and fenhexamid resistance by crossing HydR3− strains with a HydS tester strain. All HydR3− progeny strains harbored the same mutation as did the parental HydR3− strain, whereas the sensitive progeny presented the wild-type allele. Genetic crosses remain the principal approach to show genetic links between genotypes and phenotypes in B. cinerea, although they are time-consuming. Reverse genetics using site-directed mutagenesis, therefore, should be preferred, being more rapid and more precise.
In this study we wanted to take advantage of the simplicity of transformation with a replicative plasmid for functional analysis. Indeed, plasmids carrying telomeric sequences can transform filamentous ascomycetes at high frequencies (16, 17, 26). Barreau et al. (4) developed a replicative plasmid that easily transforms several fungal species and that is maintained at low copy numbers, therefore allowing rapid functional validations of dominant alleles. Indeed we were able to show for the first time a correlation between a mutation and fungicide resistance for a phytopathogenic fungus with this replicative plasmid. Although the system cannot be used for all resistance alleles (this study and our unpublished results), it may be useful for a rapid functional test in other cases.
The evaluation of risk assessment comprises the survey of field populations with respect to a given fungicide but also the characterization of resistance phenomena. Once the molecular basis of resistance is known, functional studies can be performed concerning the relationship between the fungicide and its target, or the fitness of resistant strains (5), for developing molecular diagnostics of resistance alleles in fungal populations (25) and molecules with different specificities toward the target protein.
HydR3+ strains are predominant fenhexamid-resistant isolates of French vineyards (but are also found in Germany). The mutation of a single amino acid (F412) that is responsible for resistance should be easily detectable by allele-specific PCR. Future fenhexamid resistance monitoring may make use of quantitative real-time PCR. HydR3− isolates show a highly variable erg27 sequence with at least six identified mutations. Other mutations might exist in the 5′ region of the gene that was not covered by our sequences. It is interesting that all HydR3− isolates were identical at the polymorphic nucleotide positions (2024, 2461, and 2518). They may originate from a different B. cinerea subpopulation. The practical incidence of HydR3+ and HydR3− isolates in the vineyards remains to be investigated, with respect to their competitiveness compared to that of fenhexamid-sensitive strains. The study by Ziogas et al. (31) on laboratory-isolated fenhexamid-resistant strains showed reduced pathogenicity and other fitness parameters without selective pressure. These features need to be tested on natural isolates. Competition experiments, using allele-specific quantitative PCR on characterized isolates, are suggested by the results presented here.
Supplementary Material
Acknowledgments
We are grateful to David Tepfer for correcting the English.
Footnotes
Published ahead of print on 8 September 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Akins, R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43:285-318. [DOI] [PubMed] [Google Scholar]
- 2.Albertini, C., and P. Leroux. 2004. A Botrytis cinerea putative 3-keto reductase gene (ERG27) that is homologous to the mammalian 17-beta-hydroxysteroid dehydrogenase type 7 gene. Eur. J. Plant Pathol. 110:723-733. [Google Scholar]
- 3.Albertini, C., G. Thebaud, E. Fournier, and P. Leroux. 2002. Eburicol 14α-demethylase gene (CYP51) polymorphism and speciation in Botrytis cinerea. Mycol. Res. 106:1171-1178. [Google Scholar]
- 4.Barreau, C., M. Iskandar, B. Turcq, and J. P. Javerzat. 1998. Use of a linear plasmid containig telomeres as an efficient vector for direct cloning in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 25:22-30. [DOI] [PubMed] [Google Scholar]
- 5.Brent, K. J., and D. W. Hollomon. 2007. Fungicide resistance: the assessment of risk, p. 52. FRAC monograph 2. Croplife International, Brussels, Belgium.
- 6.Büttner, P., F. Koch, K. Voigt, T. Quidde, S. Risch, R. Blaich, B. Brückner, and P. Tudzynski. 1994. Variations in ploidy among isolates of Botrytis cinerea: implications for genetic and molecular analyses. Curr. Genet. 25:445-450. [DOI] [PubMed] [Google Scholar]
- 7.Debieu, D., J. Bach, M. Hugon, C. Malosse, and P. Leroux. 2001. The hydroxyanilide fenhexamid, a new sterol biosynthesis inhibitor fungicide efficient against the plant pathogenic fungus Botryotinia fuckeliana (Botrytis cinerea). Pest Manag. Sci. 57:1060-1067. [DOI] [PubMed] [Google Scholar]
- 8.De Guido, M. A., R. M. De Miccolis Angelini, S. Pollastro, A. Santomauro, and F. Faretra. 2007. Selection and genetic analysis of laboratory mutants of Botryotinia fuckeliana resistant to fenhexamid. J. Plant Pathol. 89:203-210. [Google Scholar]
- 9.Dellaporta, S. L., J. Wood, and J. B. Hicks. 1983. A plant DNA minipreparation: version 2. Plant Mol. Biol. Rep. 1:19-21. [Google Scholar]
- 10.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 11.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 12.Faretra, F., and E. Antonacci. 1987. Production of apothecia of Botryotinia fuckeliana (de Bary) Whetz under controlled environmental conditions. Phytopathol. Mediterr. 26:29-35. [Google Scholar]
- 13.Faretra, F., E. Antonacci, and S. Pollastro. 1988. Sexual behaviour and mating system of Botryotinia fuckeliana, teleomorph of Botrytis cinerea. J. Gen. Microbiol. 134:2543-2550. [Google Scholar]
- 14.Fournier, E., T. Giraud, C. Albertini, and Y. Brygoo. 2005. Partition of the Botrytis cinerea complex in France using multiple gene genealogies. Mycologia 97:1251-1267. [DOI] [PubMed] [Google Scholar]
- 15.Fournier, E., C. Levis, D. Fortini, P. Leroux, T. Giraud, and Y. Brygoo. 2003. Characterization of Bc-hch, the Botrytis cinerea homolog of the Neurospora crassa het-c vegetative incompatibility locus, and its use as a population marker. Mycologia 95:251-261. [PubMed] [Google Scholar]
- 16.Javerzat, J. P., V. Bhattacherjee, and C. Barreau. 1993. Isolation of telomeric DNA from the filamentous fungus Podospora anserina and construction of a self-replicating linear plasmid showing high transformation frequency. Nucleic Acids Res. 21:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kistler, H. C., and U. Benny. 1992. Autonomously replicating plasmids and chromosome rearrangement during transformation of Nectria haematococca. Gene 117:81-89. [DOI] [PubMed] [Google Scholar]
- 18.Koller, W. 1992. Antifungal agents with target site in sterol functions and biosynthesis, p. 119-206. In W. Koller (ed.), Target sites of fungicide action. CRC Press, Boca Raton, FL.
- 19.Leroux, P. 2004. Chemical control of Botrytis cinerea and its resistance to chemical fungicides, p. 195-222. In Y. Elad, B. Williamson, P. Tudzynski, and N. Delen (ed.), Botrytis: biology, pathology and control. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 20.Leroux, P., J. Bach, D. Debieu, S. Fillinger, R. Fritz, and A. S. Walker. Mode of action of sterol biosynthesis inhibitors and resistance phenomena in fungi. In H.-W. Dehne (ed.), Modern fungicides and antifungal compounds V. BCPC, Alton, United Kingdom, in press.
- 21.Leroux, P., F. Chapeland, D. Desbrosses, and M. Gredt. 1999. Patterns of cross-resistance to fungicides in Botryotinia fuckeliana (Botrytis cinerea) isolates from French vineyards. Crop Prot. 18:687-697. [Google Scholar]
- 22.Leroux, P., D. Debieu, C. Albertini, A. Arnold, J. Bach, F. Chapeland, E. Fournier, R. Fritz, M. Gredt, T. Giraud, M. Hugon, C. Lanen, C. Malosse, and G. Thebaud. 2002. The hydroxyanilide botryticide fenhexamid/mode of action and mechanism of resistance, p. 29-40. In H.-W. Dehne, U. Gisi, K. H. Kuck, P. E. Russel, and H. Lyr (ed.), Modern fungicides and antifungal compounds III. AgroConcept GmbH, Bonn, Germany.
- 23.Leroux, P., M. Gredt, A.-S. Walker, and M.-L. Panon. 2006. Botrytis de la vigne et neuf catégories de fongicides. Caractéristiques et distribution des souches résistantes dans le vignoble champenois. Phytoma Def. Veg. 599:31-35. [Google Scholar]
- 24.Levis, C., D. Fortini, and Y. Brygoo. 1997. Transformation of Botrytis cinerea with the nitrate reductase gene (niaD) shows a high frequency of homologous recombination. Curr. Genet. 32:157-162. [DOI] [PubMed] [Google Scholar]
- 25.Ma, Z., and T. J. Michailides. 2005. Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi. Crop Prot. 24:853-863. [Google Scholar]
- 26.Powell, W. A., and H. C. Kistler. 1990. In vivo rearrangement of foreign DNA by Fusarium oxysporum produces linear self-replicating plasmids. J. Bacteriol. 172:3163-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1997. Restoration of wild-type virulence to Tri5 disruption mutants of Gibberella zeae via gene reversion and mutant complementation. Microbiology 143:2583-2591. [DOI] [PubMed] [Google Scholar]
- 28.Rosslenbroich, H.-J. 1999. Efficacy of fenhexamid (KBR 2738) against Botrytis cinerea and related fungal pathogens. Pflanzenschutz-Nachr. 52:127-144. [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Suty, A., R. Pontzen, and K. Stenzel. 1999. Fenhexamid—sensitivity of Botrytis cinerea: determination of baseline sensitivity and assessment of the resistance risk. Pflanzenschutz-Nachr. Bayer 52:149-161. [Google Scholar]
- 31.Ziogas, B. N., A. N. Markoglou, and A. A. Malandrakis. 2003. Studies on the inherent resistance risk to fenhexamid in Botrytis cinerea. Eur. J. Plant Pathol. 109:311-317. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.