Abstract
Eastern and Venezuelan equine encephalitis viruses (EEEV and VEEV, respectively) cause severe morbidity and mortality in equines and humans. Like other mosquito-borne viruses, VEEV infects dendritic cells (DCs) and macrophages in lymphoid tissues, fueling a serum viremia and facilitating neuroinvasion. In contrast, EEEV replicates poorly in lymphoid tissues, preferentially infecting osteoblasts. Here, we demonstrate that infectivity of EEEV for myeloid lineage cells including DCs and macrophages was dramatically reduced compared to that of VEEV, whereas both viruses replicated efficiently in mesenchymal lineage cells such as osteoblasts and fibroblasts. We determined that EEEV infection of myeloid lineage cells was restricted after attachment, entry, and uncoating of the genome. Using replicon particles and translation reporter RNAs, we found that translation of incoming EEEV genomes was almost completely inhibited in myeloid, but not mesenchymal, lineage cells. Alpha/beta interferon (IFN-α/β) responses did not mediate the restriction, as infectivity was not restored in the absence of double-stranded RNA-dependent protein kinase, RNase L, or IFN-α/β receptor-mediated signaling. We confirmed these observations in vivo, demonstrating that EEEV is compromised in its ability to replicate within lymphoid tissues, whereas VEEV does so efficiently. The altered tropism of EEEV correlated with an almost complete avoidance of serum IFN-α/β induction in vivo, which may allow EEEV to evade the host's innate immune responses and thereby enhance neurovirulence. Taken together, our data indicate that inhibition of genome translation restricts EEEV infectivity for myeloid but not mesenchymal lineage cells in vitro and in vivo. In this regard, the tropisms of EEEV and VEEV differ dramatically, likely contributing to observed differences in disease etiology.
Members of the Alphavirus genus, family Togaviridae, are enveloped, positive-sense RNA viruses, the majority of which are vectored by arthropods between avian and/or mammalian hosts (26). Three mosquito-borne alphaviruses that are geographically restricted to the Americas cause severe, potentially fatal encephalitis in humans and equines when they emerge from the enzootic transmission cycle: namely, eastern equine encephalitis virus (EEEV), western equine encephalitis virus, and Venezuelan equine encephalitis virus (VEEV). Both VEEV and EEEV cause epizootic outbreaks of severe, frequently fatal encephalitis in equines (54). However, disease etiologies in humans differ between these two viruses for reasons that are not well understood.
Although VEEV is classified as an encephalitic alphavirus, VEEV infection of humans typically causes a “flu-like” illness that ranges from mild to severe, with prominent lymphoid and reticuloendothelial involvement (16). Progression to neurological disease occurs in only 4 to 14% of symptomatic cases, and fatalities are rare (28, 53). By comparison, EEEV causes far more severe encephalitic disease in humans. Infected individuals experience the sudden onset of fever, chills, myalgia, arthralgia, retro-ocular pain, a headache of increasing severity, and decreased consciousness for several days (17). Then infection progresses rapidly to neurological disease with the possibility of paralysis, seizures, coma, and death in 30 to 80% of patients, and mild to severe long-term neurologic deficits occur in an estimated 35% of the survivors (17). Fortunately, human cases of EEEV are rare in the United States, most likely due to rare exposure to the virus, with only 254 confirmed and probable cases documented between 1964 and 2007 (12). Disturbingly, surveillance indicates that EEEV is endemic to most states in the eastern United States, increasing the likelihood that a relatively minor shift in environmental conditions could trigger an EEEV epidemic outbreak in human populations. Isolation of EEEV from Aedes albopictus mosquitoes, which have recently been introduced into areas in the United States where EEEV is endemic, has heightened concern because of the opportunistic feeding behavior of these mosquitoes as well as their apparent high vector competence for this virus (35).
Experimental VEEV infection of mice, in which virus is injected subcutaneously to mimic the bite of an infected mosquito vector, closely parallels the biphasic disease course seen in horses and severe human infections, and this model has been used extensively to elucidate steps in VEEV pathogenesis. After deposition in the skin, replicating virus is first observed in the draining lymph node (DLN) within 4 h postinfection (p.i.), preceding replication at the site of inoculation by several hours (6). VEEV targets Langerhans cells, dendritic cells (DCs), and possibly also macrophages in the skin and exploits the migratory properties of these immune system sentinel cells to spread from the site of inoculation and access the regional DLN (34; W. B. Klimstra, D. L. Browning, C. L. Gardner, and K. D. Ryman, unpublished data). A high-titer serum viremia is produced which seeds peripheral tissues beyond the inoculation site including other lymphoid and reticuloendothelial tissues such as LN, spleen, thymus, Peyer's patches, and pancreas and sporadically infects skeletal muscle (13, 24). VEEV infection induces the systemic release of type I interferon (alpha/beta interferon [IFN-α/β] [13]) and transcriptional upregulation of proinflammatory cytokine genes in the DLN and spleen as early as 6 h p.i. (23). By 48 to 72 h p.i., the virus has invaded the central nervous system (CNS) via the olfactory and trigeminal nerves (14, 45), resulting in fatal encephalitis by 7 to 10 days p.i.
Wild-type EEEV also produces a uniformly fatal encephalitis in peripherally inoculated mice (3, 33, 46, 51), but it differs from VEEV infection in at least one important regard: the DLN and spleen appear to be largely spared during EEEV infection (51). However, once morbidity is apparent, EEEV infection is rapidly fatal. Vogel et al. (51) demonstrated in vivo that EEEV favors cells of the mesenchymal lineage, particularly osteoblasts, as targets for peripheral amplification. Edelman and coworkers (32) suggested that the tropism of VEEV and EEEV might also differ in humans, demonstrating that VEEV efficiently infects human leukocytes in vitro but that these cells are relatively refractory to infection by EEEV. Thus, EEEV appears to be an anomaly among the alphaviruses in that this virus does not replicate well in lymphoid tissues but nevertheless achieves neuroinvasion and is highly neurovirulent.
We have further investigated these findings to determine the underlying reasons for differing disease etiologies. In head-to-head comparison with wild-type VEEV, clinical signs of infection with a North American strain of EEEV appear significantly later, implying that the lymphotropic phase of disease is less prominent. This is correlated with restricted tropism of EEEV for cells of the myeloid lineage both in vivo and in vitro. The apparent inability of EEEV to infect DCs and macrophages is controlled not by the virus-receptor interaction but at the level of genomic translation necessary to synthesize the nonstructural proteins (nsPs) and initiate replication. Moreover, although we have recently demonstrated that translation of the alphavirus genome is sensitive to the double-stranded RNA-dependent protein kinase (PKR)-dependent and PKR-independent antiviral activities of type I IFN (43, 49), translation of the EEEV genome in myeloid cells was not restored in the absence of PKR/RNase L- or IFN-α/β-mediated responses. Finally, we present data suggesting that sparing of the lymphoid tissues dramatically reduces the IFN-α/β inductive response in vivo, perhaps representing a unique approach to the evasion of antiviral responses within the host. Thus, we posit that disease etiology is determined by the interaction between the infecting virus and the host's immune system, coupled with the tropism of the virus for specific host tissues such as joint and lymphoid tissues versus CNS. These findings have important implications for the future design of acute-phase antiviral therapeutics and vaccines against EEEV and other alphaviruses.
MATERIALS AND METHODS
Cell lines.
Baby hamster kidney (BHK-21) and L929 murine fibrosarcoma cell lines were maintained in alpha minimum essential medium (αMEM) supplemented with 10% donor calf serum (DCS) and 10% tryptose phosphate broth. Murine RAW 264.7 monocyte-macrophage cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). All media contained 100 U/ml penicillin, 0.05 mg/ml streptomycin, and l-glutamine.
Mice.
Outbred CD-1 mice were purchased from Charles River Laboratories and housed in the Animal Resource Center at Louisiana State University Health Sciences Center—Shreveport (LSUHSC-S) under specific-pathogen-free conditions. All procedures were carried out in accordance with the guidelines of the LSUHSC-S Institutional Animal Care and Use Committee. Mice were used at 6 to 8 weeks of age, and all experiments were performed in the biosafety level 2 or 3 animal facilities at LSUHSC-S.
Primary cell cultures.
Bone marrow-derived, conventional DC (cDC) cultures were generated and maintained as described previously (30) in RPMI 1640 medium supplemented with 10% FBS, 1% nonessential amino acids, 1 mM sodium pyruvate, 5 mM HEPES buffer, 50 μM β-mercaptoethanol, 10 ng/ml granulocyte-macrophage colony-stimulating factor, and 10 ng/ml interleukin-4 (Peprotech, Rocky Hill, NJ). Briefly, bone marrow cells aspirated from femurs and tibias of CD-1 mice were plated for 1 h (37°C, 5% CO2) to allow attachment of macrophages (see below), after which nonadherent DC progenitors were removed and cultured. After 7 days, nonadherent cDCs in the culture supernatant were pelleted, counted, and resuspended in complete medium for infection. Adherent macrophages from bone marrow were cultured in Dulbecco's modified Eagle's medium supplemented with 20% L929-conditioned supernatant and 10% FBS, harvested after 7 days by scraping, pelleted, and seeded appropriately for experiments.
To generate primary murine osteoblast cultures, calvaria were dissected from 3- to 5-day-old suckling mice, scraped gently until white, and triply digested in αMEM containing 0.05% trypsin-EDTA and 10 mg/ml collagenase P, as described previously (7). After being cultured for 5 days in αMEM supplemented with 15% FBS, osteoblasts which had grown out from digested calvaria were trypsinized, strained to remove bone fragments, and seeded into 150-mm culture dishes. Once confluent, osteoblasts were harvested by trypsinization, counted, and seeded appropriately for experiments. All media contained 100 U/ml penicillin and 0.05 mg/ml streptomycin.
Viruses.
Construction of cDNA clones for the wild-type EEEV strain FL93-939 (1), VEEV strain ZPC738 (5), and another alphavirus, Sindbis virus (SB) strain TR339 (29), have been described previously. FL93-939 and ZPC738 cDNA clones were generously provided for these studies by Scott Weaver (University of Texas Medical Branch, Galveston). Capped, infectious viral RNAs were generated by in vitro transcription (mMessage mMachine; Ambion) from linearized cDNA plasmid templates and electroporated into BHK-21 cells. Virus particles were harvested from the supernatant 20 to 24 h after electroporation when cytopathic effect (CPE) was evident, clarified of cell debris by centrifugation, and stored at −80°C in single-use aliquots. Virus stock titers were determined by standard BHK-21 cell plaque assay, and titers were expressed as PFU/ml.
Replicons.
The replicon plasmid constructs used in this study were (i) replicons expressing green fluorescent protein (GFP) and (ii) replicons expressing firefly luciferase (fLUC). We have previously described the construction of SB-based replicon genomes expressing both GFP and fLUC and the packaging of these genomes into replicon particles (SBREP) by using a bipartite helper system to provide the structural proteins (40, 41, 44). The construction and packaging of analogous VEEV-based replicon particles (VREP) expressing GFP have been previously described (34, 39). These constructs were generously provided by Robert Johnston (University of North Carolina, Chapel Hill). The fLUC-expressing VEEV-based replicon genome was generated by the directional cloning of the fLUC gene immediately downstream of the 26S mRNA promoter into the multiple cloning site of the pVR21 replicon plasmid.
An analogous EEEV-based replicon system (EREP) was developed from the full-length cDNA clone of FL91-4679 (46). Shuttle vector 1 (SV1) was created by cloning the PmlI (nsP4)-NaeI (vector) fragment of the original FL91-4679 replicon with a BglII site immediately downstream of the 26S subgenomic promoter and upstream of the 3′ nontranslated region (NTR) into the ZeroBlunt vector (Promega). The GFP gene was amplified by PCR to introduce BglII termini, cloned into the BglII site in SV1, and screened for the correct orientation. A fragment spanning ClaI (nsP4) to NotI [immediately downstream of the poly(A) sequence] was reintroduced into EREP (EREP-GFP). Subsequently, a second shuttle vector (SV2) was made in which a multiple cloning site (AscI-XbaI-AgeI-PacI-BglII) was introduced in place of the BglII site by overlap PCR mutagenesis. The fLUC gene was amplified with 5′ AscI/3′ PacI termini and directionally cloned into SV2. As for the EREP-GFP, this fragment was cloned into EREP via ClaI-NotI.
To generate an EREP genome with the fLUC gene fused in frame within nsP3, a unique AgeI site was introduced by megaprimer PCR mutagenesis into the nsP3 gene at nucleotide (nt) 5175 (corresponding to amino acid positions 384 and 385 in the nsP3 variable region). The PCR fragment with the AgeI site was cloned into EREP-GFP using AvrII sites upstream (in nsP3) and downstream at nt 5175 in nsP4. AgeI sites were added to the 5′ and 3′ termini of the fLUC gene by PCR (without stop codon), cloned into EREP-nsP3LUC, and screened for directionality and fLUC production.
The EEEV capsid helper plasmid was generated by cloning a PCR-amplified C gene with initiation and termination codons and BglII termini into SV1. A fragment spanning BstEII (nsP4) to NotI [after poly(A)] was cloned into EREP from which the nsP genes had been removed between nt 1513 and nt 7183 by digestion with StuI (nsP1) and ClaI (nsP4) and blunt-end religation. The EEEV glycoprotein helper plasmid was generated by cloning PCR-amplified PE2/E1 genes with initiation and termination codons and 5′ AscI/3′ PacI termini directionally into SV2. The BstEII-NotI fragment was cloned into the StuI-ClaI-deleted EREP as described for the capsid helper.
Chimeric helpers to package EREP genomes in SB structural proteins are as described for EEEV capsid and glycoprotein helpers but with amplification of the C or PE2/E1 genes from wild-type SB strain TR339 instead of EEEV. Chimeric helpers to package the VREP genome in EEEV structural proteins are described elsewhere (Klimstra et al., submitted).
Alphavirus replicon particles expressing fLUC or GFP were produced by packaging infectious replicon RNA transcripts in viral structural proteins provided in trans by helper RNAs as previously described (44). Briefly, the replicon RNA genome encoding the viral nsP genes and expressing the heterologous gene from the viral 26S promoter, along with two defective helper RNAs providing the wild-type capsid and glycoprotein genes but lacking the virus-specific packaging signal, was coelectroporated into BHK-21 cells. Due to the lack of viral structural genes in the replicon genome, infectious replicon particles undergo only one round of infection. From each preparation, 10% of the total volume was evaluated by serial passage on BHK-21 cells for the presence of CPE-inducing, propagation-competent virus recombinants or contaminants. Replicon particles were concentrated from supernatants by centrifugation through a 20% sucrose cushion and resuspended in OptiMEM serum-free growth medium (Mediatech).
Translation reporters.
Construction and in vitro transcription conditions for the SB, EEEV, and VEEV RNA translation reporters have been described previously (43, 49). Briefly, primers were designed to truncate the nsP1 gene and fuse in frame with the fLUC gene followed by the 3′ NTR and poly(A) tail, yielding a reporter whose translation was initiated from the authentic genome initiation site of the respective virus.
Mortality and pathogenesis studies.
Virus inocula containing 103 PFU of VEEV or EEEV in a 10-μl volume (1 × 105 PFU/ml) were administered subcutaneously in each hind leg footpad using a 27-gauge needle and 100-μl gastight Hamilton syringe. Mock-infected mice received 10 μl phosphate-buffered saline (PBS)-1% DCS by the same route. Virus-infected and corresponding mock-infected mice were observed at 24-h intervals, scored for degree of sickness, and weighed where appropriate. Average survival times (ASTs) and percent mortality were then calculated.
At predetermined intervals p.i., groups of three mice per treatment were sacrificed under isoflurane anesthesia and blood was collected by cardiac puncture. Serum was separated from whole blood by using Microtainer tubes (Becton-Dickinson) and stored in single-use aliquots at −80°C. Mice were perfused with PBS-1% DCS for 10 min at 7 ml/min to flush blood-borne virus. Tissues were harvested into preweighed Kontes tubes, homogenized in PBS-1% DCS, and clarified by centrifugation (13,000 × g, 15 min, 4°C). Supernatants were assayed for virus by standard plaque assay, expressed as PFU/DLN, ml, or g.
In vivo bioluminescence imaging and data quantification.
Inoculation with fLUC-expressing replicon particles was performed as described above. In vivo imaging of fLUC activity in mice was performed on a charge-coupled device camera (Xenogen Corp., Alameda, CA). Briefly, mice were anesthetized by intraperitoneal injection of ketamine-xylazine (90 mg ketamine/kg of body weight and 10 mg/kg xylazine) and 0.7 mg/kg luciferin substrate approximately 10 min prior to imaging. Images were acquired for 2 to 10 s, depending on relative light emission from various sites of infection. Relative intensities of transmitted light from in vivo bioluminescence were represented as a pseudocolor image ranging from violet (least intense) to red (most intense) superimposed on the corresponding gray-scale photograph using LivingImage (Xenogen) image analysis software. fLUC expression was quantified by dissecting the DLNs of these animals and performing an in vitro luciferase activity assay (Stop n’ Glo; Promega). Luciferase activity is expressed as relative light units (RLU) per μg of protein.
Virus growth curves and replicon infections.
Adherent cells were infected in 24-well or 6-well plates at a multiplicity of infection (MOI) of 1 PFU/cell or 0.1 PFU/cell, as indicated in the figure legends. After incubation with virus or replicon particles for 1 h at 37°C, cells were washed three times with PBS-1% DCS diluent, appropriate medium was replaced, and cells were incubated (37°C, 5% CO2). Nonadherent cDCs were infected in suspension in 96-well plates at the desired MOI (37°C, 1 h) and washed three times in PBS-1% DCS by pelleting and resuspension. Cells were resuspended in supplemented RPMI 1640 medium and seeded into larger wells as appropriate. For virus growth curves, supernatants were harvested at time zero and subsequent time points for titration by plaque assay. For GFP-expressing replicon infections, cells were monitored by fluorescence microscopy and photographed using equal exposure times. For fLUC-expressing replicon infections, cell lysates were prepared and luciferase activity was assayed as described above.
Translation assays.
Virus-derived reporters were introduced into cells by electroporation (7.5 μg RNA per reaction mixture; Bio-Rad GenePulser). Briefly, harvested cells were pelleted twice and electroporated in OptiMEM. Cells were then plated in six-well plates or maintained in suspension for various intervals. Luciferase assays were performed on cell lysates as previously described (43).
IFN-α/β analyses.
Serum IFN-α/β was measured by standard biological assay on L929 cells as described previously (50), using a commercially prepared IFN-α/β standard (Access Biomolecular) and encephalomyocarditis virus as the indicator virus. The endpoint was defined as the dilution of IFN-α/β required to protect 50% of the cells from encephalomyocarditis virus-induced CPE, and the level of IFN-α/β was expressed as international units (IU)/ml or g.
RESULTS
Differential disease progression following VEEV and EEEV infection.
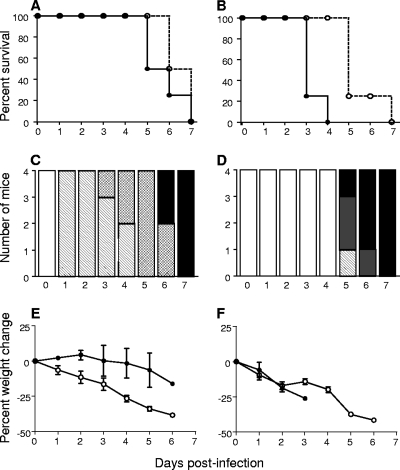
Stocks of wild-type VEEV strain ZPC738 and North American EEEV strain FL93-393 were generated from genome-length cDNA clones (1, 5). We performed a head-to-head comparison of wild-type VEEV and EEEV infections of adult CD-1 mice to explore anecdotal observations that the clinical disease course differs between these two closely related viruses (51). A peripheral route of inoculation was utilized to mimic natural infection and to facilitate an analysis of viral replication, dissemination, neuroinvasion, and neurovirulence as discrete steps. Mice were infected with 103 PFU subcutaneously in the hind leg footpad, corresponding to approximately 50 50% lethal doses of each virus determined by dose dependence studies (data not shown). Following subcutaneous administration of VEEV and EEEV at comparable doses, both viral infections were observed to be rapidly and uniformly fatal (Fig. 1A), with no significant differences in ASTs (4.8 ± 1.0 days for EEEV versus 5.5 ± 0.6 days for VEEV; P = 0.23).
FIG. 1.
Morbidity and mortality differ following infection of outbred mice with VEEV or EEEV. Adult, outbred CD-1 mice were infected with cDNA clone-derived ZPC738 VEEV or FL93-939 EEEV and monitored for morbidity and mortality as follows. (A and B) Percent survival following subcutaneous (A) or intracerebral (B) inoculation of CD-1 mice with 103 PFU of VEEV (white circles, dashed line) or EEEV (black circles, solid line). (C and D) Clinical score over the disease course of mice infected subcutaneously with 103 PFU of VEEV (C) or EEEV (D). Clinical scores to assess signs of disease in the mice were as follows: 0, healthy (white); 1, ruffled fur and behavioral changes (hatched); 2, paresis or ataxia (crosshatched); 3, moribund (gray); 5, dead (black). (E and F) Percent weight change over the disease course of mice that were infected subcutaneously (E) or intracerebrally (F) with 103 PFU of VEEV (white circles) or EEEV (black circles). Data are representative of at least two independent experiments.
During the course of these experiments, it was noted that, despite the similar ASTs, VEEV-infected animals exhibited disease signs significantly earlier than did those infected with EEEV. To quantify these observations, we recorded ensuing signs of morbidity by clinical score (Fig. 1C and D) and weight loss criteria (Fig. 1E). In the early stages of infection, VEEV-infected mice experienced increased severity of disease compared to EEEV-infected counterparts, with piloerection and weight loss invariably evident within 24 h p.i. and mice becoming ataxic by 72 h p.i. VEEV-infected mice developed evidence of CNS infection, 4 to 6 days p.i., progressing to paresis/paralysis and death. In striking contrast, EEEV-infected mice appeared healthy during the first 3 to 4 days p.i. with little weight loss and exhibited signs of disease only 12 to 24 h before death. Frequent seizures were observed in the latter stages of EEEV infection without signs of paralysis, whereas seizures were rarely observed in VEEV-infected counterparts.
We compared the neurovirulence of EEEV and VEEV in the absence of a requirement for peripheral replication and neuroinvasion, by introducing the virus inoculum intracerebrally. Although both virus infections caused the mice to lose weight within 24 h p.i. (Fig. 1F), EEEV-infected mice had a dramatically shorter AST than did VEEV-infected mice (2.3 ± 0.5 days versus 4.5 ± 1.0 days, respectively; P = 0.007; Fig. 1B), from which we infer that at similar doses EEEV is more neurovirulent than VEEV.
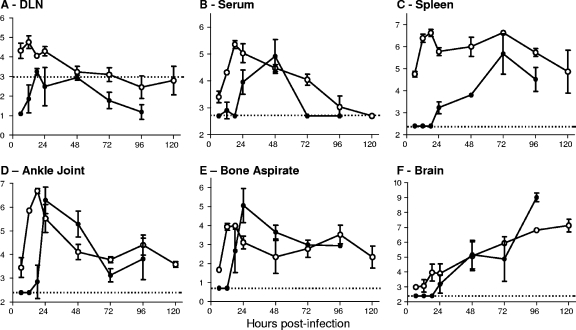
Differential tissue tropisms of VEEV and EEEV in vivo.
To reveal pathogenic correlates of the differential disease course described above, we compared the replication and dissemination of VEEV and EEEV in vivo following subcutaneous inoculation of 103 PFU per footpad. Serum and PBS-perfused tissues were collected from virus-infected mice at various times p.i. to measure serum viremia and viral load within the DLN, spleen, liver, muscle, bone aspirate, ankle joint, and brain. Both the sites and the kinetics of EEEV and VEEV replication were found to be distinctly different (Fig. 2). Compared with VEEV, which reached peak titers in serum and peripheral tissues within 6 to 12 h p.i., replication and spread of EEEV within all the tissues were delayed by 18 to 24 h. It should be noted that differential specific infectivity of VEEV versus EEEV virions for BHK cells could influence comparative plaque assay titers; however, all indications are that EEEV is equally as infectious per particle as or more infectious per particle than VEEV (Klimstra et al., unpublished), and therefore, in a direct comparison, EEEV titers tend to be slightly overestimated relative to those of VEEV.
FIG. 2.
Replication and dissemination profiles of VEEV and EEEV differ in subcutaneously inoculated mice. CD-1 mice, infected with 103 PFU of ZPC738 VEEV (white circles) or FL93-939 EEEV (black circles) subcutaneously in each rear footpad, were sacrificed at various times p.i. Titers of virus from DLN (A), serum (B), spleen (C), ankle joint (D), bone aspirate (E), and brain (F) were determined. Values represent the geometric mean virus titers (log10 PFU/ml or g) for three mice, determined on BHK cells, and are shown ± standard deviations. Dotted line denotes inoculum dose (A) or limit of detection (B to F).
(i) DLNs.
The DLN is a known site of early VEEV amplification, believed to be important for seeding of the primary serum viremia and dissemination (24, 55). As expected, in our study the VEEV ZPC738 strain replicated rapidly and extensively in the popliteal LNs following subcutaneous inoculation in the rear footpad, reaching peak titers of approximately 105 PFU/LN within 12 h p.i. and gradually decreasing after 24 h p.i. (Fig. 2A). However, on a per-LN basis, titers in EEEV-infected mice barely exceeded the initial inoculum of 103 PFU by 18 h p.i. Thus, peak titers within the popliteal DLN of VEEV-infected mice were over 100-fold higher than those in EEEV-infected counterparts and were attained at least 6 h earlier.
(ii) Serum viremia.
Viremic virus was detectable within 6 h of VEEV infection, peaking by 18 h p.i. and then gradually diminishing, to fall below the limit of detection (BLD) by 5 days p.i. (Fig. 2B). These data are consistent with previous descriptions of early release of virus from the DLN, fueling of the viremia by release from other infected tissues at later times, and clearance coincident with the development of humoral immunity (13). In contrast, the serum viremia in EEEV-infected mice was delayed and not consistently evident until 24 h p.i., peaking 24 h later and then falling BLD by 72 h p.i. Although peak levels of serum viremia were approximately equal in the two virus infections as measured by plaque assay, EEEV viremia was of considerably shorter duration. Furthermore, the magnitude and timing suggested that replication in the DLN was not a significant source of viremic virus during EEEV infection.
(iii) Peripheral tissues.
To examine replication of the two viruses in the periphery, virus titers were determined for a range of tissues (Fig. 2C to E). We initially focused on lymphoid and reticuloendothelial tissues, known to be major targets in VEEV infection (13, 24, 34), and consistent with these previous reports, the ZPC738 VEEV strain replicated extensively within the spleen (Fig. 2C) and liver (data not shown), peaking at 18 h p.i. and remaining elevated throughout the course of infection. However, EEEV replication was delayed, peaking ∼72 h p.i. Although EEEV exhibited replication levels as high as those of VEEV at 72 h p.i., these levels were not maintained and viral loads were observed to be decreasing by 96 h p.i.
Detailed immunohistochemical and in situ hybridization analyses performed by Vogel et al. (51) indicated that EEEV preferentially infects cells of the mesenchymal lineage in peripheral tissues, including osteoblasts, fibroblasts, and myocytes. Similarly in our studies, titration of virus from the epiphyses and metaphyses of the femur (Fig. 2D) and bone marrow aspirates (Fig. 2E) demonstrated efficient replication of EEEV in these tissues 1 to 2 days p.i., peaking at levels similar to those of VEEV although delayed by approximately 12 h. Both tissues would be expected to contain a proportion of osteoblasts derived from the periosteum and/or endosteum. Furthermore, VEEV and EEEV replicated to equivalent levels in the gastrocnemius muscle (data not shown).
(iv) CNS.
VEEV was detectable in the CNS as early as 6 h p.i. and continued to accumulate throughout the course of infection. EEEV was not detectable in the CNS until 24 h p.i. but amplified rapidly, equaling VEEV titers by 48 h p.i. and surpassing them by 100-fold at 96 h p.i. (Fig. 2F). Taken together, these data suggest that the lack of early disease signs in EEEV-infected mice may be due to the sparing of lymphoid tissues, with the extensive CNS replication accounting for the rapid onset of disease, seizures, and death.
Early cellular targets for virus replication are distinctly different.
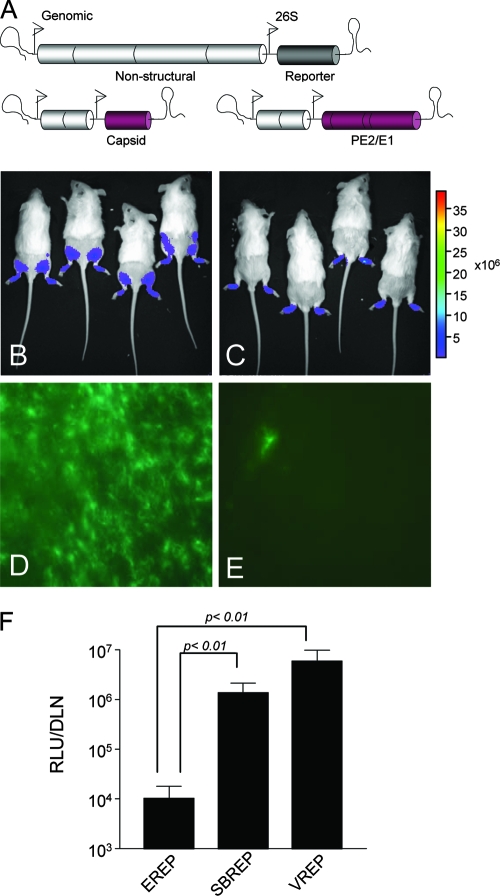
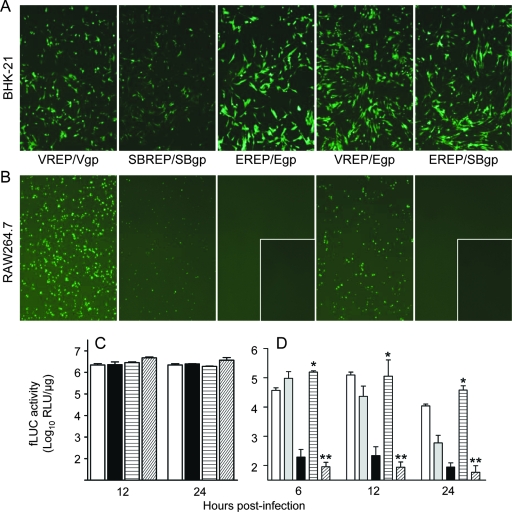
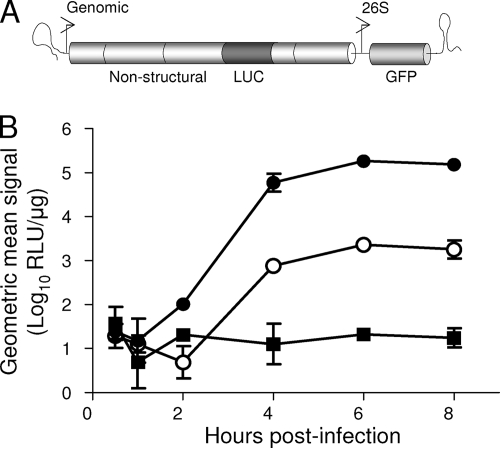
During the initial steps of VEEV spread from the site of inoculation in the skin, DCs and macrophages are primary targets for infection by inoculated VEEV particles and migrate to the DLN following infection (34; Klimstra et al., unpublished). These events have been characterized using viral replicon particles which can undergo only one round of infection and do not produce progeny virions, thus unequivocally marking early cellular targets (34, 39). While EEEV antigen is found in the DLN early after inoculation, little viral RNA is detectable, suggesting that replication is limited (51). We have further demonstrated that infectious EEEV virion titers barely surpass inoculum levels (Fig. 2A). To specifically examine and quantify infection of cells at the site of inoculation and in the DLN, CD-1 mice were inoculated subcutaneously in both rear footpads with equal doses of packaged replicon particles, representative of EEEV (EREP), VEEV (VREP), or SB (SBREP), expressing fLUC from the subgenomic promoter of the replicon genome as an infection reporter (Fig. 3A). At 8 h p.i. (Fig. 3) and 48 h p.i. (data not shown), mice were injected intraperitoneally with luciferin substrate and imaged (In Vivo imaging system; Xenogen Corp.) to reveal the location of initial cellular targets. In VREP-inoculated mice, signal appeared in areas corresponding to the footpad, the DLN (popliteal), and occasionally also the inguinal LN (Fig. 3B). Imaging of SBREP-infected animals produced qualitatively similar results, although signal intensity was generally reduced (data not shown), consistent with the avirulent phenotype of wild-type SB in adult mice (40). However, in EREP-inoculated mice, dramatically lower levels of signal were emitted from the DLN, with the majority of signal confined to the inoculation site in the footpad (Fig. 3C). When popliteal LNs were harvested and processed for in vitro fLUC activity assay, fLUC activity in EREP-infected DLNs was significantly reduced compared to that in both VREP- and SBREP-infected DLNs (∼500-fold and 100-fold lower, respectively; Fig. 3F). Thus, EREP is impaired in its ability to infect DLN cells during the first round of replication compared with VREP and even compared with replicons derived from nonpathogenic SB.
FIG. 3.
EEEV replication within DLN cells is reduced compared to that of VEEV. CD-1 mice were inoculated subcutaneously in each rear footpad with EEEV-based (EREP) or VEEV-based (VREP) replicon particles expressing a reporter of infection, GFP or fLUC. (A) Diagrammatic representation of replicon genome and bipartite helper system. (B and C) Mice infected with fLUC-expressing VREP (B) or EREP (C) were inoculated intraperitoneally with luciferin substrate as described in Materials and Methods and subjected to in vivo imaging at 8 h p.i. The intensity of the luciferase signal is depicted by a pseudocolor map. All animals were imaged for 2 s to prevent signal saturation. (D to F) DLNs were dissected from these mice as well as SBREP-inoculated controls, and fLUC activity was quantified by in vitro luciferase assay (F). Datum points are presented as RLU per DLN ± standard deviation where n = 8, and confidence limits are indicated. DLNs from mice similarly inoculated with GFP-expressing VREP (D) or EREP (E) were harvested 8 h p.i., flattened with a coverslip, and photographed using a Nikon inverted fluorescence microscope to reveal GFP-positive cells.
To further confirm these findings, popliteal LNs were collected 8 h p.i. with GFP-expressing VREP or EREP and viewed as whole organs by fluorescence microscopy (Fig. 3D and E). Rare GFP-positive infected cells with DC-like morphology were observed in the DLN of EREP-inoculated animals compared with almost confluent GFP expression in the subcapsular sinus of DLNs from VREP-inoculated counterparts. These cells have previously been shown to be primarily CD11c+, CD11b+, and/or F4/80+ myeloid lineage cells (Klimstra et al., unpublished).
Apparent restriction of virus tropism to mesenchymal, not myeloid, lineage cells.
To determine whether the relative inability of EEEV to efficiently replicate within lymphoid tissues is due to the inability to productively infect myeloid cells, we infected CD-1 bone marrow-derived cDCs (Fig. 4A) and macrophages (data not shown) or RAW 264.7 monocytes/macrophages (Fig. 4B) and measured progeny virion release over a time course. Primary cDCs and macrophages were determined to be almost completely refractory to EEEV infection but permissive to infection by VEEV. Interestingly, the RAW 264.7 cell line exhibited low permissivity to EEEV infection, although significantly less than that to VEEV infection, allowing a gradual accumulation of progeny virions in the supernatant. In striking contrast, similar analyses in primary mesenchymal lineage osteoblasts (Fig. 4C) and BHK-21 fibroblasts (Fig. 4D) revealed that both EEEV and VEEV replicated efficiently. These data suggest that EEEV has a greatly reduced capacity to productively infect myeloid lineage cells in vitro, consistent with in vivo observations. However, infectivity for mesenchymal lineage cells, including osteoblasts, which have been identified as a predominant infected cell type in vivo (51), appears to be unimpaired or even enhanced compared with that of VEEV.
FIG. 4.
EEEV productively infects mesenchymal, but not myeloid, lineage cells. Primary CD-1 bone marrow-derived DCs (A), RAW 264.7 monocytes/macrophages (B), primary CD-1-derived osteoblasts (C), and BHK-21 fibroblasts (D) were infected with VEEV (white circles) or EEEV (black circles) at equal MOIs (0.1). Titers of progeny virus released into the supernatant were determined by plaque assay. Values represent the geometric mean virus titers (log10 PFU/ml). Datum points are shown ± standard deviations where n = 3.
Replication block occurs before subgenomic RNA translation but after entry and uncoating.
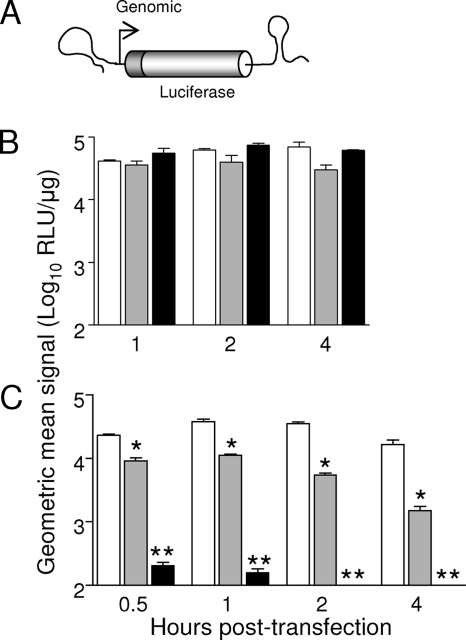
To determine the nature of this restricted tropism, we utilized alphavirus replicons to investigate infectivity and segregate steps in the virus replication cycle. Expression of the reporter protein (GFP or fLUC) from the 26S subgenomic RNA is indicative that the following steps in replication have occurred: binding/entry and uncoating of the replicon genome, translation of the replicon genome to produce the nsPs, synthesis of negative-sense template RNA, and synthesis and translation of the subgenomic RNA.
All three replicons replicated efficiently in mesenchymal lineage cells (represented by fibroblasts) as indicated by expression of GFP (Fig. 5A) and fLUC (Fig. 5C), although expression from SBREP was typically lower per cell. However, whereas both SBREP and VREP replicated within the myeloid cells (represented by RAW 264.7 macrophages), EREP infected these cells with dramatically lower efficiency (Fig. 5B and D). By 8 h p.i, expression of the reporter in SBREP-infected RAW 264.7 cells was reduced compared to that in cells infected with VREP, similar to BHK-21 cells, presumably reflecting a reduced ability of genomic replication or translation of the subgenomic RNA. We extended these observations to survey infectivity for other myeloid cell types including primary murine cDCs and macrophages (CD-1 derived), primary human DCs, and the human monocytic THP-1 cell line and observed that EEEV-based replicons possessed little to no infectivity for any myeloid lineage cell type tested, whereas VEEV replicons infected all types efficiently (data not shown). In contrast, we observed that EEEV replicons were highly infectious for all mesenchymal lineage cells tested, including primary CD-1 osteoblasts, the osteoblastic MC3T3 cell line (both undifferentiated and differentiated), DF-1 chicken fibroblasts, and Chinese hamster ovary (CHO K1) cells (data not shown).
FIG. 5.
The restricted tropism of EEEV for myeloid lineage cells is not mediated by the virus-receptor interaction. (A and B) Cells of the mesenchymal (A) (represented by BHK-21 fibroblasts) or myeloid (B) (represented by RAW 264.7 monocytes/macrophages) lineage were infected at equal multiplicities (MOI = 1) with GFP-expressing parental or chimeric replicon particles and photographed using a fluorescence microscope to reveal GFP-positive cells. Replicons are as follows: VEEV replicon genome with VEEV structural proteins (VREP/Vgp) or EEEV structural proteins (VREP/Egp), SB replicon genome with SB structural proteins (SBREP/SBgp), and EEEV replicon genome with EEEV structural proteins (EREP/Egp) or SB structural proteins (EREP/SBgp). Inset panels for EREP/Egp and EREP/SBgp depict infection at 100-fold-higher MOIs than those of other infections. (C and D) Parallel experiment in which fibroblasts (C) or CD-1-derived cDCs (D) were infected with fLUC-expressing parental and chimeric replicons: VREP/Vgp (white bars), SBREP/SBgp (gray bars), EREP/Egp (black bars), VREP/Egp (horizontally hatched bars), and EREP/SBgp (diagonally hatched bars). Cells were harvested for luciferase activity assay at various times p.i. Values represent the geometric mean fLUC activities (log10 RLU/μg). Datum points are shown ± standard deviations where n = 3. For panel D, a single asterisk indicates that VREP/Egp is not significantly reduced compared with VREP/Vgp (P > 0.5) and a double asterisk indicates that EREP/SBgp is not significantly increased versus EREP/Egp (P > 0.5); EREP/Egp values are significantly reduced compared with those for SBREP/SBgp and VREP/Vgp at all time points (P < 0.01).
To determine whether the apparent block in replication of EEEV in myeloid cells was due to binding, entry, and/or uncoating, chimeric replicon particles were produced in which the reporter-expressing replicon genome of one alphavirus was coated with the structural proteins of another (38; Klimstra et al., unpublished). The infectivities of chimeric replicons with the VREP genome encapsidated in the EEEV structural proteins (VREP-Egp) or the EREP genome encapsidated in the SB structural proteins (EREP-SBgp) were comparable to those of the respective replicon particles packaged in homologous structural proteins (VREP-Vgp and EREP-Egp) in BHK-21 fibroblasts (Fig. 5A and C), indicating that packaging in heterologous structural proteins did not produce defective virions. Furthermore, the VREP-Egp chimeric replicon was not impaired in binding, entry, and uncoating events in RAW 264.7 cells compared to VREP-Vgp, from which we infer that the VREP genome was delivered into the cytoplasm of myeloid cells with equal efficiency by the EEEV and by the VEEV structural protein coats (Fig. 5B and D). However, the infectivity of EEEV for RAW 264.7 cells was not restored by packaging EREP genomic RNA in SB structural proteins (EREP-SBgp). Similar findings were made for primary CD-1-derived DCs and macrophages (data not shown). We conclude that (i) EEEV infectivity for myeloid cells is restricted after binding, entry, and uncoating and is therefore not controlled at the level of the virus-receptor interaction; (ii) the point of restriction occurs prior to translation of the subgenomic message; and (iii) the restricted tropism phenotype of EEEV cosegregates predominantly with the 5′ and 3′ cis-acting sequence elements (CSEs) and nsP sequences, not with the structural protein genes.
Replication block occurs at the level of genome translation.
To facilitate sensitive detection of early genome translation, the next step after uncoating of the nucleocapsid, we have previously generated EEEV-, VEEV-, and SB-based RNA translation reporter constructs (43, 49). These in vitro-transcribed reporter RNAs include the m7G cap structure, the 5′ and 3′ NTRs, and the poly(A) tract of the virus genome and encode a truncated nsP1 protein (retaining important CSEs) fused in frame to the fLUC reporter protein (depicted in Fig. 6A). When transfected into permissive cells, reporter RNAs are translated, producing nsP1/fLUC fusion protein as an indicator of translation from the incoming genome (43, 49). Here, we investigated whether the infectivity of EEEV for myeloid lineage cells was restricted at the level of genome translation by transfecting equal amounts of reporter RNA into mesenchymal (BHK-21) and myeloid (RAW 264.7) lineage cells. Whereas in vitro-transcribed EEEV-, VEEV-, and SB-based reporter RNAs were all translated with similar efficiencies in BHK-21 fibroblasts (Fig. 6B), translation of the incoming EEEV-based RNA was reduced by 50- to 100-fold compared to that of VEEV- and SB-based reporters in RAW 264.7 macrophages (Fig. 6C) and in primary cDCs (data not shown).
FIG. 6.
Translation of an EEEV-based reporter RNA is dramatically reduced in myeloid lineage cells. (A) Diagrammatic representation of in vitro-transcribed reporter RNA expressing fLUC. (B and C) Cells of the mesenchymal (B) (represented by BHK-21 fibroblasts) or myeloid (C) (represented by RAW 264.7 monocytes/macrophages) lineage were electroporated with equal concentrations of RNA reporters based upon the genomes of VEEV (white bars), SB (gray bars), or EEEV (black bars). Cells were harvested for luciferase activity assay at various times p.i. Values represent the geometric mean fLUC activities (log10 RLU/μg). Datum points are shown ± standard deviations where n = 3. In panel C, a single asterisk indicates that translation of SB RNA reporter is significantly reduced compared with that of VEEV reporter (P < 0.01) and a double asterisk indicates that translation of EEEV reporter is significantly reduced compared with that of VEEV and SB reporters (P < 0.01).
To confirm these findings when the genome is introduced into the cell by infection as opposed to transfection, we generated a novel replicon genome cDNA in which the fLUC gene is inserted into the nonconserved region of nsP3 (depicted in Fig. 7A), similar to the approach used by Rice and coworkers to create an nsP3-fLUC SB virus (10). As for conventional replicons, the nsP3-fLUC replicon genome can be packaged into self-replicating, propagation-incompetent replicon particles in order to evaluate translation of the incoming genomic RNA delivered to the cytoplasm by infection. The nsP/fLUC polyprotein is immediately translated from the incoming genome without a requirement for negative-strand or subgenomic RNA synthesis. However, once the genome is replicated, additional nsP/fLUC polyprotein is translated from the progeny genomes. The nsP3-fLUC replicon also expresses GFP under the control of the subgenomic promoter which requires RNA replication for expression, and comparison to conventional GFP-expressing EREP infection demonstrated that the replication of the nsP3-fLUC EREP was not significantly impaired by insertion of the fLUC gene (data not shown). Infection of BHK-21 fibroblasts with nsP3-fLUC EREP resulted in the rapid production of fLUC, demonstrating efficient translation of incoming and subsequently progeny genomes, and a similar, albeit lower-level, accumulation of fLUC activity was observed in infected primary osteoblasts (Fig. 7B). In contrast, little or no evidence of fLUC production was detected in RAW 264.7 macrophages, further supporting the hypothesis that EEEV infection of myeloid lineage cells is dramatically restricted at the level of genome translation compared to VEEV and SB and compared to EEEV infection of mesenchymal lineage cell types.
FIG. 7.
EEEV genome translation is blocked in myeloid lineage cells following introduction of the genome by infection. (A) Diagrammatic representation of the EREP-nsP3-fLUC replicon genome. (B) EEEV replicon particles containing this genome were packaged in the standard EEEV structural protein coat (Egp) and used to infect BHK-21 fibroblasts (black circles), primary osteoblasts (white circles), or RAW 264.7 macrophages (black squares) at an MOI of 1. Cells were harvested for luciferase activity assay at various times p.i. Values represent the geometric mean fLUC activities (log10 RLU/μg). Datum points are shown ± standard deviations where n = 3.
Restricted tropism is not controlled by IFN-α/β-mediated pathways.
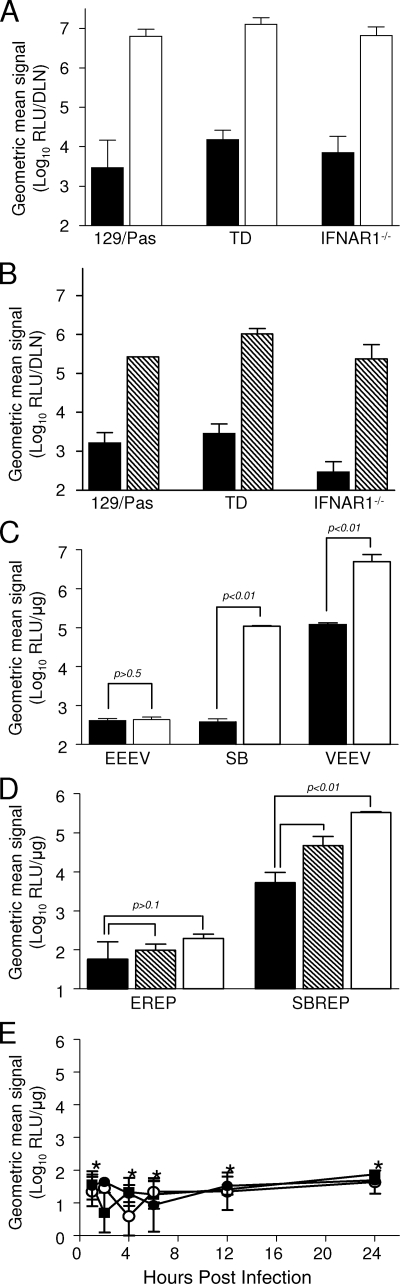
Although both VEEV and SB infect murine DCs and macrophages in vivo (20, 34, 42, 44) and in vitro (36, 42-44, 55), the permissivity of these cells to infection by SB, and to a lesser extent by VEEV, is controlled by IFN-α/β (42-44, 55). We have demonstrated previously that the ability of SB to productively infect myeloid cells in vitro and in vivo is controlled by IFN-α/β via PKR and PKR/RNase L-independent “alternative” pathways that inhibit translation of the genomic RNA (42-44, 49). Accordingly, we reasoned that the impaired translation of the EEEV genome within myeloid cells might be due to increased sensitivity to IFN-α/β and/or PKR or RNase L. Mice deficient in PKR, RNase L, and Mx (triply deficient [TD]) or the IFN-α/β receptor (IFNAR1−/−) and 129/Pas congenic controls were inoculated with EREP, VREP, or SBREP expressing fLUC, and the popliteal LNs were harvested at 8 h p.i. for in vitro fLUC assay (Fig. 8). Neither the absence of PKR/RNase L nor that of IFN-α/β signaling restored replication of EREP within the DLN (Fig. 8A and B), although extensive replication was observed at the site of inoculation in the footpad by use of the In Vivo imaging system (data not shown). VREP displayed the highest infectivity regardless of the presence or absence of PKR/RNase L or the IFN-α/β receptor (Fig. 8A), whereas the absence of PKR/RNase L caused a significant increase in SBREP fLUC signal in the DLN compared to that for control mice (Fig. 8B), in keeping with our previous findings (44).
FIG. 8.
The absence of IFN-α/β pathway components neither restores EEEV infectivity for DLN cells in vivo nor alleviates the restriction on EEEV productive infection and genome translation in myeloid lineage cells in vitro. (A and B) Mice lacking PKR, RNase L, and Mx (TD); mice lacking IFN-α/β receptor-mediated signaling (IFNAR1−/−); or normal congenic controls (129/Pas) were inoculated subcutaneously in each rear footpad with equal doses of EREP (black bars in panels A and B), VREP (white bars in panel A), or SBREP (hatched bars in panel B) replicon particles expressing fLUC. Data comparing EREP to VREP (A) and EREP to SBREP (B) are presented in separate panels as these were independent experiments. For panels A and B, DLNs were harvested at 8 h p.i. and fLUC activity was quantified by in vitro luciferase assay. Datum points are presented as RLU per DLN ± standard deviation where n = 8. (C) Primary cDCs generated from 129/Pas (black bars) or IFNAR1−/− (white bars) mice were infected with EEEV, SB, or VEEV at equal MOIs (0.1). Titers of progeny virus released into the supernatant were determined by plaque assay. Values represent the geometric mean virus titers (log10 PFU/ml) at 48 h p.i. ± standard deviation where n = 3 and confidence limits are indicated. (D) Primary cDCs generated from 129/Pas (black bars), TD (hatched bars), or IFNAR1−/− (white bars) mice were infected with fLUC-expressing EREP or SBREP conventional replicon particles (MOI, 1). Cells were harvested for luciferase activity assay at 24 h p.i. Values represent the geometric mean fLUC activities (log10 RLU/μg) ± standard deviations where n = 3, and confidence limits are indicated. (E) Primary cDCs derived from 129/Pas (black circles), TD (white circles), or IFNAR1−/− (black squares) mice were infected with EREP-nsP3-fLUC replicon particles (MOI, 1). Cells were harvested for luciferase activity assay at various times p.i. For panels C to E, values represent the geometric mean fLUC activities (log10 RLU/μg) ± standard deviations where n = 3. A single asterisk in panel E indicates that there was no significant elevation of fLUC activity in the absence of PKR/RNase L or the IFN-α/β receptor versus 129/Pas control cells (P > 0.5).
To confirm in vivo data, we generated primary DCs and macrophages deficient in components of the type I IFN pathway from IFNAR1−/− and TD mice. Like CD-1-derived cells, cDCs derived from 129/Pas mice were permissive to VEEV infection but almost completely refractory to EEEV as measured by the release of progeny virions (Fig. 8C). In this experiment, cDC cultures were infected at a low multiplicity (MOI, 0.1 PFU/cell), and consequently these cells were also refractory to productive infection by SB. In the absence of IFN-α/β receptor-mediated signaling, susceptibility to VEEV and particularly to SB infection was increased. However, progeny virion production from EEEV-infected 129/Pas cDCs versus that from IFNAR1−/− cDCs was not significantly altered. Infection of cDC cultures with fLUC-expressing SBREP and EREP particles revealed that, while SB replication and/or subgenome translation was significantly enhanced in the absence of PKR/RNase L or IFN-α/β receptor-mediated signaling, confirming our previous findings (44), these cells remained refractory to EEEV infection (Fig. 8D). Finally, we confirmed that the block on translation of incoming and progeny EEEV genomes was not alleviated in the absence of IFN-α/β-mediated signaling and/or PKR/RNase L by infecting these cells with the nsP3-fLUC EREP (Fig. 8E) and detected little production of the fLUC-fused nonstructural polyprotein through 24 h p.i. (compare to translation in mesenchymal cells in Fig. 7B).
Taken together, these data indicate that IFN-α/β signaling-dependent antiviral responses suppressed the production of progeny virions during SB, and to a lesser extent VEEV, infection of normal cells. However, infectivity of EEEV in myeloid lineage cells and lymphoid tissue was not restored by the absence of PKR, RNase L, or IFN-α/β-inducible antiviral effectors in vivo or in vitro.
IFN-α/β cytokine induction in vivo.
To investigate whether a differential IFN-α/β induction profile resulted from differential tissue and cell tropisms, sera were assayed for systemic type I IFN using a biological IFN-α/β assay. Confirming previous findings (13, 55), systemic IFN-α/β release was detectable within 12 h p.i. in VEEV-infected mice, with peak levels of 105 IU/ml of serum at 18 h p.i. (Fig. 9). In contrast, only one EEEV-infected mouse had detectable circulating IFN-α/β at 48 h p.i. and at a level barely above the limit of detection. Thus, peak IFN-α/β levels in VEEV-infected mice were over 1,000-fold higher than those in EEEV-infected counterparts.
FIG. 9.
EEEV infection does not result in systemic induction of IFN-α/β in vivo. CD-1 mice were inoculated subcutaneously with 103 PFU of EEEV (black bars) or VEEV (white bars), and serum was collected at intervals p.i. Serum levels of IFN-α/β were measured by bioassay. Values represent the geometric mean cytokine levels (log10 IU/ml of IFN-α/β) for three mice and are shown ± standard deviations where n = 3. The limit of detection for each data set is indicated by the dotted line.
DISCUSSION
Early events in the course of alphavirus infection are critical to the eventual outcome and severity of disease, involving intricate interactions between the virus, DCs, and the antiviral innate immune response. VEEV, in particular, has a strong tropism for lymphoid tissues that is the result of its predilection for infecting DCs and macrophages (34, 36; Klimstra et al., unpublished), and infection of these cells appears to be pivotal to many aspects of the VEEV pathogenesis pathway. DCs are infected at the site of inoculation and commandeered by the virus as a means to access the DLN, the earliest site of virus amplification and source of the primary serum viremia (34). VEEV mutants that are impeded in their ability to complete this step are attenuated in vivo (6, 9, 24, 34, 55). During subsequent steps in the pathogenic sequence, DCs and macrophages are believed to serve as reservoirs for virus replication and the source of the nonspecific proinflammatory response.
Over the years, studies have anecdotally alluded to the fact that EEEV does not display the same degree of lymphotropism that VEEV does in rodents (37, 51), but it preferentially infects fibroblastic and osteoblastic cells of the mesenchymal lineage (27, 51). By comparing the two virus infections directly, we have demonstrated that EEEV replication in lymphoid tissues is severely restricted compared with that of VEEV; indeed, EEEV titers in the DLN barely surpassed the initial inoculum dose. Using nonpropagative replicon particles expressing fLUC and in vivo imaging, we observed that EEEV replication in DLN tissues was greatly reduced compared to that of VEEV. Moreover, we noted that SB-based replicons, derived from a virus that is extremely sensitive to IFN-α/β and therefore avirulent for adult animals (42, 44), also were significantly more infectious for DLN cells than were EEEV-based replicons. When considered in combination with the observation made by Vogel et al. (51) that little EEEV replication was observed in the DLN by in situ hybridization, it is evident that the tropism of EEEV is altered compared to that of VEEV and SB.
We hypothesize that the relative inability of EEEV to infect migratory cells of the immune system, and thereby access and amplify within the DLN, causes a “bottleneck” in the early stages of the pathogenesis pathway and contributes to the delay in establishment of viremia and seeding of tissues distal to the inoculation site. However, low-level primary viremia from inoculation site capillaries and/or the limited replication observed in the DLN presumably is sufficient to seed mesenchymal lineage cells such as osteoblasts, which are accessible from the bloodstream and appear to be the major site of EEEV amplification in peripheral tissues (51). Since blood-borne virus titers far exceed replication levels in the lymphoid tissues, it is likely that these mesenchymal cells are the source of the amplified, secondary viremia, necessary for EEEV to neuroinvade, as speculated previously (27, 51). Interestingly, whereas VEEV primarily invades the CNS via the olfactory neuroepithelium (14, 45), EEEV appears to directly cross the blood-brain barrier, while sparing the olfactory epithelium and bulb (51). We speculate that VEEV may infect DCs lining the nasal passages as a means of accessing olfactory neurons, but the restricted tropism of EEEV may necessitate the use of an alternate neuroinvasive pathway.
In keeping with observations of in vivo tropism, EEEV replicated at least as efficiently as did VEEV in a range of primary and immortalized mesenchymal lineage cell types, including osteoblasts and fibroblasts. However, whereas VEEV also replicated efficiently in myeloid lineage cells such as cDCs and macrophages, EEEV exhibited little or no infectivity for these cell types. We considered first that the cell tropism restriction of EEEV might be controlled at the level of binding and entry, as the virus-receptor interaction is the most commonly described determinant of alphavirus tropism and virulence. Adaptation to heparan sulfate (HS) binding in vitro, for example, is associated with attenuation (9, 29, 47) and reduced infectivity for DCs in vivo (34). Further supporting this hypothesis, we have recently demonstrated that, unlike other alphaviruses, wild-type EEEV strains bind HS (Klimstra et al., unpublished). However, in chimeric replicon particles the EEEV structural proteins delivered the VEEV genome to myeloid cells as efficiently as did the VEEV or SB structural proteins, from which we infer that the tropism restriction occurs after attachment, entry, and uncoating steps have been accomplished. It remains possible, however, that while the virus-receptor interaction is not primarily responsible for the impediment to replication in DCs in vivo or at all in vitro, HS binding of EEEV virions may also affect the appearance of infected migratory cells in the DLN.
Once released into the cytoplasm, the positive-sense genome of an alphavirus is a multifunctional molecule serving first as an mRNA for the production of the nonstructural polyprotein and subsequently as a transcription template for synthesis of negative-sense RNA replicative intermediates (48). Like cellular mRNAs, capped, polyadenylated alphavirus genomes associate with the cellular cap-binding protein, eIF4E (8). Although subsequent steps have not been elucidated, we assume that translation initiation proceeds as for cellular mRNAs with assembly of the eIF4F complex, recruitment of the 40S ribosomal subunit, scanning to the initiation codon, and recruitment of the 60S ribosomal subunit. Interestingly, our data indicate that an early and profound block on EEEV infectivity for myeloid lineage cells occurs at the level of translation of the incoming viral message (and presumably also translation of progeny genomes), suggesting that the efficiency with which this essential first step in the virus life cycle is completed might contribute significantly to cell and/or tissue tropism. Specifically, we interpret our data to indicate that (i) translation of the EEEV genome occurs with much greater efficiency in mesenchymal than in myeloid cells; (ii) translation of the VEEV (and SB) genome occurs with similar efficiencies in mesenchymal and myeloid cells; and (iii) as the in vitro-transcribed translation reporter RNAs possess only the 5′ and 3′ CSEs of the genome flanking the fLUC gene, the translation potential of the genome in myeloid versus mesenchymal cells is dictated by the nucleotide sequence of these regions. Provocatively, recent studies have demonstrated that levels and activity of translation machinery components are influenced by cell type, affecting permissivity to SB infection (15), and thus, we speculate that differences in the translational environment between myeloid and mesenchymal cell types affect the stability of the EEEV genome or the efficiency with which it is translated. In contrast, translation of the VEEV and SB genomic RNAs is unaffected in the myeloid environment.
Although sequence homology between alphaviruses is not particularly evident in the genome termini, both the 5′ and 3′ CSE regions are predicted to form highly conserved, secondary structures (19, 57). In particular, the M-fold algorithm predicts four stem-loop structures within the 5′ 220 nt. Deletion or disruption of each of these predicted stem-loop structures in SB affects negative-sense RNA synthesis and the efficiency of progeny genome synthesis (19), and mutations in the first stem-loop affect IFN-α/β sensitivity of VEEV in vitro and virulence in vivo (55). However, little attention has been paid to the role of these secondary structures in genome stability, efficiency of translation, or determination of cell tropism. Perhaps differences in the folding of the stem-loops alter RNA stability, affinity for eIF4E or another translation factor, or the ability of the genome to circularize. It has been demonstrated that eIF4E binds with significantly lower affinity to the alphavirus genome than to the subgenomic capped mRNA (8) and that the requirement for specific translation initiation factors differs between these molecules (11). This suggests that the specific alphavirus RNA structure can alter interaction with translation initiation factors. Differences in infectivity among closely related strains of another mosquito-borne RNA virus, dengue virus, have been shown to correlate with efficiency of translation of input viral RNA (18).
We have demonstrated previously, and confirmed in this study, that the ability of SB to replicate in DCs and macrophages is restricted in adult mice and in cultured primary cells in vitro (42-44). This apparent restriction in cell tropism is controlled by the sensitivity of SB to the antiviral activity of type I IFN via a combination of PKR-dependent pathways and IFN-α/β-receptor signaling-dependent, PKR-independent pathways (43, 44). Thus, in the absence of the type I IFN receptor SB replicates systemically in DCs and macrophages in vivo and in primary cultures of these cells in vitro. Both constitutively expressed PKR and IFN-α/β priming independent of PKR dramatically diminish translation of the SB, VEEV, and EEEV genomes in otherwise permissive cells (49), and therefore, we tested the hypothesis that components of the type I IFN pathway might differentially inhibit EEEV translation in myeloid cells. However, our data seem to rule out the involvement of PKR or other IFN-α/β-inducible antiviral effectors in the restricted tropism, both in vivo and in vitro, as infectivity for myeloid lineage cells was not restored in the absence of PKR or IFN-α/β receptor-mediated signaling.
As both VEEV and EEEV remain virulent in the face of an intact, functional IFN-α/β system, it is clear that these viruses can evade and/or disable components of the IFN-α/β response. VEEV infection results in the rapid and high-level induction of type I IFN and proinflammatory cytokines in the DLN and spleen (23), from both infected and neighboring cells (31). Systemic type I IFN is known to rapidly induce the transcription of IFN-stimulated genes in peripheral tissues and in the CNS (52). Among the IFN-stimulated genes induced are some that we have recently demonstrated to possess strong antiviral activity against SB (56). However, although the IFN-α/β response diminishes the virulence of wild-type VEEV, slowing viral replication and dissemination and increasing the survival time of infected animals by several days, it cannot control infection sufficiently to confer protection (25, 55), indicating that VEEV possesses partial resistance to the IFN-α/β-induced antiviral state. In contrast, we have demonstrated that EEEV infection induces little or no systemic IFN-α/β release in vivo. Taken in conjunction with the findings of Aguilar et al. (3) that EEEV becomes attenuated in an environment in which IFN-α/β is induced, it appears that the induction of IFN-α/β is avoided during EEEV infection as an alternative to antagonizing IFN-α/β signaling or effector function. Furthermore, in the absence of type I IFN induction during early peripheral replication, the CNS environment may be relatively unprepared and unprotected against the invading virus, perhaps contributing to the rapid replication of EEEV in the CNS and extreme neurovirulence compared with VEEV.
It has recently been demonstrated that expression of the EEEV capsid protein results in the arrest of cellular transcription (2, 4, 22). Thus, we propose that EEEV possesses at least two mechanisms acting in concert by which the induction of type I IFN is suppressed and/or avoided: (i) failure to replicate in myeloid cells, particularly DCs, results in reduced virus replication in lymphoid tissues and therefore less activation of both infected and neighboring immune system sentinel cells and (ii) expression of the capsid protein within those cells that are productively infected including fibroblasts, osteoblasts, and the occasional DC or macrophage further suppresses responses. This hypothesis is in keeping with recent findings by Basler and colleagues in which a capsid deletion mutant of EEEV was found to be attenuated in vivo (2). EEEV antagonizes the induction of IFN-α/β from infected cells (e.g., osteoblasts) and avoids the stimulation of the major IFN-producing cells by not infecting them. Interestingly, the capsid protein of VEEV is similarly described to shut off cellular transcription in infected cells in vitro (21, 22), but nevertheless this virus induces substantial IFN-α/β secretion in vivo (Fig. 9) (13), from both infected and uninfected cells in lymphoid tissues (31).
Finally, we believe that our findings have import for both the design of acute-phase therapeutics and vaccines against EEEV. As EEEV appears to evade the type I IFN antiviral response by a program of avoiding and suppressing induction of this cytokine, it seems likely that acute-phase therapy during EEEV infection could be achieved using IFN-α/β or an inducer, particularly if given during the early stages of infection. It is anticipated that such therapies would be significantly more efficacious in ameliorating EEEV than VEEV disease. Moreover, we hypothesize that since DCs are major IFN-α/β producers and pivotal to IFN sensitivity to SB, attenuated and immunogenic virus vaccine candidates could be produced by genetically manipulating EEEV to infect these cells.
Acknowledgments
We thank Michael Farmer, Danielle Gonzalez, and DeAquanita McKinney for excellent technical assistance and Tracée Terry in the LSUHSC-S Research Core Facility for performing in vivo imaging studies. We thank Michael Parker for critical reading of the manuscript and Gerard Karsenty's group for providing protocols for the generation of primary murine osteoblast cultures. We are particularly indebted to Scott Weaver and Robert Johnston for providing us with cDNA clones for generating viruses and replicons.
This work was supported by National Institutes of Health grant R21 AI069158 (K.D.R.) and grants from NIAID/NIH through the Western Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (WRCE) U54 AI057156 (Career Development award to K.D.R. and major project subcontract to W.B.K.).
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Aguilar, P. V., A. P. Adams, E. Wang, W. Kang, A.-S. Carrara, M. Anishchenko, I. Frolov, and S. C. Weaver. 2008. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J. Virol. 824920-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, P. V., L. W. Leung, E. Wang, S. C. Weaver, and C. F. Basler. 2008. A five-amino-acid deletion of the eastern equine encephalitis virus capsid protein attenuates replication in mammalian systems but not in mosquito cells. J. Virol. 826972-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, P. V., S. Paessler, A. S. Carrara, S. Baron, J. Poast, E. Wang, A. C. Moncayo, M. Anishchenko, D. Watts, R. B. Tesh, and S. C. Weaver. 2005. Variation in interferon sensitivity and induction among strains of eastern equine encephalitis virus. J. Virol. 7911300-11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilar, P. V., S. C. Weaver, and C. F. Basler. 2007. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 813866-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anishchenko, M., S. Paessler, I. P. Greene, P. V. Aguilar, A. S. Carrara, and S. C. Weaver. 2004. Generation and characterization of closely related epizootic and enzootic infectious cDNA clones for studying interferon sensitivity and emergence mechanisms of Venezuelan equine encephalitis virus. J. Virol. 781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronson, J. F., F. B. Grieder, N. L. Davis, P. C. Charles, T. Knott, K. Brown, and R. E. Johnston. 2000. A single-site mutant and revertants arising in vivo define early steps in the pathogenesis of Venezuelan equine encephalitis virus. Virology 270111-123. [DOI] [PubMed] [Google Scholar]
- 7.Bakker, A., and J. Klein-Nulend. 2003. Osteoblast isolation from murine calvariae and long bones. Methods Mol. Med. 8019-28. [DOI] [PubMed] [Google Scholar]
- 8.Berben-Bloemheuvel, G., M. A. Kasperaitis, H. van Heugten, A. A. Thomas, H. van Steeg, and H. O. Voorma. 1992. Interaction of initiation factors with the cap structure of chimaeric mRNA containing the 5′-untranslated regions of Semliki Forest virus RNA is related to translational efficiency. Eur. J. Biochem. 208581-587. [DOI] [PubMed] [Google Scholar]
- 9.Bernard, K. A., W. B. Klimstra, and R. E. Johnston. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 27693-103. [DOI] [PubMed] [Google Scholar]
- 10.Bick, M. J., J. W. Carroll, G. Gao, S. P. Goff, C. M. Rice, and M. R. Macdonald. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 7711555-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castello, A., M. A. Sanz, S. Molina, and L. Carrasco. 2006. Translation of Sindbis virus 26S mRNA does not require intact eukaryotic initiation factor 4G. J. Mol. Biol. 355942-956. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2008. Confirmed and probable eastern equine encephalitis cases, human, United States, 1964-2007, by state. http://www.cdc.gov/ncidod/dvbid/Arbor/pdf/EEE_DOC.pdf.
- 13.Charles, P. C., J. Trgovcich, N. L. Davis, and R. E. Johnston. 2001. Immunopathogenesis and immune modulation of Venezuelan equine encephalitis virus-induced disease in the mouse. Virology 284190-202. [DOI] [PubMed] [Google Scholar]
- 14.Charles, P. C., E. Walters, F. Margolis, and R. E. Johnston. 1995. Mechanism of neuroinvasion of Venezuelan equine encephalitis virus in the mouse. Virology 208662-671. [DOI] [PubMed] [Google Scholar]
- 15.Colina, R., M. Costa-Mattioli, R. J. Dowling, M. Jaramillo, L. H. Tai, C. J. Breitbach, Y. Martineau, O. Larsson, L. Rong, Y. V. Svitkin, A. P. Makrigiannis, J. C. Bell, and N. Sonenberg. 2008. Translational control of the innate immune response through IRF-7. Nature 452323-328. [DOI] [PubMed] [Google Scholar]
- 16.de la Monte, S., F. Castro, N. J. Bonilla, A. Gaskin de Urdaneta, and G. M. Hutchins. 1985. The systemic pathology of Venezuelan equine encephalitis virus infection in humans. Am. J. Trop. Med. Hyg. 34194-202. [DOI] [PubMed] [Google Scholar]
- 17.Deresiewicz, R. L., S. J. Thaler, L. Hsu, and A. A. Zamani. 1997. Clinical and neuroradiographic manifestations of eastern equine encephalitis. N. Engl. J. Med. 3361867-1874. [DOI] [PubMed] [Google Scholar]
- 18.Edgil, D., M. S. Diamond, K. L. Holden, S. M. Paranjape, and E. Harris. 2003. Translation efficiency determines differences in cellular infection among dengue virus type 2 strains. Virology 317275-290. [DOI] [PubMed] [Google Scholar]
- 19.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 71638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner, J. P., I. Frolov, S. Perri, Y. Ji, M. L. MacKichan, J. zur Megede, M. Chen, B. A. Belli, D. A. Driver, S. Sherrill, C. E. Greer, G. R. Otten, S. W. Barnett, M. A. Liu, T. W. Dubensky, and J. M. Polo. 2000. Infection of human dendritic cells by a Sindbis virus replicon vector is determined by a single amino acid substitution in the E2 glycoprotein. J. Virol. 7411849-11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garmashova, N., S. Atasheva, W. Kang, S. C. Weaver, E. Frolova, and I. Frolov. 2007. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J. Virol. 8113552-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garmashova, N., R. Gorchakov, E. Volkova, S. Paessler, E. Frolova, and I. Frolov. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 812472-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grieder, F. B., B. K. Davis, X. D. Zhou, S. J. Chen, F. D. Finkelman, and W. C. Gause. 1997. Kinetics of cytokine expression and regulation of host protection following infection with molecularly cloned Venezuelan equine encephalitis virus. Virology 233302-312. [DOI] [PubMed] [Google Scholar]
- 24.Grieder, F. B., N. L. Davis, J. F. Aronson, P. C. Charles, D. C. Sellon, K. Suzuki, and R. E. Johnston. 1995. Specific restrictions in the progression of Venezuelan equine encephalitis virus-induced disease resulting from single amino acid changes in the glycoproteins. Virology 206994-1006. [DOI] [PubMed] [Google Scholar]
- 25.Grieder, F. B., and S. N. Vogel. 1999. Role of interferon and interferon regulatory factors in early protection against Venezuelan equine encephalitis virus infection. Virology 257106-118. [DOI] [PubMed] [Google Scholar]
- 26.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Hurst, E. 1936. Infection of the rhesus monkey (Macaca mulatta) and the guinea-pig with the virus of equine encephalomyelitis. J. Pathol Bacteriol. 42271-302. [Google Scholar]
- 28.Johnson, K. M., and D. H. Martin. 1974. Venezuelan equine encephalitis. Adv. Vet. Sci. Comp. Med. 1879-116. [PubMed] [Google Scholar]
- 29.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 727357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimstra, W. B., J. C. Williams, K. D. Ryman, and H. W. Heidner. 2005. Targeting Sindbis virus-based vectors to Fc receptor-positive cell types. Virology 3389-21. [DOI] [PubMed] [Google Scholar]
- 31.Konopka, J. L., L. O. Penalva, J. M. Thompson, L. J. White, C. W. Beard, J. D. Keene, and R. E. Johnston. 2007. A two-phase innate host response to alphavirus infection identified by mRNP-tagging in vivo. PLoS Pathog. 3e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levitt, N. H., H. V. Miller, and R. Edelman. 1979. Interaction of alphaviruses with human peripheral leukocytes: in vitro replication of Venezuelan equine encephalomyelitis virus in monocyte cultures. Infect. Immun. 24642-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, C., D. W. Voth, P. Rodina, L. R. Shauf, and G. Gonzalez. 1970. A comparative study of the pathogenesis of western equine and eastern equine encephalomyelitis viral infections in mice by intracerebral and subcutaneous inoculations. J. Infect. Dis. 12253-63. [DOI] [PubMed] [Google Scholar]
- 34.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell, C. J., M. L. Niebylski, G. C. Smith, N. Karabatsos, D. Martin, J. P. Mutebi, G. B. Craig, Jr., and M. J. Mahler. 1992. Isolation of eastern equine encephalitis virus from Aedes albopictus in Florida. Science 257526-527. [DOI] [PubMed] [Google Scholar]
- 36.Moran, T. P., M. Collier, K. P. McKinnon, N. L. Davis, R. E. Johnston, and J. S. Serody. 2005. A novel viral system for generating antigen-specific T cells. J. Immunol. 1753431-3438. [DOI] [PubMed] [Google Scholar]
- 37.Paessler, S., P. Aguilar, M. Anishchenko, H. Q. Wang, J. Aronson, G. Campbell, A. S. Cararra, and S. C. Weaver. 2004. The hamster as an animal model for eastern equine encephalitis—and its use in studies of virus entrance into the brain. J. Infect. Dis. 1892072-2076. [DOI] [PubMed] [Google Scholar]
- 38.Perri, S., C. E. Greer, K. Thudium, B. Doe, H. Legg, H. Liu, R. E. Romero, Z. Tang, Q. Bin, T. W. Dubensky, Jr., M. Vajdy, G. R. Otten, and J. M. Polo. 2003. An alphavirus replicon particle chimera derived from Venezuelan equine encephalitis and Sindbis viruses is a potent gene-based vaccine delivery vector. J. Virol. 7710394-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239389-401. [DOI] [PubMed] [Google Scholar]
- 40.Ryman, K. D., C. L. Gardner, K. C. Meier, C. A. Biron, R. E. Johnston, and W. B. Klimstra. 2007. Early restriction of alphavirus replication and dissemination contributes to age-dependent attenuation of systemic hyperinflammatory disease. J. Gen. Virol. 88518-529. [DOI] [PubMed] [Google Scholar]
- 41.Ryman, K. D., C. L. Gardner, C. L. Wyza, K. C. Meier, J. M. Thompson, and W. B. Klimstra. 2007. Heparan sulfate binding can contribute to the neurovirulence of neuroadapted and nonneuroadapted Sindbis viruses. J. Virol. 813563-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryman, K. D., W. B. Klimstra, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 743366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryman, K. D., K. C. Meier, E. M. Nangle, S. L. Ragsdale, N. L. Korneeva, R. E. Rhoads, M. R. Macdonald, and W. B. Klimstra. 2005. Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells. J. Virol. 791487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryman, K. D., L. J. White, R. E. Johnston, and W. B. Klimstra. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 1553-76. [DOI] [PubMed] [Google Scholar]
- 45.Ryzhikov, A. B., E. I. Ryabchikova, A. N. Sergeev, and N. V. Tkacheva. 1995. Spread of Venezuelan equine encephalitis virus in mice olfactory tract. Arch. Virol. 1402243-2254. [DOI] [PubMed] [Google Scholar]
- 46.Schoepp, R. J., J. F. Smith, and M. D. Parker. 2002. Recombinant chimeric western and eastern equine encephalitis viruses as potential vaccine candidates. Virology 302299-309. [DOI] [PubMed] [Google Scholar]
- 47.Smit, J. M., B. L. Waarts, K. Kimata, W. B. Klimstra, R. Bittman, and J. Wilschut. 2002. Adaptation of alphaviruses to heparan sulfate: interaction of Sindbis and Semliki Forest viruses with liposomes containing lipid-conjugated heparin. J. Virol. 7610128-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tesfay, M. Z., J. Yin, C. L. Gardner, M. V. Khoretonenko, N. L. Korneeva, R. E. Rhoads, K. D. Ryman, and W. B. Klimstra. 2008. Alpha/beta interferon inhibits cap-dependent translation of viral but not cellular mRNA by a PKR-independent mechanism. J. Virol. 822620-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trgovcich, J., J. F. Aronson, and R. E. Johnston. 1996. Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis. Virology 22473-83. [DOI] [PubMed] [Google Scholar]
- 51.Vogel, P., W. M. Kell, D. L. Fritz, M. D. Parker, and R. J. Schoepp. 2005. Early events in the pathogenesis of eastern equine encephalitis virus in mice. Am. J. Pathol. 166159-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wacher, C., M. Muller, M. J. Hofer, D. R. Getts, R. Zabaras, S. S. Ousman, F. Terenzi, G. C. Sen, N. J. King, and I. L. Campbell. 2007. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J. Virol. 81860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walton, T. E., and M. A. Grayson. 1989. Venezuelan equine encephalomyelitis, p. 203-231. In T. P. Monath (ed.), The arboviruses: epidemiology and ecology, vol. 4. CRC Press, Boca Raton, FL. [Google Scholar]
- 54.Weaver, S. C., A. M. Powers, A. C. Brault, and A. D. Barrett. 1999. Molecular epidemiological studies of veterinary arboviral encephalitides. Vet. J. 157123-138. [DOI] [PubMed] [Google Scholar]
- 55.White, L. J., J. G. Wang, N. L. Davis, and R. E. Johnston. 2001. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J. Virol. 753706-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Y., C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 8111246-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuker, M. 2000. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 10303-310. [DOI] [PubMed] [Google Scholar]