Abstract
The Tf1 retrotransposon of Schizosaccharomyces pombe represents a group of eukaryotic long terminal repeat (LTR) retroelements that, based on their sequences, were predicted to use an RNA self-primer for initiating reverse transcription while synthesizing the negative-sense DNA strand. This feature is substantially different from the one typical to retroviruses and other LTR retrotransposons that all exhibit a tRNA-dependent priming mechanism. Genetic studies have suggested that the self-primer of Tf1 can be generated by a cleavage between the 11th and 12th bases of the Tf1 RNA transcript. The in vitro data presented here show that recombinant Tf1 reverse transcriptase indeed introduces a nick at the end of a duplexed region at the 5′ end of Tf1 genomic RNA, substantiating the prediction that this enzyme is responsible for generating this RNA self-primer. The 3′ end of the primer, generated in this manner, can then be extended upon the addition of deoxynucleoside triphosphates by the DNA polymerase activity of the same enzyme, synthesizing the negative-sense DNA strand. This functional primer must have been generated by the RNase H activity of Tf1 reverse transcriptase, since a mutant enzyme lacking this activity has lost its ability to generate the self-primer. It was also found here that the reverse transcriptases of human immunodeficiency virus type 1 and of murine leukemia virus do not exhibit this specific cleavage activity. In all, it is likely that the observed unique mechanism of self-priming in Tf1 represents an early advantageous form of initiating reverse transcription in LTR retroelements without involving cellular tRNAs.
Long terminal repeat (LTR) retrotransposons are retrovirus-like elements found in a broad variety of eukaryotic cells (6, 7, 10). Their structure and mechanisms of propagation are related to retroviruses, and like them, they encode the Gag, protease, reverse transcriptase (RT), and integrase proteins. As with all retroviruses, the RTs of LTR retrotransposons are pivotal for converting the positive-sense, single-stranded RNA genome into a double-stranded and integration-competent viral DNA. This reverse transcription (RTN) process is catalyzed by two RT activities, DNA polymerase activity, which copies both RNA and DNA, and RNase H activity, which concomitantly cleaves the RNA in the DNA-RNA heteroduplex formed. In all retroviruses and LTR retrotransposons, the synthesis of negative-strand DNA is initiated from an RNA primer that is complementary to a specific genomic RNA sequence, the primer binding site (PBS), which is located near the 5′ end of the RNA genome. It is well documented that this RNA primer is a specific cellular tRNA (for examples, see references 6, 7, 16, and 27).
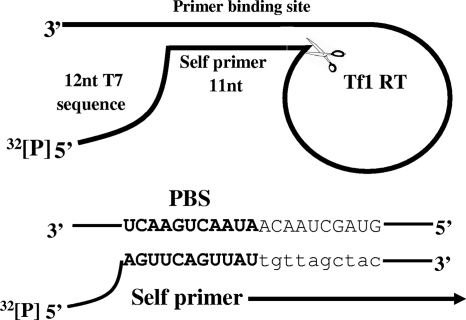
Interestingly, an alternative tRNA-independent priming mechanism was proposed for the LTR retrotransposon Tf1 of the fission yeast Schizosaccharomyces pombe by Levin (17-20). This model suggests that a self-complementarity within the RNA genome allows intramolecular base pairing near the 5′ end of the RNA, causing this end to fold back. As a result, this RNA forms an intramolecular 11-bp RNA duplex that after cleavage releases an 11-nucleotide (nt) self-primer for initiating the negative-strand cDNA synthesis (Fig. 1). Genetic studies of Tf1 have suggested that cleavage occurs between the 11th and 12th bases of the RNA transcript, providing the primer for DNA synthesis. Mutagenesis experiments have also suggested that the RNase H domain of Tf1 RT may be required for this specific cleavage (18). Hence, Tf1 RT could possibly have a novel activity, never found so far in the RTs of retroelements, to specifically nick the genomic RNA at the 5′ end of a duplexed region and thus to create an RNA primer for negative-strand cDNA synthesis.
FIG. 1.
Schematic description of the substrate used for the self-primer cleavage reactions. The in vitro-synthesized RNA that mimics the 365-nt-long 5′ end of the Tf1 RNA served as the substrate for the cleavage reactions described in the text. This RNA was fused at its 5′ end to 12 nt derived from the plasmid used for RNA synthesis and labeled at its 5′ end with 32[P]. The sequences of the 11-nt self-primer and the PBS, to which it is annealed, are both marked in bold capital letters. The first RNA nucleotides to be reverse transcribed, located 5′ of the PBS, are shown in regular capital letters, and the first deoxynucleotides incorporated into DNA are shown in lowercase letters.
From the time when a self-priming mechanism was suggested for Tf1, a similar priming mechanism has also been proposed for several LTR retroelements, such as the highly homologous Tf2 and Maggy, Skippy, Cft-1, Boty, copia of maize, and Tf1/shushi of vertebrates, which all belong to a single lineage of the T3/gypsy group of LTR retrotransposons (5, 7, 21). These elements are believed to have diverged early in the evolution of LTR retrotransposons, well before retroviruses. Therefore, it is likely that the mechanism of self-priming represents an early form of initiating RTN in LTR retroelements (16), as it was advantageous for these retroelements to use a tRNA-independent mechanism of priming cDNA synthesis.
The only RTs from LTR retrotransposons extensively studied in vitro were those of Ty1 and Ty3 that belong to different retrotransposon groups, the Pseudoviridae and the Metaviridae, respectively (4, 8, 23, 24). We have recently expressed in bacteria an enzymatically active Tf1 RT and studied its basic biochemical properties (14). This RT has all the enzymatic activities typical of RTs, namely, RNA-dependent and DNA-dependent DNA polymerase and RNase H activities. The most outstanding feature of this RT is its ability to add nontemplated deoxynucleotides to the 3′ end of the nascent DNA strand. This RT property, combined with its capacity to incorporate wrong nucleotides at the 3′ ends of the growing strand, indicates an infidelity of Tf1 RT in synthesizing DNA at the template ends.
In the present study, the ability of Tf1 RT to cleave an RNA segment that mimics the 5′ end of genomic Tf1 RNA was tested biochemically. Indeed, Tf1 RT has generated the specific RNA self-primer that was not produced by other tested retroviral RTs. An RNase H-lacking Tf1 RT mutant did not exhibit the self-primer cleaving activity. It was also shown that the produced primer is functional, as it was extended by the RT-associated DNA polymerase activity.
MATERIALS AND METHODS
Construction of the vectors for expressing Tf1 RT and its mutant.
The plasmids with the Tf1-related sequences were gifts of H. Levin from the NIH. The Tf1 RT versions used in this work are the highly purified 505-residue-long recombinant enzymes expressed in Escherichia coli (14). The wild-type RT gene was derived from the pET15b provirus-containing plasmid that was generated from the Tf1-107 NCYC 132 strain from S. pombe. The gene encoding the D362N mutant of Tf1 RT (equivalent to the D710N mutation in the Gag-Pol polyprotein of Tf1) was derived from a plasmid designated pHL915-4 (18). The RT-encoding genes were synthesized by PCR, sequenced, and cloned as described in detail in reference 14. The two plasmids were designated pT5m-6His-Tf1 RT and pT5m-6His-Tf1 RT D362N for the wild-type Tf1 RT and the D362N RT mutant, respectively.
Expression and purification of Tf1 RT.
The expression of the Tf1 RT versions, both containing a six-histidine tag at their amino termini, was induced in E. coli strain BL21(DE3)pLysS and purified as described previously (14). The wild-type RT had a distinct RNase H activity, whereas the mutant RT did not exhibit any detectable RNase H activity (data not shown). In contrast, both RT versions had substantial DNA polymerase activity.
Recombinant human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MuLV) RTs.
These enzymes were expressed in our laboratory and purified as described in detail previously (2, 26).
Construction of the plasmid used to synthesize RNA in vitro.
A portion of the 5′ LTR (from positions 188 to 540) of the Tf1 provirus was generated by PCR using suitable oligonucleotides. The two primers, used to synthesize the DNA segment, were as follows: the 5′-end sense primer containing a KpnI site upstream from the self-primer (5′-GCTCGGCGGTACCAGTTCAGTTATGAGCTATAT-3′) and the 3′-end antisense primer with a SacI site downstream (5′-CTCGACGGGAGCTCTCTTGGAATAGCGAGTTG-3′). The synthesized 365-bp DNA segment contains at its 5′ end the 11-nt putative self-primer sequence (5′-AGTTCAGTTAT-3′), the U5, and the 11-nt PBS up to the end of the 5′ LTR. This PCR product and the target plasmid Bluescript SK were first cleaved by the KpnI restriction enzyme. Then the 4-nt overhangs, generated by KpnI (5′-GTAC-3′), were removed by the 3′-to-5′ exonuclease activity of the E. coli DNA polymerase I large fragment (Klenow fragment) in the presence of all four deoxynucleoside triphosphates (dNTPs), thus forming blunt ends. This step was designed to shorten by 4 nt the T7-derived sequence that is added to the authentic Tf1 sequence while transcribing the DNA into RNA by the T7 RNA polymerase (see below). The synthetic donor DNA and the target plasmid were then cleaved by SacI, followed by ligation of both DNAs to form the appropriate vector for RNA synthesis. The resulting plasmid was designated pBSKspTf1, and its sequence was verified by DNA sequencing.
The preparation of the Tf1-derived RNA substrate.
The RNA segment that was used in the self-priming assay was synthesized in vitro from the plasmid pBSKspTf1 that was linearized by the SacI restriction enzyme (followed by digestion with proteinase K and phenol extraction of the DNA). A runoff transcription with phage T7 RNA polymerase was performed using an RNA synthesis kit (the AmpliScribe T7 high-yield transcription kit from Epicentre). In all, there is an addition of 12 T7-derived nt to the 5′ end of the Tf1-derived RNA sequence. The RNA transcription reaction was performed for 4 h with highly purified nucleoside triphosphates (NTPs) at 37°C, and the RNA was purified by spin column chromatography, using the SV total RNA isolation system kit (Promega). The sequence of this synthesized RNA contained 365 nt, including 12 nt derived from the T7 gene upstream from the 11-nt self-primer sequence (5′-GGGCGAAUUGGC-3′ and 5′-AGUUCAGUUAU-3′, respectively). In order to label radioactively the RNA segment, a dephosphorylation reaction was done using calf intestinal alkaline phosphatase for 1 h at 37°C, followed by phenol extractions. The RNA was then end-labeled at its 5′ end, using polynucleotide kinase and [γ-32P]ATP, and was purified again by the SV total RNA isolation kit (25, 26).
Self-priming assay.
The 32P-5′-end-labeled synthetic RNA in 50 mM NaCl and 50 mM Tris-HCl (pH 8.0) was self-annealed by heating to 72°C for 10 min and then slowly cooling down to 4°C. Next, acetylated bovine serum albumin, dithiothreitol (DTT), and 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS) were added to the mixture to final concentrations of 100 μg/ml, 2 mM, and 10 mM, respectively. All enzymatic reactions were carried out with this end-labeled RNA in 75 mM NaCl, 15 mM Tris-HCl (pH 8.0), 0.6 mM DTT, and 30 μg/ml bovine serum albumin with either 0.5 mM MnCl2 or 5 mM MgCl2 (in the absence or presence of 100 ng of each RT tested), all at final volumes of 10 μl. In assays containing dNTPs, each dNTP was at a final concentration of 50 μM. The reactions were performed at 37°C for 10 min and then were stopped by adding formamide loading buffer (90% formamide, 10 mM EDTA, 1 mg/ml bromophenol blue, 1 mg/ml xylene-cyanole), followed by heat denaturation for 3 min at 95°C. Finally, the reaction products were analyzed by high-voltage and high-resolution electrophoresis through 12% polyacrylamide gels with 6 M urea that were dried and underwent autoradiography.
RESULTS AND DISCUSSION
Until now, there was no direct biochemical evidence that Tf1 RT can specifically nick its cognate RNA and use the generated RNA segment as a functional primer for initiating RTN. The present in vitro study was designed to answer the following questions. (i) Can Tf1 RT cleave its related genomic RNA to generate a self-primer and utilize it for DNA synthesis in RTN? (ii) Are the formation and use of the self-primer in RTN specific to Tf1 RT, or are they catalyzed also by retroviral RTs? (iii) Is the RNase H activity of Tf1 RT involved in cleaving the self-primer?
To study the outlined issues, a 365-nt in vitro-transcribed RNA substrate that represents the first 353 nt of the genomic Tf1 RNA (and is appended at the 5′ end to 12 nt derived from the phage T7 sequence) was tested (Fig. 1). Thus, the Tf1-derived sequence starts from the 13th nucleotide of this RNA transcript. Because this transcript was 5′-end labeled, the expected length of the radiolabeled Tf1 RT-derived product is 23 nt, of which the last 11 nt represent the self-primer (with the sequence 5′-AGUUCAGUUAU-3′) and the remaining 12 nt are the T7-derived sequence.
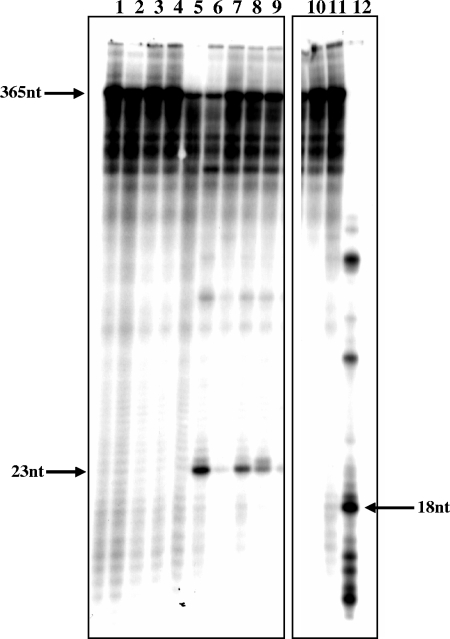
Figure 2 shows the electrophoretic pattern of the cleavage products generated by Tf1 RT and HIV-1 RT that was used as a control RT. It is apparent that Tf1 RT cleaves the RNA substrate in a Mn2+-dependent manner, generating a few products that are noticeable above background levels. The major RNA product, shown in lane 6, is 23 nt in length and comigrates with a synthetic 23-nt oligonucleotide RNA marker. This product was not seen in the reaction carried out with Mg2+ (lane 5). Interestingly, such a high Mn2+ specificity was already observed for the Tf1 RT-associated RNase H activity, in contrast to the RT-directed DNA polymerase activities that function with both divalent cations (14). This correlation of the Mn2+ specificity may suggest the involvement of the RT-associated RNase H activity in cleaving the RNA. Yet, a more direct and specific approach was to test this activity with an RNase H-deficient RT mutant, as described below.
FIG. 2.
Cleavage (and extension) of the Tf1-derived RNA sequence by Tf1 RT and HIV-1 RT. The in vitro-synthesized 365-nt RNA was 5′-end labeled with [γ-32P]ATP (see Materials and Methods). This RNA was incubated with or without 100 ng of Tf1 RT or HIV-1 RT, and each reaction was conducted in the absence of or with 5 mM MgCl2 or 0.5 mM MnCl2 (in the presence or the absence of 50 μM dNTPs). After incubating for 10 min at 37°C, the reaction products were resolved by high-voltage and high-resolution electrophoresis through 12% polyacrylamide gels with 6 M urea. All reaction products contained the RNA substrate and other components, as shown in the following gel lanes: 1, the 365-nt RNA substrate alone; 2, with Mg2+; 3, with Mn2+; 4, with Tf1 RT; 5, with Tf1 RT and Mg2+; 6, with Tf1 RT and Mn2+; 7, with Tf1 RT, Mn2+, and all four dNTPs; 8, with Tf1 RT, Mn2+, dATP, dCTP, and dGTP; 9, with Tf1 RT, Mn2+, dATP, dCTP, and dTTP; 10, with HIV-1 RT; 11, with HIV-1 RT and Mg2+; and 12, with HIV-1 RT and Mn2+. To precisely localize the position of the produced self-primer, a 23-nt 5′-end-labeled synthetic marker RNA was used (indicated by an arrow). The position of the 18-nt product, generated by HIV-1 RT (in the presence of Mn2+), was determined from the molecular ladder of the partially cleaved 23-nt RNA marker.
Like all DNA polymerases, Tf1 RT requires a primer with a preexisting 3′-OH end to create the first phosphodiester bond of the polymerized nascent DNA strand. The natural primer of Tf1 was predicted to be the 11-nt fragment released from the 5′ end of the 4.5-kb-long genomic RNA (20). This prediction was challenged in the present study by testing in vitro whether the RNA segment generated by Tf1 RT can also prime RTN (Fig. 2). Indeed, supplementing the RT-dependent cleavage reaction with all four dNTPs caused an almost complete disappearance of the 23-nt RNA band, suggesting that the RNA segment was extended by the RT-associated DNA polymerase activity (Fig. 2, lane 7). The polymerase activity of RTs is usually not highly processive (1). Likewise, Tf1 RT-associated DNA polymerase was shown by us to be only partially processive, with many stops occurring while extending the primers (14). Therefore, it is not surprising that extension products cannot be clearly discerned in the gel analysis shown in lane 7 of Fig. 2. Further support for the sequence specificity of elongating the generated 11-nt RNA primer, emerges from the fact that omitting a single dNTP from the dNTP mixture leads to partial extensions, depending on the synthesized sequence. Accordingly, the absence of dTTP resulted in no extension of the primer, since the first nucleotide to be incorporated into the nascent DNA strand is a T (Fig. 2, lane 8; Fig. 1). Likewise, omitting dGTP from the dNTP mixture resulted in a partial extension of the primer by only a single nucleotide (lane 9), as the second nucleotide expected to be incorporated after the first T is a G. In all, it is evident that Tf1 RT can generate in vitro a functional primer and then extend it by the enzyme's RNA-dependent DNA polymerase activity.
To examine how specific the self-primer cleavage activity to Tf1 RT is, the effects of the well-studied wild-type RTs of HIV-1 and MuLV were tested on the Tf1-derived RNA substrate. Like all RTs studied, these two RTs have a distinct RNase H activity; thus, the specific RT preparations used herein were confirmed to possess this activity (data not shown). HIV-1 RT is known to prefer Mg2+ over Mn2+ (11). Nonetheless, HIV-1 RT cleaved the labeled RNA substrate detectably only with Mn2+, completely degrading the 365-nt RNA substrate into several distinct RNA bands (none of which coincided with the self-primer) (Fig. 2, lane 12). The HIV-1 RT cleavage product closest in size to the Tf-1 RT-generated self-primer was 18 nt long (namely, this product has only 6 Tf1-related nt). Moreover, none of the RNA species generated by HIV-1 RT could serve as functional primers for DNA synthesis, since there was no significant extension of any of the generated RNA bands in the presence of all dNTPs (data not shown). The cleavage activity of HIV-1 RT observed in the presence of Mn2+ could be related to its reported Mn2+-dependent cleavage of double-stranded RNA (3). As the in vivo relevance of this activity is obscure, it was redesignated RNase H*, analogous to the relaxed specificity of some restriction enzymes (12). In contrast to HIV-1 RT, MuLV RT did not show any detectable RNA cleavage above the background level (Fig. 3, lane 6). MuLV RT was tested with only Mn2+, similarly to the published study on its potential RNase D/H* activity (3). Therefore, it is apparent that in our assay system, this RT does not have any detectable RNA cleaving activity.
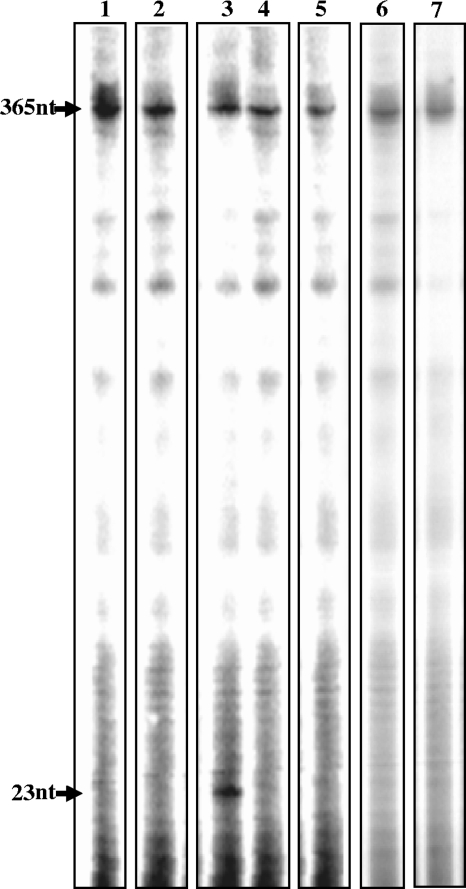
FIG. 3.
Cleavage of the Tf1-derived sequence by the RNase H-deficient mutant of Tf1 RT and by MuLV RT. The 5′-end-labeled 365-nt synthetic RNA was incubated as described in Fig. 2 with either wild-type Tf1 RT, the RNase H-deficient Tf1 RT mutant (the D362N mutant), or with MuLV RT. Lane 1, the 365-nt RNA substrate by itself; lane 2, with wild-type Tf1 RT; lane 3, with wild-type Tf1 RT and Mn2+; lane 4, with the D362N mutant of Tf1 RT; lane 5, with the D362N mutant of Tf1 RT and Mn2+; lane 6, with MuLV RT; lane 7, with MuLV RT and Mn2+. The position of the 5′-end-labeled 23-nt synthetic RNA marker is also indicated as in Fig. 2.
The self-primer cleaving activity of Tf1 RT differs from the RNase H* activity of HIV-1 RT in several aspects. First, unlike HIV-1 RT, Tf1 RT cleaves precisely 11 nt from the 5′ end of the Tf1 RNA, generating a functional primer for DNA synthesis. Second, Tf1 RT exhibits the same high preference for Mn2+ over Mg2+ in both the self-primer cleaving and RNase H activities, whereas HIV-1 RT exhibits its RNase H* activity under conditions not optimal for its RNase H activity. This proposed linkage between Tf1 RT RNase H and self-primer cleavage was further confirmed herein by the failure of the purified Tf1 RT D362N mutant, which lacks RNase H activity, to show any activity (above background levels) to produce the 23-nt self-primer segment (Fig. 3, lane 4). This Tf1 RT mutation is equivalent to the D710N mutation in the Tf1 Gag-Pol polyprotein, suggested to eliminate the RT's RNase H activity (18). Indeed, the Tf1 RT D362N mutant was confirmed to lack any detectable levels of RNase H activity, as measured by the in vitro assays previously employed by us (14), despite having a full DNA polymerase activity (data not shown). The D362 mutation in Tf1 RT is at a position homologous to D443 in HIV-1 RT, which was shown to be essential for the HIV-1 RT-associated RNase H activity (22).
The cleavage of the 5′ end of the genomic RNA for generating a self-primer for negative-strand DNA synthesis is a mechanism proposed for initiating RTN in several groups of LTR retrotransposons, which have probably diverged early in the evolution of the retroelements, well before retroviruses (16). Therefore, it is likely that this self-priming mechanism is an early form of RTN initiation without the involvement of the host tRNA. Here, the RT's RNase H activity provides also this self-primer cleavage function, thus making the RT responsible for the whole RTN process. The present report reinforces this concept by providing the first in vitro proof that the purified Tf1 RT can indeed execute this self-priming cleavage-mediated RTN process.
The sequence complementarity between the Tf1 PBS and the first 11 bases of the Tf1 RNA genome has suggested that priming requires the cleavage of the 5′-terminal 11 nt of the genome to provide the appropriate 3′-OH for extending the generated primer throughout DNA synthesis (18, 20). The data presented here support this idea experimentally. Results from a fragment ligation assay have suggested that the 11-nt RNA cleavage product generated in vivo has a 3′-OH terminus (18). Since the RT-associated RNase H activity performs primarily as an endonuclease, which cleaves the phosphodiester bonds in the RNA strand, yielding 3′-OH and 5′-PO4 termini (15, 27), the Tf1 RT-directed production of the RNA self-primer is also likely to generate such ends. Given that retroviral RTs (as opposed to RTs of some bacterial retroelements) specifically prime in vitro DNA synthesis from the 3′-OH group of the RNA primer (13, 27, 28), the finding presented in this study that the Tf1 RT-generated primer is functional in DNA synthesis implies that this in vitro-produced primer also has a 3′-OH terminus.
As mentioned above, under certain conditions the RNase H activity of HIV-1 RT can cleave in vitro double-stranded RNA (3). An examination of this HIV-1 RT-associated activity has shown that the RNA is degraded if the enzyme is artificially arrested during DNA synthesis, even when no Mn2+ was present (9). Still, this activity was 30-fold slower than the rate of hydrolysis of the RNA in RNA-DNA heteroduplexes. Accordingly, it was suggested that the RNase H active site of HIV-1 RT can slowly cleave double-stranded RNA only if it is tightly bound to the duplex RNA (9). Yet, unlike the hereby-presented activity of Tf1 RT, the in vivo biological relevance of this HIV-1 RT activity is obscure. The results obtained here indicate that, although HIV-1 RT can cleave the substrate in a Mn2+-dependent mode, this activity differs from that of Tf1 RT, as the RNA segment(s) produced by HIV-1 RT cannot prime DNA synthesis. Therefore, one can hypothesize that this HIV-1 RT feature may represent a “fossil” activity, required for self-priming early in the evolution of LTR retrotransposons. It is possible as well that the Tf1 RT-associated self-primer cleavage activity shown here represents such a missing link. Only afterward in evolution have retroelements adapted to utilize cellular tRNAs as primers instead of their own RNA, despite the presumably straightforward prediction that it might be advantageous to use self-primers over utilizing heterologous cellular components. The obviously very intriguing and so-far-unanswered questions are what are the advantages for using these different primers and why did such a switch between self-primers and tRNA primers occur?
In summary, the reported ability of Tf1 RT to cleave, generate, and also utilize an RNA self-primer, which is independent of any cellular tRNA primers, is distinctive to the novel Tf1 RT studied herein. As far as we know, such a unique activity has never been reported for any RT. One question is whether any specific RNA sequences are required for this activity. We plan to assay this novel Tf1 RT activity with different self-complementing RNA sequences and to follow the mechanism of this function. In addition, we would like to learn whether the RT-associated RNase H and self-primer cleavage activities can be segregated by mutagenizing the RT and testing these two activities.
Acknowledgments
I am extremely grateful to Henry Levin from NIH for introducing us to the field of Tf1 and for his generous gift of the plasmids containing both the wild-type and the D710N Tf1 sequences. Many thanks also to G. Kaufmann and I. Oz Gleenberg from Tel Aviv University for critically reading the manuscript and providing helpful suggestions and to N. Kirshenboim for performing the experiments.
This research was supported in part by the Israeli Science Foundation (grant no. 411/07).
A. Hizi is an incumbent of the Gregorio and Dora Shapira Chair for the Research of Malignancies at Tel-Aviv University.
Footnotes
Published ahead of print on 27 August 2008.
REFERENCES
- 1.Avidan, O., and A. Hizi. 1998. The processivity of DNA synthesis exhibited by drug-resistant variants of human immunodeficiency virus type-1 reverse transcriptase. Nucleic Acids Res. 261713-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avidan, O., S. Loya, R. R. Tonjes, Z. Sevilya, and A. Hizi. 2003. Expression and characterization of a recombinant novel reverse transcriptase of a porcine endogenous retrovirus. Virology 307341-357. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Artzi, H., E. Zeelon, S. F. Le-Grice, M. Gorecki, and A. Panet. 1992. Characterization of the double stranded RNA dependent RNase activity associated with recombinant reverse transcriptases. Nucleic Acids Res. 205115-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutabout, M., M. Wilhelm, and F. X. Wilhelm. 2001. DNA synthesis fidelity by the reverse transcriptase of the yeast retrotransposon Ty1. Nucleic Acids Res. 292217-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, M., T. Goodwin, M. Simpson, M. Singh, and R. Poulter. 2001. Vertebrate LTR retrotransposons of the Tf1/sushi group. J. Mol. Evol. 52260-274. [DOI] [PubMed] [Google Scholar]
- 6.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 7.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz. 2002. Mobile DNA II. ASM Press, Washington, DC.
- 8.Eickbush, T. H., and V. K. Jamburuthugoda. 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotte, M., S. Fackler, T. Hermann, E. Perola, L. Cellai, H. J. Gross, S. F. Le Grice, and H. Heumann. 1995. HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J. 14833-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havecker, E. R., X. Gao, and D. F. Voytas. 2004. The diversity of LTR retrotransposons. Genome Biol. 5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hizi, A., C. McGill, and S. H. Hughes. 1988. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc. Natl. Acad. Sci. USA 851218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hostomsky, Z., S. H. Hughes, S. P. Goff, and S. F. Le Grice. 1994. Redesignation of the RNase D activity associated with retroviral reverse transcriptase as RNase H. J. Virol. 681970-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, M. Y., S. G. Eagle, M. Inouye, and S. Inouye. 1992. Cell-free synthesis of the branched RNA-linked msDNA from retron-Ec67 of Escherichia coli. J. Biol. Chem. 26713823-13829. [PubMed] [Google Scholar]
- 14.Kirshenboim, N., Z. Hayouka, A. Friedler, and A. Hizi. 2007. Expression and characterization of a novel reverse transcriptase of the LTR retrotransposon Tf1. Virology 366263-276. [DOI] [PubMed] [Google Scholar]
- 15.Krug, M. S., and S. L. Berger. 1989. Ribonuclease H activities associated with viral reverse transcriptases are endonucleases. Proc. Natl. Acad. Sci. USA 863539-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Grice, S. F. 2003. “In the beginning”: initiation of minus strand DNA synthesis in retroviruses and LTR-containing retrotransposons. Biochemistry 4214349-14355. [DOI] [PubMed] [Google Scholar]
- 17.Levin, H. L. 1995. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol. Cell. Biol. 153310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin, H. L. 1996. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol. Cell. Biol. 165645-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, J. H., and H. L. Levin. 1997. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev. 11270-285. [DOI] [PubMed] [Google Scholar]
- 20.Lin, J. H., and H. L. Levin. 1997. Self-primed reverse transcription is a mechanism shared by several LTR-containing retrotransposons. RNA 3952-953. [PMC free article] [PubMed] [Google Scholar]
- 21.Malik, H. S., and T. H. Eickbush. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 735186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizrahi, V., R. L. Brooksbank, and N. C. Nkabinde. 1994. Mutagenesis of the conserved aspartic acid 443, glutamic acid 478, asparagine 494, and aspartic acid 498 residues in the ribonuclease H domain of p66/p51 human immunodeficiency virus type I reverse transcriptase. Expression and biochemical analysis. J. Biol. Chem. 26919245-19249. [PubMed] [Google Scholar]
- 23.Pandey, M., S. Patel, and A. Gabriel. 2004. Insights into the role of an active site aspartate in Ty1 reverse transcriptase polymerization. J. Biol. Chem. 27947840-47848. [DOI] [PubMed] [Google Scholar]
- 24.Rausch, J. W., M. K. Grice, M. Henrietta, M. Nymark, J. T. Miller, and S. F. Le Grice. 2000. Interaction of p55 reverse transcriptase from the Saccharomyces cerevisiae retrotransposon Ty3 with conformationally distinct nucleic acid duplexes. J. Biol. Chem. 27513879-13887. [DOI] [PubMed] [Google Scholar]
- 25.Sevilya, Z., S. Loya, N. Adir, and A. Hizi. 2003. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is modulated by residue 294 of the small subunit. Nucleic Acids Res. 311481-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevilya, Z., S. Loya, S. H. Hughes, and A. Hizi. 2001. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is affected by the thumb subdomain of the small protein subunits. J. Mol. Biol. 311957-971. [DOI] [PubMed] [Google Scholar]
- 27.Skalka, A. M., and S. P. Goff. 1993. Reverse transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 28.Varmus, H. E. 1989. Reverse transcription in bacteria. Cell 56721-724. [DOI] [PubMed] [Google Scholar]