Abstract
Sulfadoxine-pyrimethamine (SP) resistance in Plasmodium falciparum is encoded by a number of mutations in the dihydrofolate reductase (dhfr) and dihydropteroate synthetase (dhps) genes. Here, we have characterized point mutations in dhfr and dhps and microsatellite loci around dhfr on chromosome 4 and dhps on chromosome 8 as well as neutral markers on chromosomes 2 and 3 in 332 samples from Yaoundé, Cameroon. The triple mutant dhfr haplotype that originated in Southeast Asia is the most predominant in this sample set, but we also find additional independent haplotypes at low frequency and an incipient process of genetic differentiation among alleles of Southeast Asian origin. As reported for other African populations, we find evidence of a selective sweep for resistant dhfr mutants in this Cameroonian population due to drug selection. Although we find evidence for a selective sweep in dhps mutants associated with SP resistance, the dynamics of dhps mutants appear different than those observed for dhfr mutants. Overall, our results yield support for the use of microsatellite markers to track resistant parasites; however, the detection of resistant dhfr alleles in low frequency, the evidence of divergence among dhfr alleles that share a common evolutionary origin, and the distinct dynamics of resistant dhps alleles emphasize the importance of comprehensive, population-based investigations to evaluate the effects of drug selection on parasite populations.
Plasmodium falciparum resistance to the most commonly used antimalarial drugs has been detected worldwide, reaching the level of a public health emergency (15, 37). Resistance to chloroquine has led to the discontinued use of the drug in many parts of the world, and resistance to sulfadoxine-pyrimethamine (SP), an affordable and widely available alternative to chloroquine, has rendered this drug ineffective in many areas as well.
Malaria control programs around the world are turning to artemisinin-based combination therapies. However, policy decisions to delay the emergence of resistance against artemisinin-based combination therapies must be made before critical information is widely available. Thus, the fundamental understanding of how resistance against drugs such as SP and chloroquine emerges and how this resistance disseminates will provide critical information for developing strategies to identify and contain resistance to other drugs. In addition, because of its safety for pregnant women and infants and its long action, SP is the only drug recommended for intermittent preventive treatment in these vulnerable populations, and new antifolate combinations are under development (15). Thus, understanding the dynamics of mutations associated with resistance against SP is still a matter of great epidemiologic and public health importance.
SP acts as an inhibitor of the P. falciparum folic acid pathway, and point mutations in the genes encoding dihydrofolate reductase (DHFR) and dihydropteroate synthetase (DHPS) have been implicated in SP resistance (16). Point mutations at dhfr codons 50, 51, 59, 108, and 164 act synergistically to increase resistance to pyrimethamine. Specifically, the S108N mutant has a low level of resistance or tolerance, the N51I/S108N and C59R/S108N double mutants have moderate levels of resistance, the N51I/C59R/S108N triple mutant has a higher level, and the N51I/C59R/S108N/I164L quadruple mutant parasite is considered to be resistant to the effects of pyrimethamine (11, 28). C50R aids to increase the level of resistance but only recently has been detected outside of South America (22). Similarly, mutations at dhps codons 436, 437, 540, 581, and 613 act synergistically to increase the level of resistance to sulfadoxine. Simply, the mutations S436A and A437G alone confer a low level of resistance, and when these are in a combination with K540E, A581G, and/or A613S, the parasite has an increased level of resistance to sulfadoxine (15, 38).
There are few but compelling studies that address the genetic consequences of drug selection on mutations associated with resistance in the gene encoding DHFR. Natural selection can act to increase the frequency of a beneficial allele to fixation in the population, and this process is usually referred to as a selective sweep (33). By such processes, mutations conferring resistance to antimalarial drugs remove linked neutral variation while they sweep through a parasite population, a process termed “genetic hitchhiking” (33). Selective sweeps of highly resistant dhfr alleles have been described for samples from Southeast Asia, Venezuela, South Africa, and Tanzania (21, 23, 27). All of these studies emphasize the lack of variation around dhfr, which is expected under the model of a selective sweep (33). However, only one study, carried out with samples from Venezuela, has examined the consequences of selection on dhps alleles and has shown a similar lack in variation surrounding dhps as a consequence of selection (21). In addition, no studies of dhfr have been carried out in western or central Africa.
Recent studies have shown a single microsatellite haplotype for highly pyrimethamine-resistant dhfr alleles in Southeast Asia, with subsequent spread to African populations (23, 30, 31). This Southeast Asian type is consistently seen in African samples as either the only haplotype or the most predominant haplotype, underlining the importance of gene flow of resistant parasites into P. falciparum populations (18, 22). If just one or a few haplotypes are present in a population, control programs could use molecular markers as part of their surveillance system to track the development and spread of drug-resistant alleles in populations (2). However, additional independent haplotypes have been found in low frequency in Kenyan populations (22).
In this investigation, we have examined the variation around two genes involved in SP resistance in Yaoundé, Cameroon, a central African nation with intense or perennial malaria transmission. In Cameroon, chloroquine was officially removed from the drug policy in 2002 and amodiaquine and SP became the first- and second-line drugs, respectively, for the treatment of uncomplicated P. falciparum. The combination therapy artesunate-amodiaquine was officially adopted as the recommended therapy for uncomplicated malaria in 2004 but became widely available only in 2007. Since 2004, SP has been the recommended policy for the intermittent preventive treatment of malaria during pregnancy and also is available unofficially to patients for self-treatment; thus, selection pressure for SP-resistant mutants still remains present (34). The selection pressure and increase in frequency of SP-resistant parasites across Cameroon have been documented extensively (3-9, 35, 36).
Here, we present evidence for selection on dhfr and dhps alleles in Cameroon and examine the relationship between alleles present in the population. As our previous study in Kenya has shown, we found additional low-frequency novel haplotypes for the triple mutant dhfr alleles in Yaoundé (22). We also found evidence for selective sweeps of dhps alleles, with several alleles associated with sulfadoxine resistance. Finally, we found evidence of linkage disequilibrium (LD) between dhps and dhfr mutations, an association that could be the result of drug selection.
MATERIALS AND METHODS
Study subjects.
We analyzed 332 random samples collected in Yaoundé, Cameroon, in 2001, 2002, 2004, and 2005. Study details have been described previously (5, 36). Whole-blood samples were collected from symptomatic patients ≥12 years old with uncomplicated P. falciparum malaria. Patients were treated with chloroquine, amodiaquine, SP, amodiaquine-sulfadoxine-pyrimethamine, or quinine. The study was approved by the Cameroonian National Ethics Committee and the Cameroonian Ministry of Public Health. Yaoundé is the capital of Cameroon and is in a tropical rain forest. Malaria transmission is typically intense or perennial, with rainy seasons in March through May and September through November. The entomological inoculation rate for Yaoundé has been estimated to be approximately 3 to 33 infective bites per person per year depending on the district (14, 19, 20).
DNA isolation and genotyping methods.
DNA was isolated as previously described (8). One hundred fifty samples were analyzed for mutations in codons 16, 51, 59, 108, and 164 of dhfr by direct DNA sequencing, as described previously (36). One hundred eighty-two samples were analyzed for mutations in codons 50, 51, 59, 108, and 164 of dhfr by pyrosequencing, as described previously (40). All samples were analyzed for mutations in dhps codons 436, 437, 540, 581, and 613 by pyrosequencing (40).
Microsatellite analysis.
Microsatellite characterization was conducted on all samples. Samples were assayed for 18 microsatellite loci that span 148 kb on chromosome 4 around dhfr (23, 30, 31), 15 loci that span 122 kb on chromosome 8 around dhps (21, 27), 4 loci on chromosome 2 that span 78 kb, and 4 loci on chromosome 3 that span 94 kb (21). The microsatellites used around dhfr are at −58, −30, −17, −10, −7.5, −5.3, −4.5, −4.4, −3.8, −1.2, −0.3, 0.2, 0.52, 5.87, 20, 50, 70, and 90 kb, where negative numbers refer to positions 5′ to the gene and positive numbers refer to positions 3′ to the gene. The microsatellites used around dhps are at −50, −17, −10, −7.4, −2.5, −1.6, −0.8, 0.06, 0.144, 1.59, 6.19, 9.8, 17.5, 33.1, and 71.6 kb. The microsatellites used on chromosome 2 are at 302, 313, 319, and 380 kb. The microsatellites used on chromosome 3 are at 335, 363, 383, and 429 kb. Single-reaction PCR and thermal cycling conditions were detailed by Nair et al. (23), and nested PCRs and thermal cycling conditions were detailed by Roper et al. (30). PCRs were performed using PCR Master Mix (Promega, Madison, WI), with a total reaction volume of 15 μl. PCR products were separated with an Applied Biosystems 3130XL capillary sequencer and scored using GeneMapper software v3.7 (Applied Biosystems, Foster City, CA).
In this article, we use the term “allele” for different forms in the dhfr and dhps coding sequences and also for different fragment sizes of microsatellite loci. A sensitive dhfr or dhps allele is one that has no drug-resistant mutations in the coding region. The term “haplotype” refers to a particular multilocus genotype characterized by 11 microsatellite loci spanning 11.5 kb around dhfr or 9 loci spanning 20 kb around dhps. Haplotypes were classified as different if they contained >1 allelic change across microsatellite loci (see Tables S1 and S2 in the supplemental material). Samples for which there were no mixed infections detected by pyrosequencing were used for haplotype characterization.
Statistical analysis.
The genetic variation for each microsatellite locus was measured by calculating the expected heterozygosity (He) and the number of alleles per microsatellite locus (A). He was calculated for each locus as follows: He = [n/(n − 1)](1 − ∑pi2), where n is the number of isolates sampled and pi is the frequency of the ith allele. The sampling variance for He was calculated as follows: 2(n − 1)/n3{2(n − 2)[∑pi3 − (∑pi2)2]} (24). An Excel Microsatellite tool kit was used to compute allele frequencies and A (26).
The genetic relationships among haplotypes were explored using eBURST v3 (13). eBURST identifies mutually exclusive groups of related genotypes and then identifies the founding genotype for each group. eBURST does not allow for any missing data; therefore, haplotypes with missing information for any locus were removed from the analysis. As for haplotype characterization, samples for which there were no multiple infections detected by pyrosequencing were used for eBURST analysis. In the case of multiple infections, where a microsatellite locus may have more than one allele present, the highest peak was used (1).
Population genetic structure was measured using Wright's F statistics (39). The statistic FST (Wright's fixation index) measures genetic differentiation between populations. For the FST analysis, the microsatellite loci from −10 kb to 1.47 kb were used for dhfr and the loci from −2.5 kb to 17.5 kb were used for dhps. FST calculations were computed using Arlequin, version 3.01 (12). An Excel Microsatellite tool kit was used to format data for use in Arlequin (26).
LD between loci along the chromosomes and also between dhfr and dhps point mutations was assessed by using an exact test of LD (29). For the tests of LD among chromosomal positions, loci from chromosomes 4 (around dhfr), 8 (around dhps), 2, and 3 were used. Samples for which there were multiple alleles at any locus were removed from the analysis; this was done for dhfr, dhps, and the neutral markers independently. Similarly, samples for which multiple infections were detected at any site were removed from the LD analysis testing pairs of point mutations in dhfr and dhps; this was also done independently for dhfr and dhps for a given sample. Only polymorphic loci or sites were used for the analysis. Associations between pairs of loci or sites were tested by using 10,000 Monte Carlo steps in Arlequin, version 3.01 (12). To correct for multiple testing, we used a Bonferroni correction.
RESULTS
dhfr and dhps alleles in the population.
Table 1 shows the frequencies of the dhfr and dhps alleles in the Cameroonian population. For both dhfr and dhps, the sensitive alleles are present at only 5% in this population; therefore, 95% of the dhfr and dhps alleles are alleles with mutations associated with drug resistance. Triple mutant (51I/59R/108N) dhfr alleles are present in 88% of the samples. There are multiple dhps alleles present in the population, and dhps mutant alleles with the substitution 437G were observed in 52% of the samples.
TABLE 1.
Frequencies of dhfr and dhps alleles in the sample set from Cameroon
| Type of allelea | No. of occurrences | Frequency |
|---|---|---|
| dhfr | ||
| Sensitive | 14 | 0.05 |
| 108N | 4 | 0.01 |
| 51I/108N | 3 | 0.01 |
| 59R/108N | 14 | 0.05 |
| 51I/59R/108N | 252 | 0.88 |
| dhps | ||
| Sensitive | 13 | 0.05 |
| 436A | 70 | 0.28 |
| 437G | 131 | 0.52 |
| 613S | 4 | 0.02 |
| 436A/437G | 30 | 0.12 |
| 436A/613S | 1 | 0.00 |
| 437G/613S | 3 | 0.01 |
Only alleles from single parasite infections, as determined by pyrosequencing, are represented here.
Microsatellite haplotypes in Yaoundé.
Microsatellite haplotypes were characterized for regions of reduced variation immediately surrounding both dhfr and dhps. A total of 12 loci for dhfr and 10 loci for dhps, both totaling approximately 30 kb, were used for haplotype characterization. Haplotypes for both dhfr and dhps alleles are presented in Tables S1 and S2 in the supplemental material. Haplotypes were characterized for samples for which no multiple parasite infections were detected by pyrosequencing; therefore, we can be more confident in the haplotype reconstruction. A total of 45 (13.5%) of the samples for dhfr and 80 (24%) of the samples for dhps had multiple parasites as detected by pyrosequencing. For dhfr, we found 12 haplotypes for the sensitive allele, 3 for the 108N allele, 3 for the 51I/108N allele, 13 for the 59R/108N allele, and 77 for the 51I/59R/108N allele. Haplotype frequencies for dhfr and dhps alleles are shown in Fig. S1 and S2 in the supplemental material. Haplotype frequencies are shown only for alleles where the sample size is greater than 12. The sensitive and 59R/108N double mutant alleles each had a haplotype frequency distribution that was relatively equalized, i.e., there was no predominant haplotype for these alleles. However, for the triple mutant allele, which is presumably greatly advantageous in the presence of SP, the haplotype frequency distribution shows one predominant haplotype (haplotype 31) and multiple rare variants. Haplotype 31, which is the haplotype that has been described previously from Southeast Asia, is present for 73.5% of the triple mutant alleles. The vast majority of the remainder of the triple mutant haplotypes are variants of haplotype 31 (see Table S1 in the supplemental material); we consider these variants of haplotype 31 because they are the result of changes in 1 or 2 loci. Such new variants could be the result of mutations and/or recombination. Nevertheless, there are at least three truly novel haplotypes. Haplotypes 75, 76, and 77 (no. of occurrences = 1 for each) are different from haplotype 31 at 10, 10, and 11 out of 12 loci, respectively.
Haplotype analysis for dhps revealed 12 haplotypes for the sensitive allele, 45 for the 436A single mutant allele, 65 for the 437G allele, 3 for the 613S allele, 17 for the 436A/437G allele, 1 for the 437G/613S allele, and 1 for the 436A/613S allele. The haplotypes for the 437G/613S double mutant are found for 437G alleles, and the haplotype for the 436A/613S allele is found for 436A alleles. The frequency spectrum (see Fig. S2 in the supplemental material) shows no predominant haplotypes for the sensitive or 436A allele, but we do see one predominant haplotype with multiple minor variants for the 437G and 436A/437G alleles. dhps haplotypes 82 and 127 are predominant for the 437G and 436A/437G alleles, respectively.
Genetic differentiation and relationships among alleles.
Wright's fixation index, FST, was used to measure genetic differentiation between the alleles of dhfr and dhps with the microsatellite data. For both genes, the microsatellite loci used for haplotype characterization (i.e., those close to the genes) were also used to test for genetic differentiation between alleles of each gene. The FST analysis was conducted only for alleles that had a sample size greater than 12. The FST values for the comparisons among the three alleles of dhfr and dhps were high and significant (Table 2).
TABLE 2.
FST values for population/allele comparisons, determined using microsatellites around dhfr and dhpsa
| Type of allele |
P value
|
||||||
|---|---|---|---|---|---|---|---|
| Sensitive | 59/108 | 51I/59/108 | Sensitive | 436 | 437 | 436/437 | |
| dhfr | |||||||
| Sensitive | |||||||
| 59/108 | 0.23571 | ||||||
| 51I/59/108 | 0.40023 | 0.66262 | |||||
| dhps | |||||||
| Sensitive | |||||||
| 436 | 0.06204 | ||||||
| 437 | 0.18897 | 0.19028 | |||||
| 436/437 | 0.17319 | 0.23288 | 0.24038 | ||||
All P values shown represent significance at a P value of <0.01. The samples used for this analysis were separated into populations based on dhfr or dhps alleles. Data were used only for samples where genotyping for mutations in dhfr or dhps did not reveal multiple infections. Microsatellite loci used for the FST analysis are the same as those used for haplotype characterization.
The application eBURST was used to discern relationships among the dhfr and dhps microsatellite haplotypes. eBURST groups haplotypes based on a simple evolution model which assumes that one lineage or founding haplotype comes to high frequency in the population and then begins to differentiate, producing closely related haplotypes; this is depicted as a cluster (13). We used data from 12 microsatellite loci around dhfr and 10 loci around dhps (as for haplotype characterization) to depict genetic relationships in eBURST. It should be noted that eBURST does not allow for missing data and that samples with incomplete haplotypes were removed; therefore, there were fewer samples utilized for the eBURST analysis than for the haplotype characterization.
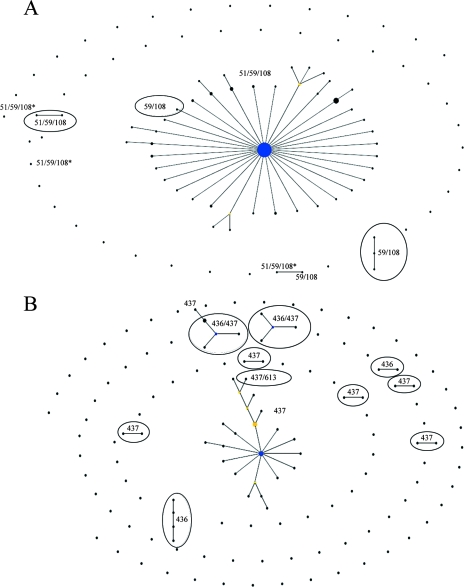
Figure 1A shows the “population snapshot” for dhfr alleles in the Cameroonian population. The majority of the samples shown here are triple mutant alleles (171 out of 191 total); therefore, it is difficult to make strong assessments about the relationships between the alleles of lower frequencies. We do see a clear relationship among the majority of the triple mutant alleles, represented by the central complex. The central complex is composed of haplotype 31, the Southeast Asian type, with minor variants of this haplotype radiating from haplotype 31. The minor haplotypes 75, 76, and 77 (Fig. 1) are not connected to the main triple mutant complex. Haplotypes 75 and 76 are not connected to any other haplotype, but haplotype 77 is connected to a 59R/108N haplotype.
FIG. 1.
Relationships among dhfr (A) and dhps (B) haplotypes in Cameroon, as determined by eBURST analysis. Each line connects haplotypes that are identical at 11 out of 12 loci. Haplotypes shown as single points differ from other haplotypes by alleles in at least two loci. The blue circles represent complex founders, and the yellow circles represent subgroup founders. The size of the circles is proportional to the number of isolates of the given haplotype. (A) One hundred ninety-one 12-locus dhfr microsatellite haplotypes. The central complex is composed mostly of haplotypes from 51I/59R/108N alleles. Triple mutant alleles marked with an asterisk are those that differ significantly from the Southeast Asian haplotype. A total of 7 samples with the sensitive dhfr allele, 2 with the 108N allele, 1 with the 51I/108N allele, 10 with the 59R/108N allele, and 171 with the 51I/59R/108N allele were used for this analysis. (B) One hundred forty-nine 10-locus dhps microsatellite haplotypes. The central complex is composed mostly of haplotypes from 437G mutant alleles. A total of 8 samples with the sensitive dhps allele, 35 with the 436A allele, 80 with the 437G allele, 1 with the 613S allele, 21 with the 436A/437G allele, 3 with the 437G/613S allele, and 1 with the 436A/613S allele were used for this analysis.
The majority of the dhps haplotypes are also grouped based on alleles (Fig. 1B), further demonstrating that most of the alleles are distinct from one another. Interestingly, there are two 436A/437G groups that are distinct from one another rather than one large double mutant allele cluster. The 436A/437G cluster on the left in Fig. 1B is composed mostly of haplotype 127, the most frequent haplotype for the allele, and the cluster on the right is composed mostly of haplotype 134, the second most frequent haplotype for the double mutant allele. The main 437G cluster is derived from haplotype 82, the most frequent for the 437G allele. This cluster analysis also shows the close relationship between 437G/613S and 437G alleles.
Levels of variation and selective sweeps.
We characterized microsatellite loci spanning approximately 150 kb around dhfr along chromosome 4 and 122 kb around dhps along chromosome 8 to document the levels of variation present for two genes under selection from drug pressure. We also analyzed microsatellite loci spanning approximately 80 and 90 kb on chromosomes 2 and 3, respectively, to assess putative levels of neutral variation in the genome. We used gene diversity, or He, as a measure of variation at microsatellite loci. The mean He for the neutral loci was 0.9258 (Table 3), which is similar to estimates from other African populations (1).
TABLE 3.
A and He values per locusa
| Chromosome and locus position | A | He ± SD |
|---|---|---|
| Chromosome 4 | 37 | 0.9211 ± 0.01 |
| −58 | 22 | 0.7893 ± 0.02 |
| −30 | 26 | 0.6720 ± 0.03 |
| −17 | 13 | 0.3354 ± 0.03 |
| −10 | 13 | 0.3315 ± 0.04 |
| −7.5 | 17 | 0.4280 ± 0.03 |
| −5.3 | 12 | 0.4653 ± 0.03 |
| −4.5 | 17 | 0.6096 ± 0.02 |
| −4.4 | 21 | 0.3620 ± 0.04 |
| −3.8 | 15 | 0.3318 ± 0.03 |
| −1.2 | 12 | 0.3931 ± 0.03 |
| −0.30 | 15 | 0.3632 ± 0.04 |
| 0.20 | 14 | 0.4278 ± 0.03 |
| 0.52 | 9 | 0.2323 ± 0.03 |
| 5.87 | 12 | 0.2308 ± 0.03 |
| 20 | 21 | 0.8986 ± 0.01 |
| 50 | 12 | 0.7043 ± 0.02 |
| 70 | 9 | 0.6459 ± 0.02 |
| 90 | ||
| Chromosome 8 | ||
| −50.1 | 18 | 0.8520 ± 0.01 |
| −17 | 30 | 0.8621 ± 0.01 |
| −10 | 20 | 0.8558 ± 0.01 |
| −7.4 | 18 | 0.7352 ± 0.02 |
| −2.5 | 20 | 0.7619 ± 0.02 |
| −1.6 | 10 | 0.7014 ± 0.02 |
| −0.8 | 24 | 0.7929 ± 0.02 |
| 0.06 | 6 | 0.5583 ± 0.02 |
| 0.14 | 23 | 0.8703 ± 0.01 |
| 1.59 | 18 | 0.6177 ± 0.03 |
| 6.19 | 20 | 0.8073 ± 0.01 |
| 9.8 | 9 | 0.5603 ± 0.01 |
| 17.5 | 10 | 0.6497 ± 0.02 |
| 33.1 | 14 | 0.6380 ± 0.02 |
| 72 | 41 | 0.9384 ± 0.01 |
| Chromosome 2 | ||
| 302 | 25 | 0.9292 ± 0.00 |
| 313 | 40 | 0.9549 ± 0.00 |
| 319 | 24 | 0.9366 ± 0.00 |
| 379 | 32 | 0.9500 ± 0.00 |
| Chromosome 3 | ||
| 335 | 29 | 0.9428 ± 0.00 |
| 363 | 27 | 0.9218 ± 0.00 |
| 383 | 26 | 0.8532 ± 0.02 |
| 429 | 40 | 0.9179 ± 0.01 |
The means were as follows: for chromosome 2, A of 30.25 and He of 0.9427; for chromosome 3, A of 30.5 and He of 0.9089; and for chromosomes 2 and 3, A of 30.38 and He of 0.9258.
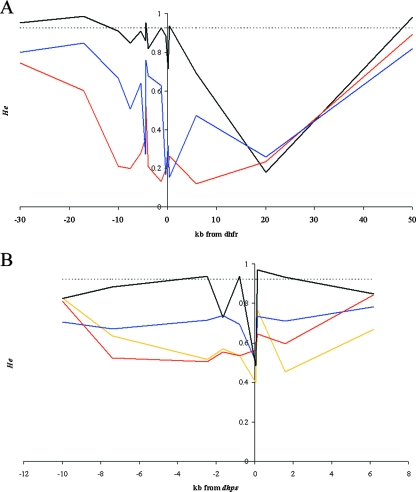
Considering the entire population of alleles, there is a clear diminution in heterozygosity at the region immediately surrounding dhfr for approximately 40 kb (Table 3; also see Fig. S3 in the supplemental material). The lack of variation surrounding dhps is less pronounced but extends for approximately 16 kb. This lack of variation is characteristic of a possible selective sweep, and if a sweep has occurred, then there should be a sharp reduction in variation around drug-resistant alleles versus drug-sensitive alleles (27). Indeed, there is a striking relationship between the amount of variation surrounding resistant double and triple mutant dhfr alleles and that surrounding sensitive (wild-type) alleles (Fig. 2A). The sensitive alleles exhibit variation similar to that seen for neutral markers for most of the loci surrounding dhfr. The locus at 20 kb exhibits lowered variation for both the sensitive and the mutant alleles, which indicates that this particular locus may have a lower mutation rate. Nevertheless, we can clearly see the greatest reduction in variation for triple mutant alleles, followed by the 59R/108N double mutant alleles.
FIG. 2.
Expected heterozygosity around dhfr (A) and dhps (B) alleles. Only alleles that had a sample size greater than 12 are plotted. The horizontal dashed line across the top of the graph represents the mean He observed at neutral loci on chromosomes 2 and 3. (A) Sensitive alleles are represented by the black line, 59R/108N alleles are represented by the blue line, and 51I/59R/108N alleles are represented by the red line. (B) Sensitive alleles are represented by the black line, 436A alleles by the blue line, 437G alleles by the yellow line, and 436A/437G alleles by the red line.
We also see a reduction in He surrounding mutant dhps alleles compared to that for the sensitive alleles for many of the loci on chromosome 8 (Fig. 2B). The lowest curves are seen for the 437G and 436A/437G mutant alleles; both of these alleles also had skewed haplotype frequency spectrums, with one predominant haplotype.
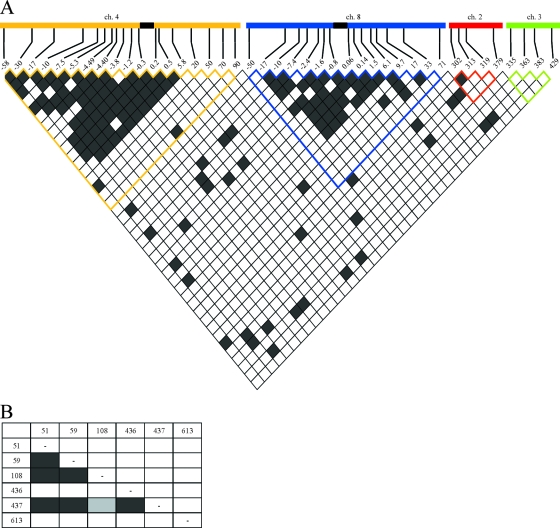
An alternative way of observing such selective sweeps is to explore the pattern of LD. Figure 3A shows the patterns of pairwise LD between all pairs of microsatellite loci used in this study (total sample sizes of 180 for loci on chromosome 4, 147 for chromosome 8, and 164 for chromosomes 2 and 3). We observed a clear increase in the amount of LD in the region surrounding the genes dhfr and dhps compared to that in the regions covered by the neutral markers on chromosomes 2 and 3. In addition, we tested for LD between the codons involved in drug resistance of dhfr and dhps. Again, only polymorphic codons could be tested; thus, dhfr codons 51, 59, and 108 and dhps codons 436, 437, and 613 were included in the pairwise tests. We observed significant LD between codons 51, 59, and 108 in dhfr and codon 437 of dhps (Fig. 3B).
FIG. 3.
Pairwise LD between microsatellite loci on different chromosomes (A) and between sites in dhfr and dhps (B). Each box represents one comparison between polymorphic pairs of loci; nonpolymorphic pairwise comparisons are not included. Bonferroni's correction for multiple comparisons was conducted for each comparison. Dark-gray cells represent significance at a P value of 0.01, light-gray cells represent significance at a P value of 0.05, and white cells represent values that were not significant. (A) The location of each microsatellite locus is at the top of the matrix (loci are named according to their positions relative to dhfr or dhps or along chromosome [ch] 2 or 3 according to the 3D7 genome sequence available from NCBI). The black boxes within chromosomes 4 and 8 represent the positions of dhfr and dhps, respectively. (B) Pairwise LD between sites in dhfr (codons 51, 59, and 108) and dhps (codons 436, 437, and 613).
DISCUSSION
We have found that only 5% of both dhfr and dhps alleles were sensitive alleles. Given that SP has been used both as a recommended governmental policy since the 1990s and also unofficially as self-medication, we know that drug selection has been strong in this population; therefore, the lack of sensitive alleles for both genes is not surprising. The majority of the dhfr alleles (88%) belong to the triple mutant class; these data alone suggest a possible selective sweep of the triple mutant allele.
In the case of dhfr alleles, the data from Yaoundé are similar to data from western Kenya (22) and show that the Southeast Asian triple mutant allele is prevalent in this central African population; this also is consistent with recent studies of African parasites (18, 30). A single triple mutant haplotype has appeared to have spread across Africa after its introduction from Southeast Asia (31), as previous studies with limited sampling have suggested (18, 30, 31). We do find additional rare novel variants for this highly resistant triple mutant allele, which reemphasizes the importance of local ecology and evolution in the generation of rare variants. Nevertheless, it appears that gene flow has allowed the widespread dispersion of the Southeast Asian haplotype, shaping the pattern of drug-resistant alleles across multiple African populations. Interestingly, however, such strong evidence of a single drug-resistant allele sweeping across Africa appears to be limited solely to dhfr.
Although there is a predominant group of haplotypes, several minor haplotypes of dhps in this Cameroonian population are genetically distinct and evolved independently from one another. This result can be explained by de novo origination and maintenance of distinct dhps allelic lineages in the Yaoundé population and/or by gene flow of particular dhps lineages into the Yaoundé population. Nevertheless, it is interesting that although the dhps mutation 437G is linked to all dhfr mutations, there is not a single dhfr allele that is particularly linked to a dhps allele in the population. A first obvious observation is that recombination is important across the continent and so both dhfr and dhps alleles segregate independently. However, a second observation is that the linkage between dhps mutation 437G and dhfr mutations could be due to drug selection. Selection acts on phenotypes, in this case, mutations changing the primary structure of the proteins so that the linkage between dhps mutation 437 and dhfr mutations is maintained regardless of the fact that the dhps and dhfr alleles, as defined by neutral markers, segregate independently. Our observations support previous studies indicating that drug selection acts differently on these two loci (25, 32). Strong selection on dhfr together with gene flow has allowed the successful dispersion of a particular dhfr haplotype across Africa. On the other hand, drug selection on dhps alleles acts differently; thus, regardless of the existence of gene flow, there is no evidence of a single dhps allele sweeping in central Africa, but there is evidence that at least one amino acid-changing mutation (437G) is linked with dhfr mutations. Additional studies with samples from across Africa are needed to address this matter.
Our results also indicate that there is an increase in divergence among dfhr triple mutant alleles. Although the high frequency of this group of closely related triple mutant alleles of Southeast Asian origin lends support for the use of molecular markers to monitor drug resistance, even in areas where there is a great deal of genetic diversity, it should be noted that over time an increase in divergence between alleles together with recombination could break down linkage groups. The genetic differentiation among dhfr triple mutant alleles in Yaoundé is not necessarily representative of the entire African continent; therefore, further investigation of the relationships and dispersion of resistant alleles on the continent and among different regions is needed.
We have shown evidence of selection acting on both dhfr and dhps. We see skewed haplotype frequencies for the triple mutant dhfr allele and the 437G and 436A/437G dhps alleles. Skewed haplotype frequency spectrums are characteristic of selective sweeps: the spectrum is dominated by an advantageous allele and linked neutral variation at high frequency along with many low-frequency variants (10, 17). We also show lack of variation and strong LD immediately surrounding both dhfr and dhps mutant alleles, classic signatures of genetic hitchhiking due to strong selection (33).
Genetic hitchhiking and selective sweeps have been reported for dhfr in Southeast Asia, Venezuela, and South Africa and for dhps only in Venezuela (21, 23, 27). Under the genetic hitchhiking model, the extent of the reduction in variation is determined by the strength of selection and the amount of recombination (33). Within the population from Yaoundé, we can see the effects of the strength of selection. There is an expectation that the triple mutant dhfr allele would yield a level of resistance to the parasite in the presence of pyrimethamine greater than that of the double mutant and thus would have a stronger selection coefficient. This strong selection coefficient would produce a greater extent of hitchhiking around dhfr. Indeed, this is what we observed from the Yaoundé population in both width and depth of the variation surrounding dhfr. By extension, the curves for dhps indicate a stronger strength of selection for the 437G and 436A/437G alleles than for the 436A and sensitive alleles. This is supported by the haplotype frequency data, which show a predominant haplotype for the 437G and 436A/437G alleles, indicating strong selection for a single haplotype and thus for the alleles.
In addition, we do not see such a strong selective sweep on dhps alleles. This could indicate that the population of dhps alleles may have been captured in the early stages of a selective sweep. This observation is consistent with those made by others, where mutations associated with sulfadoxine resistance in dhps appear after mutations in dhfr have reached high frequencies (25, 32). Why SP selection acts asymmetrically in dhfr and dhps is still a matter that deserves further investigation. It would be interesting to assay these populations again in samples to observe whether the shape and depth of the heterozygosity curve have changed more recently as a result of continued selection on dhps alleles. It will also be important to assess how gene flow may affect the observed pattern of dhps alleles in Africa.
In summary, we have conducted the first investigation using microsatellite markers around dhfr and dhps to understand the effects of selection for SP resistance in a large sample set from a central African population. We have shown local origination of alleles highly resistant to pyrimethamine, genetic differentiation among dhfr and dhps alleles, and effects of strong drug pressure on genetic patterns. Although we show evidence that a molecular surveillance system using microsatellite markers in an area of moderate transmission is feasible, this study also highlights the need to complement surveillance studies with comprehensive population-based investigations. It is possible that recombination or point mutations could affect our capacity for tracking particular alleles. Although this seems to have limited relevance in the case of dhfr, where a single allele appeared to sweep across Africa, it could be critical in the case of dhps or other loci involved in drug resistance in the future. It is important to continue to study natural selection with Plasmodium populations in order to understand the effects of drug pressure in different ecologies.
Supplementary Material
Acknowledgments
Financial support was provided by the Antimicrobial Resistance Working Group, Centers for Disease Control and Prevention. The Atlanta Research and Education Foundation helped support this work. A. A. Escalante is supported by grant NIH R01 GM084320.
We thank Vincent Hill and Jothikumar Narayanan of the Parasitic Diseases Branch, Division of Parasitic Diseases, NCZVED, CDC, and the CDC Core Facility for allowing us to use the PSQ MA96 system and vacuum prep station for pyrosequencing.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 2 September 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Anderson, T. J., B. Haubold, J. T. Williams, J. G. Estrada-Franco, L. Richardson, R. Mollinedo, M. Bockarie, J. Mokili, S. Mharakurwa, N. French, J. Whitworth, I. D. Velez, A. H. Brockman, F. Nosten, M. U. Ferreira, and K. P. Day. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467-1482. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, T. J., and C. Roper. 2005. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 94:269-280. [DOI] [PubMed] [Google Scholar]
- 3.Basco, L. K. 2003. Molecular epidemiology of malaria in Cameroon. XVI. Longitudinal surveillance of in vitro pyrimethamine resistance in Plasmodium falciparum. Am. J. Trop. Med. Hyg. 69:174-178. [PubMed] [Google Scholar]
- 4.Basco, L. K., M. Ndounga, M. Tejiokem, V. F. Ngane, J. C. Youmba, P. Ringwald, and G. Soula. 2002. Molecular epidemiology of malaria in Cameroon. XI. Geographic distribution of Plasmodium falciparum isolates with dihydrofolate reductase gene mutations in southern and central Cameroon. Am. J. Trop. Med. Hyg. 67:378-382. [DOI] [PubMed] [Google Scholar]
- 5.Basco, L. K., and P. Ringwald. 2007. Molecular epidemiology of malaria in Cameroon. XXIV. Trends of in vitro antimalarial drug responses in Yaounde, Cameroon. Am. J. Trop. Med. Hyg. 76:20-26. [PubMed] [Google Scholar]
- 6.Basco, L. K., and P. Ringwald. 1998. Molecular epidemiology of malaria in Yaounde, Cameroon. I. Analysis of point mutations in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 58:369-373. [DOI] [PubMed] [Google Scholar]
- 7.Basco, L. K., and P. Ringwald. 1998. Molecular epidemiology of malaria in Yaounde, Cameroon. II. Baseline frequency of point mutations in the dihydropteroate synthase gene of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 58:374-377. [DOI] [PubMed] [Google Scholar]
- 8.Basco, L. K., and P. Ringwald. 1999. Molecular epidemiology of malaria in Yaounde, Cameroon. IV. Evolution of pyrimethamine resistance between 1994 and 1998. Am. J. Trop. Med. Hyg. 61:802-806. [DOI] [PubMed] [Google Scholar]
- 9.Basco, L. K., and P. Ringwald. 2000. Molecular epidemiology of malaria in Yaounde, Cameroon. VI. Sequence variations in the Plasmodium falciparum dihydrofolate reductase-thymidylate synthase gene and in vitro resistance to pyrimethamine and cycloguanil. Am. J. Trop. Med. Hyg. 62:271-276. [DOI] [PubMed] [Google Scholar]
- 10.Braverman, J. M., R. R. Hudson, N. L. Kaplan, C. H. Langley, and W. Stephan. 1995. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140:783-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortese, J. F., and C. V. Plowe. 1998. Antifolate resistance due to new and known Plasmodium falciparum dihydrofolate reductase mutations expressed in yeast. Mol. Biochem. Parasitol. 94:205-214. [DOI] [PubMed] [Google Scholar]
- 12.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 13.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fondjo, E., V. Robert, G. Le Goff, J. C. Toto, and P. Carnevale. 1992. Le paludisme urbain à Yaoundé (Cameroun). 2. Etude entomologique dans deux quartiers peu urbanisés. Bull. Soc. Pathol. Exot. 85:57-63. [PubMed] [Google Scholar]
- 15.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 16.Hayton, K., and X. Z. Su. 2004. Genetic and biochemical aspects of drug resistance in malaria parasites. Curr. Drug Targets Infect. Disord. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y. 2006. Allele frequency distribution under recurrent selective sweeps. Genetics 172:1967-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiga, O., A. A. Djimde, V. Hubert, E. Renard, A. Aubouy, F. Kironde, B. Nsimba, K. Koram, O. K. Doumbo, J. Le Bras, and J. Clain. 2007. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J. Infect. Dis. 196:165-172. [DOI] [PubMed] [Google Scholar]
- 19.Manga, L., E. Fondjo, P. Carnevale, and V. Robert. 1993. Importance of low dispersion of Anopheles gambiae (Diptera: Culicidae) on malaria transmission in hilly towns in south Cameroon. J. Med. Entomol. 30:936-938. [DOI] [PubMed] [Google Scholar]
- 20.Manga, L., V. Robert, J. Messi, M. Desfontaine, and P. Carnevale. 1992. Le paludisme urbain à Yaoundé, Cameroun. 1. Etude entomologique dans deux quartiers centraux. Mémoires de la Société royale belge d'Entomologie 35:155-162. [Google Scholar]
- 21.McCollum, A. M., K. Mueller, L. Villegas, V. Udhayakumar, and A. A. Escalante. 2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51:2085-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCollum, A. M., A. C. Poe, M. Hamel, C. Huber, Z. Zhou, Y. P. Shi, P. Ouma, J. Vulule, P. Bloland, L. Slutsker, J. W. Barnwell, V. Udhayakumar, and A. A. Escalante. 2006. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 194:189-197. [DOI] [PubMed] [Google Scholar]
- 23.Nair, S., J. T. Williams, A. Brockman, L. Paiphun, M. Mayxay, P. N. Newton, J. P. Guthmann, F. M. Smithuis, T. T. Hien, N. J. White, F. Nosten, and T. J. Anderson. 2003. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol. Biol. Evol. 20:1526-1536. [DOI] [PubMed] [Google Scholar]
- 24.Nash, D., S. Nair, M. Mayxay, P. N. Newton, J. P. Guthmann, F. Nosten, and T. J. Anderson. 2005. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc. Biol. Sci. 272:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nzila, A. M., E. K. Mberu, J. Sulo, H. Dayo, P. A. Winstanley, C. H. Sibley, and W. M. Watkins. 2000. Towards an understanding of the mechanism of pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: genotyping of dihydrofolate reductase and dihydropteroate synthase of Kenyan parasites. Antimicrob. Agents Chemother. 44:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, S. D. E. 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis, University of Dublin, Dublin, Ireland.
- 27.Pearce, R., A. Malisa, S. P. Kachur, K. Barnes, B. Sharp, and C. Roper. 2005. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Mol. Biol. Evol. 22:1834-1844. [DOI] [PubMed] [Google Scholar]
- 28.Plowe, C. V., J. G. Kublin, and O. Doumbo. 1998. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist. Updat. 1:389-396. [DOI] [PubMed] [Google Scholar]
- 29.Raymond, M., and F. Rousset. 1995. An exact test for population differentiation. Evolution 49:1280-1283. [DOI] [PubMed] [Google Scholar]
- 30.Roper, C., R. Pearce, B. Bredenkamp, J. Gumede, C. Drakeley, F. Mosha, D. Chandramohan, and B. Sharp. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174-1181. [DOI] [PubMed] [Google Scholar]
- 31.Roper, C., R. Pearce, S. Nair, B. Sharp, F. Nosten, and T. Anderson. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. [DOI] [PubMed] [Google Scholar]
- 32.Sibley, C. H., J. E. Hyde, P. F. Sims, C. V. Plowe, J. G. Kublin, E. K. Mberu, A. F. Cowman, P. A. Winstanley, W. M. Watkins, and A. M. Nzila. 2001. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 17:582-588. [DOI] [PubMed] [Google Scholar]
- 33.Smith, J. M., and J. Haigh. 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23:23-35. [PubMed] [Google Scholar]
- 34.Tahar, R., and L. K. Basco. 2007. Molecular epidemiology of malaria in Cameroon. XXVII. Clinical and parasitological response to sulfadoxine-pyrimethamine treatment and Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase alleles in Cameroonian children. Acta Trop. 103:81-89. [DOI] [PubMed] [Google Scholar]
- 35.Tahar, R., and L. K. Basco. 2006. Molecular epidemiology of malaria in Cameroon. XXII. Geographic mapping and distribution of Plasmodium falciparum dihydrofolate reductase (dhfr) mutant alleles. Am. J. Trop. Med. Hyg. 75:396-401. [PubMed] [Google Scholar]
- 36.Tahar, R., and L. K. Basco. 2007. Molecular epidemiology of malaria in Cameroon. XXVI. Twelve-year in vitro and molecular surveillance of pyrimethamine resistance and experimental studies to modulate pyrimethamine resistance. Am. J. Trop. Med. Hyg. 77:221-227. [PubMed] [Google Scholar]
- 37.Talisuna, A. O., P. Bloland, and U. D'Alessandro. 2004. History, dynamics, and public health importance of malaria parasite resistance. Clin. Microbiol. Rev. 17:235-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triglia, T., P. Wang, P. F. Sims, J. E. Hyde, and A. F. Cowman. 1998. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J. 17:3807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright, S. 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395-420. [Google Scholar]
- 40.Zhou, Z., A. C. Poe, J. Limor, K. K. Grady, I. Goldman, A. M. McCollum, A. A. Escalante, J. W. Barnwell, and V. Udhayakumar. 2006. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J. Clin. Microbiol. 44:3900-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.