Abstract
We investigated the basis of the carbapenem resistance of 17 multidrug-resistant Acinetobacter baumannii clinical isolates collected from 2004 to 2005 at the Saint George University Hospital in Beirut, Lebanon. A. baumannii isolates were clonally related and were susceptible to colistin and trimethoprim-sulfamethoxazole, susceptible or intermediate to ampicillin-sulbactam and meropenem, and resistant to all other antimicrobials. Conjugation experiments demonstrated that resistance to imipenem could be transferred along with a plasmid containing the carbapenem-hydrolyzing oxacillinase blaOXA-58 gene. The plasmid that we called pABIR was 29,823 bp in size and showed a novel mosaic structure composed of two origins of replication, four insertion sequence (IS) elements, and 28 open reading frames. The blaOXA-58 gene was flanked by IS18 and ISAba3 elements at the 5′ and 3′ ends, respectively. The production of the carbapenem-hydrolyzing oxacillinase OXA-58 was apparently the only mechanism for carbapenem resistance in A. baumannii isolates causing the outbreak at the Lebanese Hospital.
Acinetobacter baumannii is an important opportunistic pathogen responsible for a variety of nosocomial infections, especially in intensive care unit (ICU) patients (11, 18). These organisms are frequently resistant to multiple antimicrobial agents including broad-spectrum β-lactams, carbapenems, aminoglycosides, and fluoroquinolones (11, 18, 31). A. baumannii may develop resistance to carbapenems through various mechanisms including decreased permeability because of porin modifications or reduced expression, the overexpression of efflux pumps, and the production of carbapenemases, such as metallo-β-lactamase or carbapenem-hydrolyzing oxacillinases (CHDLs) (3, 17-24).
The emergence of carbapenem resistance in A. baumannii has been reported worldwide (11, 18) and has been correlated in Europe with the acquisition of CHDLs (10, 18-23). Three main acquired CHDL gene clusters have been identified in A. baumannii, represented by the blaOXA-23-, blaOXA-24-, and blaOXA-58-like genes (13). Plasmid-borne blaOXA-23 and blaOXA-58 genes have been shown to contribute significantly to carbapenem resistance in A. baumannii (3, 10, 13, 18, 21, 22). In particular, the blaOXA-58 gene has been identified in carbapenem-resistant A. baumannii isolates worldwide (3, 13, 20, 23, 30). Recent studies have shown that the flanking insertion sequence (IS) elements ISAba1, ISAba2, ISAba3, and IS18 regulate blaOXA-58 gene expression (22), and ISAba3 possibly regulates its acquisition (19).
An outbreak of multidrug-resistant Acinetobacter baumannii was observed between November 2004 and October 2005 in the Saint George University Hospital of Beirut, Beirut, Lebanon. The aim of the present study was to (i) assess the genetic relatedness and the antimicrobial susceptibility of A. baumannii isolates in the hospital, (ii) study the horizontal gene transfer of the carbapenem resistance of the A. baumannii isolates, and (iii) analyze plasmid DNA sequences involved in the acquisition of carbapenem resistance of the A. baumannii isolates.
MATERIALS AND METHODS
Microbiological methods.
A. baumannii isolates were identified as being A. baumannii spp. by using the Vitek 2 automatic system with an ID-GNB card for the identification of gram-negative bacilli according to the manufacturer's instructions (bioMerieux, Marcy-l'Etoile, France). Species identification was confirmed by PCR amplification and sequence analysis of the 16S-23S rRNA intergenic spacer region (6).
Antimicrobial susceptibility testing.
MICs were determined by a microdilution method according to Clinical and Laboratory Standards Institute (CLSI) document M7-A6 (8). Breakpoint values were those recommended by the CLSI (8). Breakpoints for colistin were those from the British Society for Antimicrobial Chemotherapy (4). Etest MBL strips (AB Biodisk, Solna, Sweden) were used to evaluate the presence of metallo-beta-lactamase (MBL) activity according to the manufacturer's procedures (16). Pseudomonas aeruginosa ATCC 27853 was used as an MBL-negative reference strain, and A. baumannii AC-54/97 producing IMP-2 MBL (24) was used as the MBL-positive reference strain. The relative contribution of oxacillinases to carbapenem resistance was assessed by analyzing carbapenem MICs with and without 200 mM of NaCl (23) through a liquid microdilution method. A. baumannii isolates of pulsed-field gel electrophoresis (PFGE) type 1 carrying blaOXA-58 (30) and one sporadic A. baumannii isolate of PFGE type 2 negative for blaOXA-58 (30) were used as CHDL-positive and CHDL-negative reference strains, respectively. The contribution of AmpC beta-lactamase was tested by determining carbapenem MICs with and without 200 mg/liter of cloxacillin (23) through a liquid microdilution method.
PFGE analysis and sequencing typing.
DNA macrorestriction of A. baumannii isolates, PFGE, and dendrogram analysis were performed as previously reported (30). Sequencing typing (ST) was performed as described previously (27).
Mating experiments.
Filter mating was performed using A. baumannii isolates Ab 1 or Ab 8 of PFGE type A, resistant to imipenem and susceptible to trimethoprim-sulfamethoxazole, and Ab F isolate of PFGE type B, susceptible to imipenem while resistant to trimethoprim-sulfamethoxazole, as donor and recipient cells, respectively. Transconjugants were selected on brain heart infusion agar plates containing imipenem (16 mg/liter) plus trimethoprim-sulfamethoxazole (250 mg/liter). The frequency of transfer was calculated as the number of transconjugants divided by the number of surviving recipients.
Plasmid DNA characterization and PCR analysis.
Plasmid DNA preparations were performed by using the QIAfilter Plasmid Purification Maxi kit adapted for low-copy-number plasmids (Qiagen Corporation, Milan, Italy) according to the manufacturer's procedure. Southern hybridization of plasmid profiles was performed as described previously by Sambrook et al. (25). PCR analysis for carbapenemase-encoding genes in Acinetobacter spp. (blaIMP, blaVIM, blaSIM, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58) was performed as previously described (30). PCR characterization of the blaOXA-58-surrounding IS was performed as described previously (22). The colinearity between IS elements and the blaOXA-69 gene was analyzed using primers for IS elements described previously by Poirel and Nordmann (22) and for the blaOXA-51-like gene described previously by Turton et al. (28). PCR amplification of the complete carO gene was performed as described previously (17).
Outer membrane protein analysis.
Outer membrane protein fractions were prepared by sonication and solubilization in 2% sodium lauroyl sarcosinate and analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions as previously described (14). N-terminal sequence analysis by automated Edman degradation and protein analysis by matrix-assisted laser desorption-ionization mass spectrometry or liquid chromatography online tandem mass spectrometry were performed as previously described (5, 9).
Plasmid DNA sequencing and computer analysis of sequencing data.
A “walking primer” approach starting with primers derived from the 5′ and 3′ ends of the blaOXA-58 gene was adopted to obtain the complete DNA sequence of the plasmids. Direct sequencing of Qiagen-purified plasmid DNA was performed using the ABI Prism BigDye Terminator v3.1 Ready Reaction cycle sequencing kit and the 3730 DNA analyzer (Applied Biosystems, Foster City, CA). DNA sequences were assembled using the program Autoassembler, version 1.4 (Applied Biosystems, Foster City, CA), and annotated using the BLAST program (1) and the sequence annotation tools integrated into the Sequin program, version 7.9 (available at http://www.ncbi.nlm.nih.gov/Sequin/index.html). The graphic view of plasmid DNA sequences was generated using the program VectorNti, version 10 (Invitrogen Corporation, Carlsbad, CA).
Nucleotide sequence accession numbers.
The nucleotide sequences of A. baumannii plasmid pABIR from A. baumannii Ab1:AbF transconjugant 1 and of carO genes from A. baumannii isolates Ab 1 and Ab F have been deposited in the GenBank nucleotide database under accession numbers EU294228, DQ642020, and DQ642021, respectively. The annotation of plasmid pABIR performed by the National Center for Biotechnology Information is also available in the GenBank genome database under accession number NC_010481.
RESULTS AND DISCUSSION
Molecular epidemiology of A. baumannii in the hospital.
The molecular epidemiology of a clonal outbreak of carbapenem-resistant A. baumannii infection was studied in Saint George University Hospital, Beirut, Lebanon, from November 2004 to October 2005. In the previous 12 months, only sporadic A. baumannii strains that were susceptible to carbapenems were isolated in the hospital. From November 2004 to October 2005, A. baumannii was isolated from 17 patients: 11 from the medical-surgical ICU and 6 from other wards. Ten patients had ventilator-associated pneumonia, three had wounds or abscesses, one had hospital-acquired pneumonia, one had pleural effusion, one had bacteremia, and one had urinary tract infection. Crude mortality was 35% (6/17).
Molecular typing by PFGE and dendrogram analysis identified one major PFGE pattern in all 17 A. baumannii isolates during the outbreak and one additional PFGE pattern in one sporadic isolate from the ICU of the hospital 6 months ahead that differed in the migration of more than six bands and exhibited <70% similarity, which we named A and B, respectively. Of the 17 outbreak A. baumannii isolates, 12 showed an identical macrorestriction pattern, which we named pattern A, and 5 showed two- to three-fragment variations in the macrorestriction pattern with a similarity of more than 80% by dendrogram analysis and were classified into three subtypes, A1 to A3. ST analysis assigned the 17 outbreak isolates of PFGE type A to ST group 2 and the sporadic isolate of PFGE type B to ST group 3. During the epidemic, A. baumannii isolates of PFGE type A were also obtained from one humidifier and one sink of two rooms of the ICU ward. Data indicate that the A. baumannii outbreak in the hospital was caused by the spread of a single epidemic clone.
Antimicrobial susceptibility patterns of A. baumannii isolates.
All A. baumannii isolates of PFGE type A showed an identical multiresistant antibiotype. In particular, they were resistant or intermediate to imipenem, susceptible or intermediate to meropenem and ampicillin-sulbactam, and susceptible to colistin and trimethoprim-sulfamethoxazole (Table 1). In contrast, the sporadic isolate of PFGE type B was susceptible to imipenem and meropenem and resistant to trimethoprim-sulfamethoxazole.
TABLE 1.
Antibiotic susceptibility profile of A. baumannii outbreak isolatesa
| Antibiotic | MIC50 (mg/liter) | MIC90 (mg/liter) | MIC range (mg/liter) |
|---|---|---|---|
| Sulbactam-ampicillin | 8 | 16 | 8-16 |
| Piperacillin-tazobactam | 125 | 250 | 125->250 |
| Ceftazidime | 250 | 250 | 125->250 |
| Cefepime | 16 | 32 | 16-32 |
| Imipenem | 16 | 16 | 8-16 |
| Meropenem | 8 | 8 | 4-8 |
| Amikacin | 64 | 125 | 64-125 |
| Gentamicin | 125 | 250 | 125->250 |
| Ciprofloxacin | 64 | 250 | 32->250 |
| Trimethoprim-sulfamethoxazole | 0.5 | 0.5 | 0.5-1 |
| Colistin | 2 | 4 | 1-4 |
A. baumannii isolates of PFGE type A were analyzed by a microdilution method for MIC determination according to CSLI guidelines.
To study the mechanism of carbapenem resistance, the MIC of imipenem and the presence of MBL activity were evaluated for all A. baumannii isolates of PFGE type A by using Etest MBL strips. All isolates were resistant or intermediate to imipenem (MICs from 8 to 16 mg/liter) but negative for MBL production (imipenem-EDTA MIC, 8 to 4 mg/liter). To further characterize the carbapenem resistance, we studied the relative contribution of oxacillinases and AmpC beta-lactamase to imipenem resistance by analyzing imipenem MICs in the presence of 200 mM NaCl or 200 mg/liter cloxacillin for A. baumannii isolates of PFGE type A through a microdilution method. These experiments showed that imipenem MICs (16 mg/liter) were inhibited by up to eightfold in the presence of NaCl (2 mg/liter) but not in the presence of cloxacillin. No changes in ceftazidime MICs (250 mg/liter) were observed in the presence of NaCl or cloxacillin. The above-described data suggested that oxacillinase activity, but not AmpC activity, contributed to imipenem resistance in epidemic A. baumannii isolates.
Molecular analysis of carbapenem resistance in A. baumannii isolates.
PCR and sequence analysis identified a blaOXA-58 gene in DNA from all imipenem-resistant A. baumannii isolates of PFGE type A but not from the imipenem-susceptible isolate of PFGE type B. No amplification products were obtained using primers for blaIMP-type, blaVIM-type, or blaSIM-type MBLs or blaOXA-23 or blaOXA-24 CHDLs. This suggested that OXA-58 was the oxacillinase contributing to carbapenem resistance. Also, PCR experiments failed to identify any IS element upstream of the naturally occurring blaOXA-69 gene in imipenem-resistant A. baumannii isolates of PFGE type A, thus excluding that IS-mediated overexpression of this oxacillinase may account for the resistance to imipenem (28).
Molecular analysis of the outer membrane protein profile of A. baumannii isolates.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis showed an outer membrane profile composed of two major protein bands with apparent molecular masses of 24 and 37 kDa in the imipenem-resistant A. baumannii isolates Ab 1 and Ab 8 and 27 and 37 kDa in the imipenem-susceptible A. baumannii isolate Ab F. Automated Edman degradation analysis of the 24- and 27-kDa proteins from the imipenem-resistant and imipenem-susceptible isolates showed identical N-terminal sequences that corresponded to those of the outer membrane protein CarO, whose loss has been associated with carbapenem resistance (17). Mass spectrometry analysis of the CarO protein from imipenem-resistant isolate Ab 1 identified the peptides expected from the hydrolysis of the deduced amino acid sequence that was identical to the deduced amino acid sequence reported previously by Mussi et al. (GenBank accession number AY684798) (17) except for a glutamine replacing the lysine at residue 197. This demonstrated that the full-length CarO protein was expressed in imipenem-resistant isolate Ab 1 and ruled out that the apparent lower molecular weight of the CarO protein from imipenem-resistant isolates was caused by a truncation at the C terminus. Therefore, data excluded that carbapenem resistance in epidemic A. baumannii isolates was contributed by modifications in the outer membrane protein profile.
Conjugative transfer of imipenem resistance.
To further study the mechanism responsible for imipenem resistance, we asked whether imipenem resistance might have been transferred through conjugation. Filter-mating experiments demonstrated that resistance to imipenem was transferred from isolates Ab 1 and Ab 8 of PFGE type A and ST group 2 to imipenem-susceptible isolate Ab F of PFGE type B and ST group 3 at a frequency ranging from 2 × 10−5 to 1.5 × 106. All the transconjugants showed the PFGE profile and the ST group of the recipient isolate Ab F (data not shown). The antimicrobial susceptibility profile of the transconjugants was identical to that of the recipient isolate Ab F with the exclusion of imipenem and meropenem, being susceptible to colistin, intermediate to ampicillin-sulbactam and meropenem, and resistant to all other antimicrobials including imipenem. Imipenem MICs for transconjugants were similar (16 mg/liter) to those for donor isolates and were inhibited by up to eightfold in the presence of 200 mM NaCl, thus suggesting that imipenem resistance was contributed by oxacillinase activity in the transconjugants also.
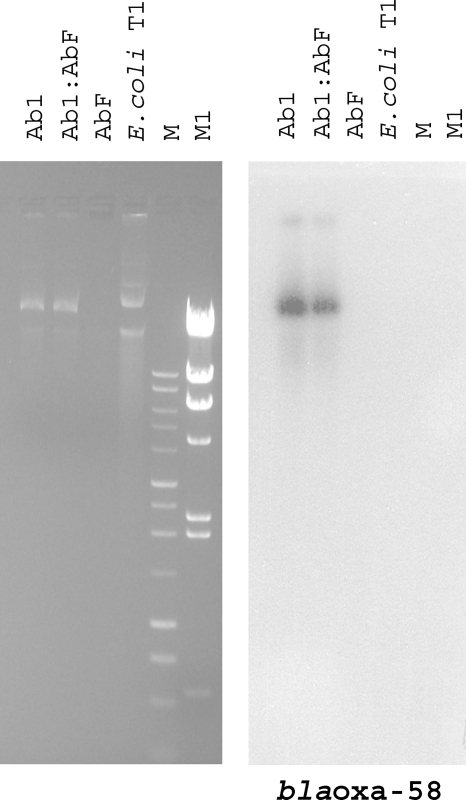
Both imipenem-resistant A. baumannii isolate Ab 1 and the Ab 1:Ab F transconjugant were shown to carry a single plasmid molecule that migrated faster than an approximately 80-kb plasmid extracted from a Klebsiella pneumoniae-Escherichia coli transconjugant (2) and that hybridized with a PCR-generated probe for blaOXA-58; in contrast, no plasmid DNA was isolated from imipenem-susceptible A. baumannii isolate Ab F (Fig. 1).
FIG. 1.
Plasmid profiles and identification of the plasmid carrying the blaOXA-58 gene in A. baumannii isolates. Agarose (0.8%) gel electrophoresis in 1× Tris-acetate-EDTA buffer of plasmid preparations from A. baumannii isolates Ab 1, Ab 1:Ab F, and Ab F and from a K. pneumoniae-E. coli transconjugant (E. coli T1) stained with ethidium bromide and visualized under UV light and Southern blot hybridization with the blaOXA-58 probe are shown. M is a 1-kb DNA ladder (Promega, Milan, Italy), and M1 is HindIII-digested lambda DNA (Invitrogen, Milan, Italy).
Genetic structure of plasmid pABIR.
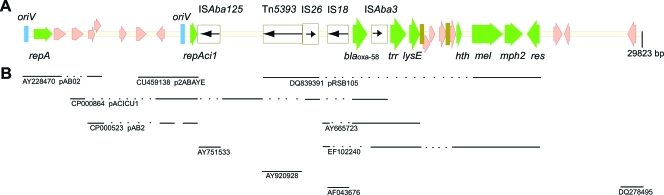
The direct sequence of plasmid preparations from isolate Ab 1 and one Ab1:AbF transconjugant identified an identical plasmid that was designated pABIR, for Acinetobacter baumannii imipenem resistance. Plasmid pABIR was 29,823 bp in size, with a G+C content of 36.8%. The analysis of pABIR identified 28 open reading frames (ORFs). Of these ORFs, 22 were transcribed in a clockwise orientation, while the remaining 6 were transcribed counterclockwise. The main features of pABIR sequences and the regions of similarity with other known sequences are depicted in Fig. 2. Table 2 lists the putative functions, the characteristics, and the closest relatives for the predicted product of each ORF. The origin of replication (oriV), a repeat region composed of four imperfect direct iterons, and the repA gene, coding for RepAB replicase, were located at the beginning of the pABIR sequences and were homologous with those found in the A. baumannii plasmid pAB02 replicon, the partial sequence for which is available (GenBank accession number AY2284790). An additional origin of replication, five direct-repeat iterons, and a gene coding for a DNA replicase, homologous to the repAci1 gene identified in A. baumannii plasmid pACICU1 (15), were also found in plasmid pABIR, where the coding sequences of the repAci1 gene were truncated by the insertion of an ISAb125 element. The additional replicon of pABIR showed high homology with those found in three other A. baumannii plasmids, pACICU1, a 28-kb plasmid containing the blaOXA-58 gene isolated from A. baumannii strain ACICU, the sequence of which has been recently reported (15); pAB2, an 11-kb plasmid from A. baumannii ATCC 17978 (CP000523); and p2ABAYE, an approximately 10-kb plasmid isolated from multidrug-resistant A. baumannii strain AYE (29) (Fig. 2 and Table 2). The presence of two replicons suggests that pABIR is a mosaic plasmid originating from the fusion of two separate molecules. Similarly, two replicons have also been found in plasmid pACICU1 from A. baumannii strain ACICU (15) and in the erythromycin resistance plasmid pRSB105 isolated from activated sludge bacteria, where they have been postulated to represent a cointegrate of two formerly separate replicons (26).
FIG. 2.
Schematic map of plasmid pABIR. (A) Linear map of pABIR with relevant features. ORFs are represented by arrow-shaped boxes. IS elements are represented by empty rectangle boxes filled with black arrows indicating the transposase gene and the direction of the transcription. Repeat regions are indicated by vertical bars. Names of various features are reported below or above the map. (B) Regions of identity or of high similarity with other sequences reported in the GenBank/EMBL database are indicated by continuous lines.
TABLE 2.
Genetic regions and ORFs of plasmid pABIRa
| Feature | Position | Gene product (no. of amino acids) | Properties and/or putative function | Homology (GenBank/EMBL accession no. match) |
|---|---|---|---|---|
| oriV | 1-200 | Origin of DNA replication | pAB02 (AY228470) | |
| Repeat region | 217-312 | Imperfect 4-repeat iterons; control of DNA replication | ||
| repA | 357-1244 | 295 | RepA AB; replicase | pAB02 (AY228470) |
| ORF | 1321-1884 | 188 | Unknown | |
| ORF | 2179-2721 | 180 | Hypothetical protein similar to COG 0790 FOG; TPR repeat, SEL1 subfamily | pMAC (AY541806), pACICU1 (CP000864) |
| ORF | 2964-3260 | 98 | Putative inner membrane protein | pAB02 (AY228470), pAB2 (CP000523) |
| ORF | 3247-3561 | 104 | Putative cytoplasmic protein | pAB02 (AY228470), pAB2 (CP000523) |
| ORF | 4455-4766 | 103 | Unknown | |
| ORF | 5235-5714 | 153 | Unknown | p2ABAYE (CU459138) |
| ORF | 6210-6581 | 123 | Unknown | pACICU1 (CP000864), p2ABAYE (CU459138), pAB2 (CP000523) |
| oriV | 7450-7650 | Origin of DNA replication | ||
| Repeat region | 7651-7760 | 5-repeat iterons; control of DNA replication | ||
| repAci1 | 7815-8150 | 112 | Plasmid replication protein; | |
| 9238-9855 | 205 | truncated by ISAb125 | ||
| ISAb125 | 8160-9230 | 322 | ISAb125/ISAb125 transposase | ISAba125 (AY751533) |
| Tn5393 | 11279-13148 | 611 | Tn5393 transposase disrupted by IS26 insertion | Salmonella enterica serovar Paratyphi A (AM412236); uncultured bacterium pRSB105 (DQ839391) |
| IS26 | 13329-13916 | 195 | IS26 transposase | pACICU1 (CP000864), uncultured bacterium pRSB105 (DQ839391) |
| IS18 | 14349-15422 | 320 | IS18/IS18 transposase | Acinetobacter sp. strain BM2716 (AF043676) |
| blaOXA-58 | 15553-16395 | 280 | Carbapenem-hydrolyzing oxacillinase | AY665723, pACICU1 (CP000864), EF102240 |
| ISAba3 | 16433-17222 | 145 | IS element ISAba3/ISAba3 transposase | AY665723, pACICU1 (CP000864), EF102240 |
| trr | 17366-18193 | 275 | Arac1 binding protein; arabinose operon control protein; transcriptional regulator | AY665723, EF102240 |
| lysE | 18243-18848 | 201 | Threonine efflux protein | AY665723, EF102240 |
| Recombination point | 18843-18869 | Repeat region 1/Re27-1 | AY665723, EF102240 | |
| ORF | 18914-19243 | 109 | Unknown | |
| ORF | 19244-19525 | 93 | Putative transcriptional regulator | Geobacter metallireducens GS-15 (CP000148) (73% identity) |
| ORF | 19605-20030 | 141 | Unknown | |
| Recombination point | 20062-20088 | Repeat region 2/Re27-2 | AY665723, EF102240 | |
| ORF | 20164-20484 | 106 | Unknown | EF102240 |
| ORF | 20477-20749 | 90 | Helix-turn-helix XRE family cds | EF102240 |
| mel | 21242-22717 | 491 | Macrolide efflux protein | EF102240, uncultured bacterium pRSB105 (DQ839391) |
| mph2 | 22773-23657 | 294 | Macrolide 2′-phosphotransferase | |
| Resolvase | 23847-24449 | 200 | Resolvase | |
| ORF | 25126-25533 | 135 | Unknown | |
| ORF | 25629-25877 | 82 | Unknown | |
| ORF | 28643-29032 | 129 | Unknown | Acinetobacter venetianus pAV1 (DQ278485) |
Sequences in the GenBank/EMBL database showing identity or high similarity are indicated. Microorganisms from which DNA sequences have been isolated are indicated when different from A. baumannii. COG, conserved domain in bacteria; cds, coding sequences.
pABIR contained a single copy of the blaOXA-58 gene that was flanked by IS18 and ISAba3 elements at the 5′ and 3′ ends, respectively, as previously described for A. baumannii isolate CH29 (22). Two additional ISs, Tn5393 and IS26, flanked IS18 in pABIR. IS26 was identical to the element found in plasmid pACICU1 from A. baumannii strain ACICU (15) but was inserted in the opposite orientation with respect to the blaOXA-58 gene. In accordance with our data, a single copy of the blaOXA-58 gene in plasmid pOUR from A. baumannii strain 183 showing an imipenem MIC of 16 mg/liter was described previously (3), while two copies of the blaOXA-58 gene were found in plasmid pACICU1 (15), and three copies of the gene were found in plasmids from A. baumannii isolates showing higher MICs (3). Downstream of the ISAba3 element, the trr and lysE genes and two sequences similar to those defined as the Re27-1 and Re27-2 structures in A. baumannii pMAD and hypothesized to be involved in homologous recombination processes (22) were identified. A genetic region containing genes involved in macrolide resistance and the res gene encoding a resolvase site-specific recombinase was identified and was identical to that found on an A. baumannii plasmid carrying the blaOXA-97 carbapenemase gene (GenBank accession number EF102240) (20) and on plasmid pRSB105, isolated from a sewage treatment plant (26).
No genes coding for the conjugative apparatus, the secretion system, or any mobilization protein were found in plasmid pABIR, thus suggesting that the plasmid is not self-conjugative and that the genes that mediate the transfer have been provided in trans. Although the genetic structure of the blaOXA-58 gene and of IS flanking regions of several plasmids have been characterized (3, 22, 26), only two complete plasmids carrying the blaOXA-58 gene have been described so far (7, 15). One of these, pTVICU53, is an 11-kb plasmid that contains a predicted origin-of-transfer DNA region and a gene coding for a mobilization protein (7) and should be transconjugated through a helper plasmid, as previously demonstrated for its homologous plasmid pMAC (12). The other, pACICU1, is a 28-kb plasmid that does not contain sequences for conjugation or mobilization but has been postulated to be mobilized in trans by a complete tra locus, encoding a conjugative apparatus and type IV secretion system, carried by plasmid pACICU2, of 64 kb, which coresides in bacteria with pACICU1 (15). The presence of a single plasmid molecule in donor cells of the isolates described herein led us to exclude that trans-mobilization occurred through a conjugation process promoted by another plasmid coresident within the same cell. trans-Mobilization mediated by chromosomally located transfer systems can also be hypothesized. In further support of this, the loci responsible for the conjugative transfer have been reported to have a chromosomal location in multidrug-resistant A. baumannii strain AYE, which belongs to ST group 2 as A. baumannii donor cells of the conjugation experiments described herein (29).
In conclusion, the acquisition of resistance to carbapenems in A. baumannii from the Lebanese hospital was caused by the spread of plasmid pABIR, carrying the blaOXA-58 gene. The mosaic genetic structure of pABIR might have been generated by multiple recombination events mediated by IS elements.
Acknowledgments
We thank G. M. Rossolini (Università di Siena, Siena, Italy) for kindly providing the A. baumannii AC-54/97 strains, A. Carattoli (Istituto Superiore di Sanità, Rome, Italy) for scientific advice in plasmid characterization, and A. Flagiello (CEINGE Biotecnologie Avanzate, Naples, Italy) for technical support in outer membrane protein studies. We also thank Pier Paolo Di Nocera for critical reading of the manuscript.
This work was supported in part by grants from the Ministero dell'Istruzione, dell'Università e della Ricerca Scientifica e Tecnologica, Italy (PRIN 2004 to R.Z.), and from the Agenzia Italiana del Farmaco (AIFA2007 contract no. FARM7X9F8K). A.U.K. was supported by a grant from the BOYSCAT fellowship program of DST Government of India, New Delhi, India.
Footnotes
Published ahead of print on 25 August 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagattini, M., V. Crivaro, A. Di Popolo, F. Gentile, A. Scarcella, M. Triassi, P. Villari, and R. Zarrilli. 2006. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit. J. Antimicrob. Chemother. 57:979-982. [DOI] [PubMed] [Google Scholar]
- 3.Bertini, A., L. Poirel, S. Bernabeu, D. Fortini, L. Villa, P. Nordmann, and A. Carattoli. 2007. Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:2324-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.British Society for Antimicrobial Chemotherapy. 2006. British Society for Antimicrobial Chemotherapy methods for antimicrobial susceptibility testing, version 6, January 2006. British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom.
- 5.Busiello, I., R. Acquaviva, A. Di Popolo, T. G. Blanchard, V. Ricci, M. Romano, and R. Zarrilli. 2004. Helicobacter pylori γ-glutamyltranspeptidase upregulates COX-2 and EGF-related peptides expression in human gastric cells. Cell. Microbiol. 6:255-267. [DOI] [PubMed] [Google Scholar]
- 6.Chang, H. C., Y. F. Wei, L. Dijkshoorn, M. Vaneechoutte, C. T. Tang, and T. C. Chang. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, T. L., R. C. Wu, M. F. Shaio, C. P. Fung, and W. L. Cho. 2008. Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. Approved standard M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Cobucci-Ponzano, B., F. Conte, D. Benelli, P. Londei, A. Flagello, M. Monti, P. Pucci, M. Rossi, and M. Moracci. 2006. The gene of an archaeal α-L-fucosidase is expressed by translational frameshifting. Nucleic Acids Res. 34:4258-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coelho, J. M., J. F. Turton, M. E. Kaufmann, J. Glover, N. Woodford, M. Warner, M.-F. Palepou, R. Pike, T. L. Pitt, B. C. Patel, and D. M. Livermore. 2006. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J. Clin. Microbiol. 44:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 12.Dorsey, C. W., A. P. Tomaras, and L. A. Actis. 2006. Sequence and organization of pMAC, an Acinetobacter baumannii plasmid harboring genes involved in organic peroxide resistance. Plasmid 56:112-123. [DOI] [PubMed] [Google Scholar]
- 13.Heritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez-Alles, S., S. Alberti, D. Alvarez, A. Domenech-Sanchez, L. Martinez-Martinez, J. Gil, J. M. Tomas, and V. J. Benedì. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 15.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. P. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole genome pyrosequencing of an epidemic multidrug resistant Acinetobacter baumannii of the European clone II. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, K., D. Yong, J. H. Yum, Y. S. Lim, A. Bolmström, A. Qwärnström, Å. Karlsson, and Y. Chong. 2005. Evaluation of Etest MBL for detection of blaIMP-1 and blaVIM-2 allele-positive clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 43:942-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., E. Lebessi, C. Heritier, A. Patsoura, M. Foustoukou, and P. Nordmann. 2006. Nosocomial spread of OXA-58-positive carbapenem-resistant Acinetobacter baumannii isolates in a pediatric hospital in Greece. Clin. Microbiol. Infect. 12:1138-1141. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, L., W. Mansour, O. Bouallegue, and P. Nordmann. 2008. Carbapenem-resistant Acinetobacter baumannii isolates producing the OXA-58-like carbapenem-hydrolyzing oxacillinase OXA-97 from Tunisia. Antimicrob. Agents Chemother. 52:1613-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., S. Marqué, C. Heritier, C. Segonds, G. Chabanon, and P. Nordmann. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel, L., and P. Nordmann. 2006. Genetic structure at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pournaras, S., A. Markogiannakis, A. Ikonomidis, L. Kondyli, K. Bethimouti, A. N. Maniatis, N. J. Legakis, and A. Tsakris. 2006. Outbreak of multiple clones of imipenem-resistant Acinetobacter baumannii isolates expressing OXA-58 carbapenemase in an intensive care unit. J. Antimicrob. Chemother. 57:557-561. [DOI] [PubMed] [Google Scholar]
- 24.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Schulter, A., R. Szczepanowski, N. Kurz, S. Schneiker, I. Krahn, and A. Puhler. 2007. Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbors a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Appl. Environ. Microbiol. 73:1952-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turton, J. F., S. N. Gabriel, C. Valderrey, M. E. Kauffmann, and T. L. Pitt. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807-815. [DOI] [PubMed] [Google Scholar]
- 28.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 29.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Arbegel, J.-M. Claverie, D. Raoult, C. Médigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS ONE 3:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarrilli, R., R. Casillo, A. Di Popolo, M.-F. Tripodi, M. Bagattini, S. Cuccurullo, V. Crivaro, E. Ragone, A. Mattei, N. Galdieri, M. Triassi, and R. Utili. 2007. Molecular epidemiology of a clonal outbreak of multidrug-resistant Acinetobacter baumannii in a university hospital in Italy. Clin. Microbiol. Infect. 13:481-489. [DOI] [PubMed] [Google Scholar]
- 31.Zarrilli, R., M. Crispino, M. Bagattini, E. Barretta, A. Di Popolo, M. Triassi, and P. Villari. 2004. Molecular epidemiology of sequential outbreaks of Acinetobacter baumannii in an intensive care unit shows the emergence of carbapenem resistance. J. Clin. Microbiol. 42:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]