Abstract
In the generation of flavivirus particles, an internal cleavage of the envelope glycoprotein prM by furin is required for the acquisition of infectivity. Unlike cleavage of the prM of other flaviviruses, cleavage of dengue virus prM is incomplete in many cell lines; the partial cleavage reflects the influence of residues at furin nonconsensus positions of the pr-M junction, as flaviviruses share basic residues at positions P1, P2, and P4, recognized by furin. In this study, viruses harboring the alanine-scanning and other multiple-point mutations of the pr-M junction were generated, employing a dengue virus background that exhibited 60 to 70% prM cleavage and a preponderance of virion-sized extracellular particles. Analysis of prM and its cleavage products in viable mutants revealed a cleavage-suppressive effect at the conserved P3 Glu residue, as well as the cleavage-augmenting effects at the P5 Arg and P6 His residues, indicating an interplay between opposing modulatory influences mediated by these residues on the cleavage of the pr-M junction. Changes in the prM cleavage level were associated with altered proportions of extracellular virions and subviral particles; mutants with reduced cleavage were enriched with subviral particles and prM-containing virions, whereas the mutant with enhanced cleavage was deprived of these particles. Alterations of virus multiplication were detected in mutants with reduced prM cleavage and were correlated with their low specific infectivities. These findings define the functional roles of charged residues located adjacent to the furin consensus sequence in the cleavage of dengue virus prM and provide plausible mechanisms by which the reduction in the pr-M junction cleavability may affect virus replication.
Dengue viruses are members of the genus Flavivirus in the family Flaviviridae. A single-stranded genomic RNA of positive polarity encodes an open reading frame which is translated into a precursor of three structural proteins and at least seven nonstructural proteins (27). During the replication of flaviviruses, proteolytic cleavages of the polyprotein by viral protease and a number of cellular enzymes occur in distinct subcellular compartments (27, 32). Three structural proteins, C, prM/M, and E, are associated with the membrane of the rough endoplasmic reticulum (ER) by C-terminal membrane-spanning regions. Cleavages distal to the membrane-spanning regions of C and the two envelope glycoproteins prM and E by host signalase in the lumen of the rough ER and a cleavage proximal to the membrane-spanning region of C by viral protease in the cytoplasm are important to the assembly of viral particles (32). During the export of immature particles through the secretory pathway, cleavage of prM by the trans-Golgi apparatus resident furin allows reorganization of the receptor-binding E protein that is required for the acquisition of infectivity (9, 43).
Assembly of flavivirus particles in the rough ER results in two types of particles: virions and subviral particles (32, 40). An immature virion, about 60 nm in diameter, is composed of an outer envelope and a genome-containing internal structure. Its surface consists of 60 spikes, each of which is formed by three prM-E heterodimers (52, 53). Just prior to release, the proteolytic cleavage of prM by furin triggers the conversion of the immature virions into slightly smaller (about 50-nm) mature particles with smooth surfaces and high infectivity (26). Subviral particles are only about 32 nm in diameter (11). These coreless particles also undergo the proteolytic cleavage of prM, but the number of E homodimers and their arrangement are different from those of mature virions (11). Thirty E homodimers assume the T = 1 icosahedral configuration in the subviral particles, whereas in mature virions, 90 E homodimers are clustered in groups of three parallel dimers that distribute icosahedrally in a “herringbone” pattern (11, 26, 31). Apart from their differences in size and E dimer arrangements, small and large particles are distinguishable by other structural and functional properties, including the N-glycosylation pattern of the E protein (2) and the ability to agglutinate red blood cells (20, 25). Among flaviviruses, the proportion of the two types of particles generated during viral infection is quite variable. The majority of particles released from dengue virus-infected Vero cells and C6/36 mosquito cells are virion-sized particles (24, 34). These large particles predominate in Japanese encephalitis virus (JEV)-infected Vero cell cultures, whereas subviral particles are more abundant in cultures of infected C6/36 cells (20, 23, 25, 29). Both types of particles are equally common following infection of COS-1 cells with tick-borne encephalitis virus (TBEV) (1). The molecular determinant(s) that affects the proportion of extracellular viral particles remains poorly understood.
During the past two decades, it has been consistently observed that the extracellular particles of dengue virus contain some uncleaved prM molecules. Partial cleavage of dengue virus prM was detected in particles released from infected mosquito cells (7, 10, 14, 33-36, 39, 47), Vero cells (3, 12, 33, 36, 49), and LLC-MK2 cells (8). As in other flaviviruses, the dengue virus pr-M junction contains three highly conserved basic residues at cleavage positions P1, P2, and P4 that are required for cleavage by furin (46, 54), so the underlying basis for partial prM cleavage in dengue virus is not readily apparent. In our previous study, the influence of a short sequence just proximal to the pr-M junction on prM cleavage was assessed by exchanging the 13-amino-acid segment of dengue virus prM with the homologous segments from other flaviviruses, representing three distinct antigenic complexes: JEV, yellow fever virus (YFV), and TBEV (22). Cleavage of prM in the first two chimeric viruses was enhanced over that in the parent dengue virus but was slightly suppressed in the last chimera (22). Because these chimeras and the dengue virus share the furin consensus sequence Arg-Xaa-(Lys/Arg)-Arg (where Xaa is any amino acid) at the pr-M junction, the results are consistent with the notion that residues at nonconsensus positions, which vary among different flaviviruses, are capable of modifying prM cleavage efficiency (22). An outstanding sequence variation that may be responsible for partial prM cleavage in dengue virus has been pointed out previously (8). Among flaviviruses with known insect vectors, the presence of an acidic residue at the P3 cleavage position appears to be unique to all four dengue virus serotypes. The P3 acidic residue is highly conserved among dengue viruses, and the only other known example of such a residue is found in the cell fusing agent virus, which also exhibits minimal prM cleavage (6, 8). In this study, we examined the influence of the P3 Glu residue and other nonconsensus charged residues on the efficiency of dengue virus prM cleavage. The results led us to further determine how various prM cleavages affect the proportions of the two types of extracellular viral particles and other associated changes in the replicative ability of the resultant particles.
MATERIALS AND METHODS
Virus, cell lines, and antibodies.
Dengue serotype 2 virus strain 16681 was kindly provided by Bruce Innis and Ananda Nisalak, Department of Virology, Armed Forces Research Institute for Medical Sciences, Bangkok, Thailand. JEVpr/16681, a chimeric virus containing the 13-amino-acid sequence proximal to the pr-M junction from JEV in the 16681 background, was described previously (22). Propagation of dengue virus in the C6/36 mosquito cell line, quantitation of infectious virus, and determination of focus size in the PS clone D cell line, employing the focus immunoassay, were carried out as described previously (22, 42). Two monoclonal antibodies, 4G2 and 3H5, specific for flavivirus and dengue serotype 2 virus E protein, respectively (14, 15), were used for the focus immunoassay titration, the immunoblot analysis, and immunoprecipitation in conjunction with an anti-dengue virus prM antibody, clone prM-6.1, and the anti-dengue virus C antibodies, clones Cap 2 S.1-D2-C1 (2G11) and D2-C2 (1A9) (C. Puttikhunt, W. Kasinrerk, and P. Malasit, unpublished results). The specificity of this latter set of antibodies was determined by immunoblot analysis (data not shown) and immunoprecipitation of viral structural proteins (Fig. 1). The clone prM-6.1, an immunoglobulin G2a (IgG2a) antibody, recognizes a linear epitope in the prM protein of dengue virus serotypes 1, 2, and 4. Both of the anti-dengue virus C antibodies belong to the IgG1 subclass; 2G11 is specific for the serotype 2 virus, while 1A9 reacts with virus serotypes 2 and 4. The plasmacytoma proteins MOPC 21 and UPC 10 were obtained from Sigma (St. Louis, MO).
FIG. 1.
Immunoprecipitation of dengue virus structural proteins with monoclonal antibodies. (Left panel) 35S-labeled extracellular viral proteins of the parent virus, 16681Nde(+), were reacted with the monoclonal antibodies specific for the E (3H5, lane E), prM (prM-6.1, lane prM), and C (1A9, lane C) proteins or for irrelevant proteins (MOPC 21, lane M; UPC 10, lane U) and then precipitated with protein G-Sepharose beads. (Middle panel) Labeled particles were treated with 2% Triton X-100 and 0.1% SDS prior to immunoprecipitation. (Right panel) Labeled particles were pretreated with 2% Triton X-100 and 2% SDS. (A) Following electrophoresis, gels were fixed, soaked in an enhancer solution (Amplify reagent; Amersham), dried, and then exposed to XAR film. The positions of the marker proteins, as indicated at the left, correspond to the following apparent molecular masses: 170, 130, 100, 72, 55, 40, 33, 24, 17, and 11 kDa. (B) The gel was fixed and dried prior to exposure to a phosphor screen. Note that 3H5 precipitated all three structural proteins (E, prM/M, and C) from untreated samples, whereas prM-6.1 also recognized the pr peptide, which, by its absence from 3H5-derived precipitate, appeared not to be associated with particles. Both C and pr proteins were present as doublets, but the pr doublet was more closely spaced than the C doublet. When C was liberated from virions by Triton X-100 treatment, it appeared to bind to Sepharose beads (middle panel, lanes C and M). The reactivity of the anti-C antibody, 1A9, with untreated particles in the left panel was unclear but may be due either to a specific reaction of 1A9 with C molecules in partially disrupted particles or to the binding of C in such particles with Sepharose beads. The specificities of 3H5 and prM-6.1 were clearly demonstrated by disrupting prM-E dimers with 2% SDS prior to the precipitation (right panel, lanes E and prM).
Generation of the pr-M junction mutant viruses.
A full-length cDNA clone of strain 16681 (42) was employed in the construction of pr-M junction mutant viruses. For generating alanine-scanning mutants, a plasmid subclone containing the silent mutation of the PstI site at the base position 402 (44) served as the template in a PCR-based in vitro mutagenesis scheme (QuikChange; Stratagene, La Jolla, CA). For the construction of multiple point mutations, an additional plasmid subclone in which the NdeI and BamHI sites were introduced on the 5′ and 3′ sides of the pr-M junction, respectively, was employed as the recipient of double-stranded oligonucleotides encoding mutated pr-M junction sequences as described previously (22). Sequences of the oligonucleotides employed in the generation of mutants, reverse transcription (RT), and PCR and nucleotide sequence analyses are available upon request. Construction of the 5′ half-genome and full-length cDNA clone containing the intended mutations, in vitro transcription, and Lipofectin-mediated transfection of C6/36 cells with capped in vitro transcripts were performed essentially as described previously (22). With the exceptions of the R201A and R202A mutants, mutant viruses were generally expanded in C6/36 cells for two or three passages. Due to its low titer, the R201A mutant had to be amplified in four consecutive passages. Following two transfection attempts, a large fraction of the R202A mutant virus preparations was found to contain additional mutations in the prM coding region. The original R202A mutant virus was not recovered by terminal dilution, and a mutant with two unintended mutations was amplified instead (Table 1). As determined by RT-PCR and nucleotide sequence analysis of the prM coding region, other mutant viruses contained only the introduced mutation(s) at the pr-M junction.
TABLE 1.
Sequences of the the pr-M junction and some characteristics of the dengue virus pr-M junction chimera and mutants
| Virus | Amino acid sequence and positiona
|
Positions P/1 to P/13
|
Titer (FFU/ml) | Focus size (no. infected cells/focus ± SD)b | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + charge | − charge | Net charge | Setc | ||||||||||||||||||||
| 15d | 10 | 5 | 1 | ↓ | 1′ | ||||||||||||||||||
| 16681Nde(+) | Y | G | T | C | T | T | M | G | E | H | R | R | E | K | R | S | 4 | 2 | +2 | 1 | 1.40 × 108 | 222.2 ± 77.4 | |
| JEVpr/16681 | Y | G | R | C | T | R | T | R | H | S | K | R | S | R | R | S | 7 | 0 | +7 | 2 | 1.54 × 106 | 17.7 ± 9.6 | |
| 16681pr(+4,−0) | Y | G | T | C | T | T | M | G | H | H | R | R | S | K | R | S | 4 | 0 | +4 | 1 | 8.18 × 106 | 42.3 ± 21.2 | |
| 16681pr(+7,−2) | Y | G | R | C | T | R | M | R | E | H | R | R | E | K | R | S | 7 | 2 | +5 | 2 | 9.09 × 106 | 22.6 ± 12.6 | |
| 16681pr(+7,−0) | Y | G | R | C | T | R | M | R | H | H | R | R | S | K | R | S | 7 | 0 | +7 | 2 | 7.32 × 105 | 17.8 ± 8.6 | |
| 16681prE199A | Y | G | T | C | T | T | M | G | A | H | R | R | E | K | R | S | 4 | 1 | +3 | 1 | 1.26 × 107 | 140.3 ± 74.5 | |
| 16681prH200A | Y | G | T | C | T | T | M | G | E | A | R | R | E | K | R | S | 4 | 2 | +2 | 1 | 4.59 × 106 | 87.1 ± 72.6 | |
| 16681prR201A | Y | G | T | C | T | T | M | G | E | H | A | R | E | K | R | S | 3 | 2 | +1 | 1 | 1.93 × 106 | 11.6 ± 23.8 | |
| 16681prR202A*e | Y | G | T | C | T | T | M | G | G | H | R | A | E | K | R | S | 3 | 1 | +2 | 0 | 2.81 × 106 | 6.1 ± 4.9 | |
| 16681prE203A | Y | G | T | C | T | T | M | G | E | H | R | R | A | K | R | S | 4 | 1 | +3 | 1 | 3.36 × 107 | 107.2 ± 65.7 | |
| 16681prK204A | Y | G | T | C | T | T | M | G | E | H | R | R | E | A | R | S | 3 | 2 | +1 | 1f | Not viable | ||
| 16681prR205A | Y | G | T | C | T | T | M | G | E | H | R | R | E | K | A | S | 3 | 2 | +1 | 0 | Not viable | ||
| 16681prS206A | Y | G | T | C | T | T | M | G | E | H | R | R | E | K | R | A | 4 | 2 | +2 | 1 | 1.51 × 107 | 189.7 ± 150.8 | |
| 191g | 196 | 201 | 205 | 206 | |||||||||||||||||||
Underlined amino acids represent charged residues at non-P1, -P2, and -P4 furin consensus positions between the cleavage positions P1 and P13. The downward arrow indicates the prM cleavage site. Boldface type indicates alanine-substituting mutations.
Data are mean standard deviations (SD) as determined by direct microscopic counting of unselected 23 to 26 foci.
The sets of nonoverlapping sequences Arg-Xaa-(Lys/Arg)-Arg and Arg-Xaa-Xaa-Arg.
Numbers refer to the position of amino acids relative to the prM cleavage site in the proximal direction (without apostrophe) and distal direction (with apostrophe).
This mutant also contains the GIn131-to-Arg and Glu199-to-Gly mutations and is designated 16681prR202A*.
The consensus furin sequence was mutated into the minimal furin motif.
Numbers refer to the positions of amino acids in the polyprotein according to reference 5.
Immunoblot analysis of viral structural proteins.
Subconfluent monolayers of C6/36 cells in 25-cm2 tissue culture flasks were infected with dengue viruses at multiplicities of infection (MOI) of up to 5 focus-forming units (FFU)/cell, washed, and cultured in Leibovitz's L-15 medium supplemented with 1.5% fetal bovine serum (FBS) for 48 to 72 h. Culture medium contents were collected and centrifuged at 10,000 rpm for 3 min in a microcentrifuge to remove detached cells and debris. Clarified culture medium was adjusted to 50 mM Tris (pH 6.8)-5% glycerol-2% sodium dodecyl sulfate (SDS)-0.01% bromophenol blue and incubated for 30 min at room temperature and loaded onto a 0.1% SDS-13% polyacrylamide gel without boiling. Following electrophoretic separation, proteins were transferred to 0.45-μm nitrocellulose membranes (Schleicher and Schuell, Keene, NH) by using a semidry electroblotter. Nonspecific binding sites on the membrane were blocked with 5% skim milk in phosphate-buffered saline (PBS) for 1 h, and then the membrane was reacted successively with 3H5, an anti-E protein antibody, and prM-6.1, an anti-prM protein antibody. Following the incubation with each primary antibody, the membrane was washed three times with 0.05% Tween 20 in PBS and then reacted with horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulins antibody (Dako, Glostrup, Denmark) at a dilution of 1:1,000 for 1 h at room temperature. The membrane was again washed three times, and the target proteins were visualized by using a chemiluminescent substrate, luminol (Perkin Elmer, Boston, MA), and exposure to X-ray film. The sizes of the protein bands were determined by comparing them to a prestained protein ladder (Fermentas, Hanover, MD).
To assess the presence of C protein in each type of viral particles, particles were concentrated from the culture medium by precipitation with 7 g% polyethylene glycol (PEG) 8000 and 2.3 g% NaCl and then separated by rate-zonal centrifugation (25,000 rpm for 3 h at 4°C, using a Beckman SW40 rotor) in a 5 to 25 g% sucrose gradient (44). About 20 μl of the 1-ml fractions collected from the bottom of the centrifuge tube was mixed with 6 μl of 200 mM Tris (pH 6.8)-20% glycerol-8% SDS-0.04% bromophenol blue, adjusted with either 10 mM Tris (pH 7.2)-2 mM EDTA-150 mM NaCl (TES) buffer or 55 g% (wt/wt) sucrose in TES buffer to equalize the sucrose concentrations in different fractions to about 15 g% (wt/wt) and subjected to electrophoresis in a 0.1% SDS-15% polyacrylamide gel. Following an electrophoretic transfer to nitrocellulose membrane, the presence of C was assessed by employing a mixture of Cap 2 S.1 (2G11), anti-dengue virus C antibody, and 3H5 in the first step. Detection of bound primary antibodies was then performed as described above.
Labeling and purification of virus by isopycnic and rate-zonal centrifugation methods.
Subconfluent monolayers of C6/36 cells in 25-cm2 flasks were infected with dengue viruses at an MOI of 0.2 FFU/cell for 48 h. Infected cells were starved for 1 h at 37°C in methionine- and cysteine-free Dulbecco's modified Eagle medium containing 1.5% dialyzed FBS and subsequently supplemented with l-[35S]methionine and l-[35S]cysteine in vitro cell-labeling mixtures (Redivue Pro-mix; Amersham, Buckinghamshire, England) to a final concentration of 50 μCi/ml. After 16 h of labeling, the culture medium was harvested and centrifuged at 14,000 rpm in a microcentrifuge for 10 min to remove cells and cellular debris. Clarified medium samples were combined with one-half volume of 21 g% PEG-6.9 g% NaCl, and the mixture was gently agitated in a rotator for 18 h at 4°C. The precipitates were collected by centrifugation at 14,000 rpm in a refrigerated microcentrifuge for 30 min and then resuspended in TES buffer for 1 to 2 days at 4°C. Concentrated virus suspension was layered onto a 5 to 55 g% sucrose gradient and centrifuged at 35,000 rpm at 4°C in an SW40 rotor for 24 h. Fractions (0.7 ml) were collected from the bottom by an upward-displacement method and tested for the presence of viral particles by lysing 15 μl of each fraction with 5 μl of 200 mM Tris (pH 6.8)-20% glycerol-8% SDS-0.04% bromophenol blue, followed by electrophoresis in a 0.1% SDS-15% polyacrylamide gel, and fluorography as described previously (22). Fractions 1 to 5 from the bottom usually contained viral particles, as indicated by the simultaneous presence of the E, prM, and M protein bands; they were pooled and centrifuged at 25,000 rpm at 4°C in an SW40 rotor for 3 h. The viral pellet was then resuspended with TES, adjusted to 50 mM Tris (pH 6.8)-5% glycerol-2% SDS-0.01% bromophenol blue, and subjected to polyacrylamide gel electrophoresis. The gel was fixed with an acetic acid-isopropanol (acetic acid/isopropanol/water = 10:25:65) mixture, dried under a vacuum, and exposed to a storage phosphor screen, which was then scanned with a phosphorimager (model Typhoon 6410; Amersham). Recorded radioactivity signals were analyzed by employing ImageQuant software (Amersham).
Following the precipitation of viral particles with PEG-NaCl, the separation of virions and subviral particles was performed by subjecting concentrated virus suspension to rate-zonal centrifugation. Virus suspension was layered onto a 5 to 25 g% sucrose gradient and centrifuged in an SW40 model rotor at 25,000 rpm at 4°C for 3 h (44). Fractions (1 ml) were collected from the bottom of the centrifuge tube, with an upward-displacement method, and stored at 4°C until used in the immunoprecipitation assay and for quantitation of infectious viruses.
Immunoprecipitation.
Unfractionated virus suspension or sucrose fractions were reacted with fixed amounts of 3H5 or prM-6.1 in the form of ascites or purified IgG preparations at 4°C for 4 h or more. Virus-antibody complexes were captured by adding protein G-Sepharose 4 Fast Flow beads (Amersham), rotating the mixture at 4°C for 4 h, and spinning it at 14,000 rpm in a refrigerated microcentrifuge for 5 min. Pelleted beads were washed once with 50 mM Tris-HCl (pH 7.4)-4 mM EDTA (pH 8.0)-300 mM NaCl, and the virus-antibody complexes were dissociated with an equal volume of 5% 2-mercaptoethanol-3% SDS-80 mM Tris-HCl (pH 6.8)-0.04% bromophenol blue and incubated at 37°C for 30 min. Equal volumes of the eluted materials were electrophoresed in a 0.1% SDS-15% polyacrylamide gel, and the gel was fixed sequentially with an acetic acid-isopropanol mixture and a methanol-glycerol (methanol/glycerol/water = 40:2:58) mixture for 1 h and then wrapped with cellophane for air drying and subsequent exposure to a phosphorimager screen. Radioactivity signals were collected with a phosphorimager and analyzed using ImageQuant software. In many instances, virus particles were captured with antibody-coated protein G-Sepharose beads to facilitate the manipulation of multiple samples, yielding essentially the same results as that obtained with the capture of virus-antibody complexes with uncoated beads.
Quantitation of prM cleavage.
Radioactivity signals from the E, prM, and M protein bands were subtracted, with local background signals derived from equal areas of the gel image by using ImageQuant software. The E, prM, and M protein signals were divided by 33, 16 (15 for the JEVpr/16681 prM protein), and 5, respectively, to adjust for the differences in methionine-plus-cysteine contents of these proteins (5, 22). The extent of prM cleavage was calculated by using the following formula: prM cleavage (%) = [M adjusted signal/(prM adjusted signal + M adjusted signal)] × 100. The proportion of prM-containing virions was determined from the following calculation: prM adjusted signal/E adjusted signal.
Negative staining and electron microscopy.
C6/36 cells were infected with dengue viruses at an MOI of 0.5 FFU/cell overnight, washed, and then cultured in L-15 medium containing 1.5% FBS at 29°C. Culture medium from 20 75-cm2 flasks was removed starting on day 2 (parent virus) or day 3 (strain H200A) after infection for 3 consecutive days, pooled, and clarified by centrifugation at 3,000 rpm (1,490 × g) for 10 min. Viral particles were concentrated by precipitation with PEG-NaCl at 4°C overnight, as described above, and collected by centrifugation at 12,000 rpm (16,260 × g) at 4°C for 25 min. Precipitates were resuspended in a small volume of 12 mM Tris (pH 8.0)-1 mM EDTA-120 mM NaCl for 1 to 2 days at 4°C and pooled. Aggregates were removed by centrifugation at 10,000 rpm (9,700 × g) for 1 min at 4°C. Virus suspension was then underlayered with a 1.5-ml cushion of 24 g% sucrose in the same buffer. Following centrifugation at 32,000 rpm at 4°C for 4 h using a model SW40 rotor, the viral pellet was resuspended and subjected to rate-zonal centrifugation in a 5 to 25 g% sucrose gradient, as described above. Fractions (1 ml) were removed from the bottom of the tube by using an upward-displacement method. Two separate pools of fractions, 2 to 4 and 9 to 10, were then concentrated by using a centrifugal filter device (Ultracel 100K; Amicon, Millipore, Cork, Ireland) and 24 ml of 12 mM Tris (pH 8.0)-1 mM EDTA-120 mM NaCl to obtain a final volume of 50 to 100 μl. For negative staining, 3.5 μl of the concentrated virus preparation was adsorbed onto a glow-discharged, Formvar/carbon-coated copper grid (Electron Microscopy Sciences, Hatfield, PA), washed with 1 ml of deionized water, fixed with 1% glutaraldehyde, and then stained twice with 7 μl of 1 g% uranyl acetate. Air-dried grids were examined using a transmission electron microscope (JEM-1200EXII; JEOL, Tokyo, Japan) at a magnification of ×50,000 at 80 kV. For each particle, a mean diameter was determined from the distances measured in two perpendicular directions.
Determination of specific infectivity and kinetics of virus replication.
The specific infectivity of a virus sample was determined by quantitating infectious virus titer, employing the focus immunoassay titration and comparing that value with the concentration of genome-containing viral particles that was assessed by using a quantitative, real-time RT-PCR essentially as described previously (22). In this study, the RT-PCR was performed in the capillary tube format using a thermocycler with a real-time display of the amplification results (Roche, Mannheim, Germany), and the specific infectivity was expressed as FFU/1,000 genomic RNA copies. A comparison of infectivity was performed between C6/36 and PS (C6/36-PS) cells and PS and PS (PS-PS) cells, according to the cellular sources employed in virus preparation and the quantitation of infectious virus titer, respectively. The determination of kinetics of virus replication was performed in PS cells employing the single-step model and the multistep model as described previously (22).
Statistical analysis.
Data analysis was performed using STATA software (version 8.2; StataCorp, TX). The t test was employed in the comparisons of specific infectivity, the proportion of proteins, and the infectious virus titer between the pr-M junction mutants and the parent virus. P values equal to or less than 0.01, 0.02, or 0.05 were considered significance levels.
RESULTS
Opposing cleavage modulatory influences of charged residues at the cleavage positions P3, P5, and P6 of the dengue virus pr-M junction.
In our previous study of the influence of the amino acid sequence just proximal to the pr-M junction on the efficiency of prM cleavage, a set of chimeric viruses was generated by replacing the 13-amino-acid segment in a dengue virus, strain 16681Nde(+), with those from JEV, TBEV, and YFV (22). Although the chimeric viruses contained basic residues at the furin consensus positions P1, P2, and P4, cleavage of prM was enhanced in JEVpr/16681 and YFVpr/16681, which contained JEV and YFV sequences, respectively, indicating that the cleavage up-modulatory influence of nonfurin consensus residues derived from these viruses (22). Within this 13-amino-acid segment, three types of sequence variations differentiated JEV from dengue virus: a lack of acidic side chains at positions P3 and P7, the presence of basic residues at positions P8, P10, and P13, and the substitution of uncharged residues at positions P6 (Ser versus His) and P9 (Thr versus Met) (Table 1). In the present study, mutants bearing each type of these changes, pr(+4,−0), pr(+7,−2), and pr(+7,−0), were constructed to separately assess their influences on prM cleavage (Table 1). The mutant designations refer to the total positive and negative charges, respectively, in the P1 to P13 segment just proximal to the pr-M junction. Mutant viruses were viable, but their titers and focus sizes were reduced. In the two mutants with an entire dengue virus background, pr(+4,−0) lacked the P3 Glu and P7 Glu residues, whereas pr(+7,−2) contained Arg at positions P8, P10, and P13.
Following infection of C6/36 cells, immunoblot analyses of the E and prM proteins in the culture medium revealed that, at similar levels of E, less prM was detected in pr(+4,−0) and JEVpr/16681 than in the parent virus and pr(+7,−2) (Fig. 2A, left panel). The reduction of prM in pr(+4,−0) and JEVpr/16681 was accompanied by an increase in the levels of pr peptide. M protein was not detected by prM-6.1 or by six other anti-prM monoclonal antibodies or in pooled human convalescent-phase sera (data not shown). As E and prM are present in newly assembled particles in equal proportions, this result suggested that prM had been cleaved to a greater extent in pr(+4,−0) than in pr(+7,−2). However, a weaker signal was detected for pr than for prM by prM-6.1 in 16681Nde(+), which was incompatible with our previous estimates of 66 to 77% cleavage of prM for this parent virus (22). Detection of an unexpectedly low level of pr was not unique to prM-6.1, as other anti-prM antibodies and a pool of human convalescent-phase sera revealed similar findings (data not shown). Also, the reciprocal increase in pr signals in pr(+4,−0) and JEVpr/16681 was less than expected from the combined signals of prM and pr in 16681Nde(+).
FIG. 2.
Cleavage of prM in pr-M junction mutants. (A) Culture medium from virus-infected C6/36 cells was lysed with SDS, electrophoresed in a 0.1% SDS-13% polyacrylamide gel, and blotted onto a nitrocellulose membrane. Three viral proteins, E and prM and pr, were detected successively by using 3H5 and then prM-6.1, followed by horseradish-peroxidase-conjugated anti-mouse immunoglobulin antibody and a chemiluminescent substrate, luminol. Light signals were captured on X-ray film. (B and C) Virus-infected C6/36 cells were metabolically labeled with [35S]methionine and [35S]cysteine. Viral proteins in the culture medium were immunoprecipitated with prM-6.1 (B) or 3H5 (C), eluted from Sepharose beads with SDS-containing buffer, and then electrophoresed in a 0.1% SDS-15% polyacrylamide gel. (D) Virus-infected C6/36 cells were metabolically labeled with [35S]methionine and [35S]cysteine. Particles in the culture medium were concentrated by precipitation with PEG and then partially purified by isopycnic centrifugation using a 5 to 55 g% (wt/wt) sucrose gradient. Viral particles in a pool of several lower fractions were pelleted by centrifugation, lysed with SDS, and electrophoresed in a 0.1% SDS-15% polyacrylamide gel. Viral structural proteins are indicated. Following subtraction with local background signals and adjustment for the differences in methionine-plus-cysteine contents, the radioactivity signals of the E, prM and M protein bands in panels B to D are shown numerically in Table 2.
To distinguish between the reduced reactivity of prM-6.1 with pr in the immunoblot format and the partial degradation of pr, virus-infected cells were metabolically radiolabeled, and the viral proteins were immunoprecipitated separately from the culture medium with prM-6.1 and 3H5, an anti-E protein antibody. As shown in Fig. 2B (left panel), prM-6.1 pulled down a higher level of pr than prM from the 16681Nde(+)-infected culture medium, consistent with the low sensitivity of prM-6.1 for detecting pr in the immunoblot analysis. Similarly, the reciprocal increases in the signals of pr in pr(+4,−0) and JEVpr/16681 were more evident with immunoprecipitation than with immunoblot analysis (Fig. 2B, left panel). Immunoprecipitation with prM-6.1 and 3H5 (Fig. 2B and C, left panel) also revealed the reciprocal increase of M in pr(+4,−0) and JEVpr/16681, although the visual changes were not as obvious as in the case of pr. The disparity likely reflected a higher level of radiolabeling of pr than M (10 or 11 versus 5 methionine-plus-cysteine residues per molecule), which resulted in a relatively greater increase in the signal of the pr band. In addition, there was relatively more pr available than M in the immunoprecipitation with anti-prM antibody (Fig. 2B), as pr's that were liberated from particles undergoing either complete or partial prM cleavages would be structurally similar and thereby would be recognized as efficiently by the antibody, whereas M could not be precipitated from particles that had achieved complete prM cleavage. When the signal intensities of prM and M bands were adjusted for their differences in methionine-plus-cysteine content, a higher level of prM cleavage was detected in pr(+4,−0) than in pr(+7,−2), regardless of the antibodies employed (Table 2).
TABLE 2.
Quantitation of viral structural proteins in virus particles obtained by various methods by a phosphorimager
| Figure | Method | Virus | Adjusted signala
|
Ratio of protein to prM-plus-M
|
Avg content (molecules/ virion)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| E | prM | pr | M | E | M | prM | M | |||
| 2B, left panel | Immunoprecipitation | 16681Nde(+) | 1256117.0 | 364631.2 | 1441780.7 | 607547.1 | 1.29 | 0.62 | 67.5 | 112.5 |
| with prM-6.1 | pr(+4,−0) | 1286969.8 | 212790.4 | 3192401.4 | 683755.6 | 1.43 | 0.76 | 42.7 | 137.3 | |
| pr(+7,−2) | 1415817.6 | 319494.6 | 2141837.9 | 638610.7 | 1.48 | 0.67 | 60.0 | 120.0 | ||
| JEVpr/16681 | 735938.8 | 89114.6 | 9345678.3 | 781852.3 | 0.84 | 0.90 | 18.4 | 161.6 | ||
| 2C, left panel | Immunoprecipitation | 16681Nde(+) | 1351403.9 | 402368.7 | 754095.8 | 1.17 | 0.65 | 62.6 | 117.4 | |
| with 3H5 | pr(+4,−0) | 1632291.2 | 231728.6 | 1031865.0 | 1.29 | 0.82 | 33.0 | 147.0 | ||
| pr(+7,−2) | 1507093.9 | 325042.9 | 718824.1 | 1.44 | 0.69 | 56.0 | 124.0 | |||
| JEVpr/16681 | 1678672.8 | 62215.4 | 1088936.7 | 1.46 | 0.95 | 9.7 | 170.3 | |||
| 2D, left panel | Isopycnic centrifugation | 16681Nde(+) | 4971617.6 | 2026770.6 | 3355778.4 | 0.93 | 0.62 | 67.8 | 112.2 | |
| pr(+4,−0) | 5397920.4 | 644768.6 | 4639059.4 | 1.02 | 0.88 | 22.0 | 158.0 | |||
| pr(+7,−2) | 7044915.8 | 1892181.6 | 4349778.8 | 1.13 | 0.69 | 54.6 | 125.4 | |||
| JEVpr/16681 | 6108800.9 | 268908.4 | 6296970.2 | 0.93 | 0.95 | 7.4 | 172.6 | |||
| 2B, right panel | Immunoprecipitation | 16681Nde(+) | 2163015.5 | 1073930.4 | 11585028.2 | 1218178.0 | 0.94 | 0.53 | 84.3 | 95.7 |
| with prM-6.1 | E199A | 2163353.3 | 754755.9 | 5297646.6 | 1195762.7 | 1.11 | 0.61 | 69.7 | 110.3 | |
| H200A | 2683048.1 | 1973920.9 | 3872756.8 | 553912.6 | 1.06 | 0.22 | 140.6 | 39.4 | ||
| R201A | 2845058.6 | 2203588.8 | 6504194.0 | 570788.3 | 1.02 | 0.21 | 143.0 | 37.0 | ||
| R202A* | 2658742.5 | 2208796.8 | 3806341.5 | 319235.0 | 1.05 | 0.13 | 157.3 | 22.7 | ||
| E203A | 3335999.3 | 1192224.2 | 35385072.2 | 1852398.0 | 1.10 | 0.61 | 70.5 | 109.5 | ||
| S206A | 3262718.0 | 1821336.8 | 12785999.3 | 1471035.3 | 1.00 | 0.45 | 99.6 | 80.4 | ||
| 2C, right panel | Immunoprecipitation | 16681Nde(+) | 4243501.6 | 1859684.9 | 2802733.7 | 0.91 | 0.60 | 71.8 | 108.2 | |
| with 3H5 | E199A | 3508763.1 | 1282781.7 | 2000932.4 | 1.07 | 0.61 | 70.3 | 109.7 | ||
| H200A | 3235397.3 | 2767551.6 | 934022.1 | 0.87 | 0.25 | 134.6 | 45.4 | |||
| R201A | 4042596.7 | 3459968.4 | 1218725.9 | 0.86 | 0.26 | 133.1 | 46.9 | |||
| R202A* | 4682545.7 | 5272599.9 | 836287.7 | 0.77 | 0.14 | 155.4 | 24.6 | |||
| E203A | 3652792.4 | 736231.9 | 3330977.4 | 0.90 | 0.82 | 32.6 | 147.4 | |||
| S206A | 3338105.1 | 1589490.9 | 2044307.3 | 0.92 | 0.56 | 78.7 | 101.3 | |||
| 2D, right panel | Isopycnic centrifugation | 16681Nde(+) | 187254.8 | 74825.1 | 167675.4 | 0.77 | 0.69 | 55.5 | 124.4 | |
| E199A | 142434.2 | 42132.9 | 123088.4 | 0.86 | 0.74 | 45.9 | 134.1 | |||
| H200A | 163736.5 | 121547.8 | 71117.7 | 0.85 | 0.37 | 113.6 | 66.4 | |||
| R201A | 137867.0 | 109674.2 | 81430.1 | 0.72 | 0.43 | 103.3 | 76.7 | |||
| R202A* | 131589.5 | 149103.4 | 44316.9 | 0.68 | 0.23 | 138.8 | 41.2 | |||
| E203A | 141833.0 | 22263.5 | 224796.2 | 0.57 | 0.91 | 16.2 | 163.8 | |||
| S206A | 232504.1 | 89775.0 | 201979.8 | 0.79 | 0.69 | 55.4 | 124.6 | |||
Signals from equal areas of E, prM, pr and M bands were subtracted from the background level, and then corrected with the following methionine-plus-cysteine contents: E, 33; prM of 16681Nde(+) and other mutants, 16; prM of JEVpr/16681, 15; pr of 16681Nde(+) and other mutants, 11; pr of JEVpr/16681,10; and M, 5. The original data contained four decimal points, but are shown here with only one decimal point.
The influence of pr-M junction sequences on prM cleavage was next assessed in partially purified viral particles. Viral particles released from metabolically radiolabeled, virus-infected cells were separated from nonparticulate components in the culture medium by isopycnic centrifugation. Following the disruption of particles and electrophoresis, phosphorimager analyses of the levels of prM and M confirmed a higher level of cleavage of prM in pr(+4,−0) than in pr(+7,−2) (Fig. 2D, left panel, and Table 2). In three separate experiments, the level of prM cleavage in pr(+4,−0) was comparable to that in JEVpr/16681 (means ± SD of 89.6% ± 3.1% versus 91.3% ± 7.1%, respectively). Cleavage of prM in pr(+7,−2) (67.4% ± 2.2%) differed slightly from that in the parent virus (60.7% ± 3.0%). The pr(+7,−0) mutation, which was constructed by replacing P6 Ser and P9 Thr in the JEVpr/16681 chimera with dengue virus counterparts, exhibited enhanced prM cleavage similar to that of JEVpr/16681 (data not shown). Thus, prM cleavage was augmented in mutants lacking both of the P3 Glu and P7 Glu residues, pr(+4,−0) and pr(+7,−0), but not in pr(+7,−2), which contained substituted basic residues at P8, P10, and P13. These results indicated the suppressive effect of acidic residues at position P3 and/or position P7 on the cleavage of the dengue virus pr-M junction.
In dengue virus prM, the 13-amino-acid segment proximal to the pr-M junction also contains an Arg residue at position P5, as well as a His residue at P6 (Table 1). It was documented that the P6 His residue in a furin substrate allowed cleavage by furin in the absence of a basic residue at position P4 (4). In the presence of P4 Arg, it was intriguing to determine whether P6 His and the conserved P5 Arg could exert any influence on the cleavage of the dengue virus pr-M junction. To examine the effect of each of these residues, a set of alanine-scanning mutant viruses was constructed for positions P1′ to P7. Viable viruses were recovered only for the mutated positions P1′ and P3 to P7 (Table 1). As expected, infectious P1 and P2 Ala mutant viruses were not detected at any time during the 4-week period after transfection. In two separate attempts, the P4 Ala mutant initially replicated at a lower level (<103 FFU/ml) than other mutant viruses, and then replication abruptly increased to a higher level of over 104 FFU/ml, beginning at the fifth week after transfection. Sequence analysis of the P4 Ala mutant virus preparations with high titer revealed additional mutations involving P3 Glu (E203K) in one instance and P7 Glu (E199G), as well as the Q131R substitution, in another. Only the latter P4 Ala mutant virus, designated the R202A* mutant, was expanded for subsequent studies.
Analysis of prM cleavage in the alanine-scanning mutants was performed by comparing the levels of E and prM/M proteins in the culture medium, with immunoblot analysis (Fig. 2A, right panel), as well as metabolic radiolabeling, followed by either immunoprecipitation (Fig. 2B and C, right panel, and Table 2) or isopycnic centrifugation (Fig. 2D, right panel, and Table 2). The results obtained by these methods were generally comparable, but there was a tendency toward lower prM cleavage levels in the immunoprecipitation with an anti-prM antibody. Based on three separate determinations of the prM cleavage efficiency after radiolabeling and isopycnic centrifugation, an enhancement of prM cleavage (90.9% ± 0.4%) was observed for the P3 E203A mutant, and, to a much lesser extent (73.0% ± 3.1%), for the P7 E199A mutant. In contrast, the P5 R201A and P6 H200A mutants displayed markedly reduced levels of prM cleavage (30.8% ± 12.9% and 32.1% ± 11.5%, respectively). The prM protein in the P4 R202A* mutant was cleaved even less efficiently (16.2% ± 6.9%). Replacement of P1′ Ser with Ala in S206A did not affect prM cleavage (61.9% ± 7.2%) compared with the baseline level of 66.0% ± 5.1% in the parent virus in this set of experiments. These results indicated that the P3 Glu residue exerted a strong suppressive effect on prM cleavage, and the P6 His and P5 Arg residues were able to up-modulate the cleavage of prM, even in the presence of basic residues at the three furin consensus positions. The importance of the P4 Arg residue was indicated by the poor cleavability of prM in the R202A* mutant that could not be compensated by mutations at two other positions of this protein.
Changes in the proportions of extracellular viral particles.
Extracellular particles of flaviviruses are composed of large, infectious particles and smaller subviral particles that are devoid of capsids and genomic RNA (32). The proportion of these two types of particles can be affected by changes in the cleavage at the prM N terminus (28), as well as the pr-M junction (2). In the latter case, expression of the TBEV prM+E genes in transfected cells resulted in the production of mainly subviral particles, whereas large particles became more abundant when prM cleavage was abolished by deleting the P2 basic residue (2). Whether this shift in particle size classes occurs during virus infection remains unclear. The availability of the viable pr-M junction mutants with altered prM cleavage enabled us to address the influence of prM cleavage on the proportion of particles released from virus-infected cells. Viable alanine-scanning mutant viruses and the parent strain were metabolically radiolabeled, and the viral particles were subjected to rate-zonal centrifugation in 5 to 25 g% (wt/wt) sucrose gradients. From each fraction, particles were immunoprecipitated with anti-E or anti-prM antibodies and dissociated with SDS, and the viral proteins were separated with polyacrylamide gel electrophoresis.
For the parent virus, immunoprecipitation with an anti-E antibody revealed the three structural proteins, E, prM/M, and C, mainly in fractions 4 and 5 from the bottom (Fig. 3A). These two fractions, which accounted for 69.0% of the total E protein signals combined from all 12 fractions, corresponded with the peak of infectious viruses (Fig. 4A), indicating the presence of virions in fractions 4 and 5. Each of the remaining upper fractions contained 6.5% or less of the total E signal, with a small peak in fractions 10 to 11. Consistent with the findings of previous reports (20, 34), the prM proteins in the upper sucrose fractions were enriched with the faster-migrating species, whereas those in the lower fractions contained the faster- and slower-migrating species in approximately equal amounts (Fig. 3A). This finding suggested that less abundant particles in the upper fractions were distinct from infectious virions, likely in the N-glycosylation pattern of prM. When immunoprecipitation was performed with prM-6.1, essentially the same distribution of viral structural proteins was detected, but with one exception (Fig. 3B and 4A). The anti-prM antibody also precipitated pr in fraction 12, especially when high quantities of labeled materials were employed (Fig. 3B, lane 12, and Fig. 5A, lane 12). Therefore, the majority of extracellular viral particles generated by the parent virus were rapidly sedimenting virions. The predominance of virions observed for the 16681Nde(+)-infected C6/36 cultures was in accordance with previous results obtained from dengue virus-infected C6/36 and Vero cells (24, 34). A similar distribution of particles was observed with the E199A and S206A mutants, in which prM cleavage was minimally affected (Fig. 3C and H and 4B and G).
FIG. 3.
Rate-zonal separation of viral particles. Virus-infected C6/36 cells were metabolically labeled with [35S]methionine and [35S]cysteine. Viral particles in the culture medium were concentrated by precipitation with PEG and then subjected to rate-zonal centrifugation using a 5 to 25 g% (wt/wt) sucrose gradient. Viral particles in the unfractionated preparation (lane U) and each of the 12 sucrose fractions (lanes 1 to 12) removed from the bottom of the centrifuge tube were immunoprecipitated with 3H5 or prM-6.1 and protein G-Sepharose beads. Following dissociation of the virus-antibody complexes from beads with SDS-containing buffer, the mixture was electrophoresed in a 0.1% SDS-15% polyacrylamide gel. The radioactivity signals were detected using a phosphorimager. Viral structural proteins are indicated.
FIG. 4.
Distribution of viral particles and infectious viruses in sucrose fractions following rate-zonal centrifugation. Radiolabeled viruses were separated by rate-zonal centrifugation and immunoprecipitated from each sucrose fraction with antibodies specific for dengue virus E (filled squares) or prM (open squares) proteins. The relative quantity of viral particles that was present in each fraction was expressed as the percentage of total E signal combined from all fractions. Infectious virus titer (filled circles) was determined by using the focus immunoassay titration.
FIG. 5.
Rate-zonal separation of viral particles and sequential immunoprecipitation with anti-prM and anti-E antibodies. 35S-labeled particles of the 16681Nde(+) (A) and H200A viruses (B) were concentrated by precipitation with PEG and subjected to rate-zonal centrifugation using a 5 to 25 g% (wt/wt) sucrose gradient. Viral particles in the unfractionated preparation (lane U) and each of the 12 sucrose fractions (lanes 1 to 12) removed from the bottom of the centrifuge tube were precipitated with prM-6.1-coated protein G-Sepharose beads (lane P). Unbound materials were subsequently reacted with 4G2-coated beads (lane E). Following dissociation of the virus-antibody complexes, the mixture was electrophoresed in a 0.1%SDS-15% polyacrylamide gel, and the radioactivity signals were detected using a phosphorimager. Viral structural proteins are indicated. For H200A virus, the total radioactivity employed for immunoprecipitation was about fourfold greater than that shown in Fig. 3D. Note that following precipitation with PEG and rate-zonal centrifugation, pr signals were relatively less intense in the immunoprecipitation with prM-6.1 than those in the direct immunoprecipitation of radiolabeled virus preparation with the same antibody in Fig. 1. This reflects a relatively inefficient precipitation of pr by PEG compared with that of the viral particles. Also, low levels of E, prM, and M were detected in fraction 12, likely as a result of employing larger amounts of radiolabeled particles in this rate-zonal centrifugation than those shown in Fig. 3.
In four alanine-scanning mutants, the proportions of the two types of extracellular particles differed from that of the parent virus. The H200A mutant exhibited two peaks of particles following precipitation with anti-prM or anti-E antibodies (Fig. 3D and 4C). The rapidly sedimenting peak, found in fractions 4 and 5, as in the parent virus, corresponded with the peak of infectious virus, whereas the slowly sedimenting peak, detected in fractions 10 and 11, was associated with the background level of infectivity (Fig. 4C). Based on the intensity of E signals, fractions 4 and 5 accounted for 43.7% of the combined E signals, which were comparable to 41.4% of those in fractions 10 and 11. For the H200A mutant, the C protein doublet was visible in fraction 4 but not in fractions 10 and 11 when large amounts of total precipitated radioactivity were employed (Fig. 5B), suggesting that the slowly sedimenting, noninfectious particles in fractions 10 and 11 did not contain C and, therefore, might represent subviral particles. The presence of the M band in fractions 10 and 11, as in the virions in fractions 4 and 5, distinguished the nominal subviral particles in these upper fractions from soluble E or prM proteins, which would have been precipitated without M. Moreover, the contribution of soluble E and prM should be detected in fraction 12, as in the case of pr in the analysis of the parent virus. The bimodal distribution of the H200A mutant particles closely resembled the separation of two types of TBEV particles in a similar (5 to 30 g%) sucrose gradient (1).
Two additional strains, the R201A and R202A* mutants, with low levels of prM cleavage displayed the highest accumulation of particles in fractions 9 to 11 (Fig. 3E and F), which did not correspond to the infectious viruses present in the lower fractions (Fig. 4D and E). These three fractions contained 86.7% and 80.6% of the combined E signals in the R201A and R202A* mutants, respectively. As observed for the parent virus, the nominal subviral particles present in fractions 10 and 11 of the three mutants with reduced levels of prM cleavage were enriched with the faster migrating prM species. The pr peptide was not detected in the immunoprecipitation of the R201A and R202A* mutants with anti-prM antibody, in agreement with a low prM cleavability of these two mutants.
In contrast to the bimodal distribution or the predominance of subviral particles in mutants with reduced prM cleavage, the E203A mutant was deprived of subviral particles, as the E signals in fractions 10 and 11 were almost undetectable (Fig. 3G and 4F). The peak of rapidly sedimenting particles in fractions 4 and 5 accounted for 82.5% of the total E protein present in all 12 fractions, and fraction 4 was associated with the same peak of infectious virus as in the parent virus (Fig. 4F).
In the separation of viral particles by rate-zonal centrifugation assay, particles were first precipitated with PEG, resuspended for several hours, and then centrifuged through the sucrose gradient. During these manipulations, it was possible that some virions were partially damaged and/or aggregates of virions could not be completely removed; these damaged virions and aggregates might have become associated with the upper sucrose fractions. Due to a poor metabolic labeling of C with radioactive methionine and cysteine, the extent to which aggregates and damaged virions contributed to the E and prM/M signals in the upper fractions was difficult to assess in previous experiments. In an attempt to address this possibility, the presence of C in various sucrose fractions was determined with an immunoblot analysis using a combination of anti-E and anti-C antibodies (Fig. 6). For the parent virus, stronger signals of E were detected in fractions 4 and 5 than in fractions 9 to 11, in a pattern similar to that identified by immunoprecipitation of radiolabeled viruses (Fig. 6A). In contrast, C was detected only in fractions 4 and 5 but not the upper fractions, despite overexposure of the blot to X-ray film. A similar pattern of C reactivity was observed for the H200A, R201A, and E203A mutants employed (Fig. 6B to D). These results indicated that partially damaged virions and aggregates with intact C protein did not contribute to the E and prM/M signals in the upper sucrose fractions.
FIG. 6.
Immunoblot analysis of sucrose gradient fractions with anti-C antibody. Culture medium from virus-infected C6/36 cells was precipitated with PEG, resuspended, and subjected to rate-zonal centrifugation in 5 to 25 g% (wt/wt) sucrose gradient. Equal volumes of each of the 1-ml fractions that were removed from the bottom of the centrifuge tube were adjusted to the final concentration of about 15 g% (wt/wt) sucrose, electrophoresed in a 0.1% SDS-15% polyacrylamide gel, and blotted onto the nitrocellulose membrane. The membrane was incubated with a mixture of 3H5 and Cap2 S.1 (2G11), an anti-C antibody, washed, and then reacted with horseradish peroxidase-conjugated rabbit anti-mouse Ig antibody and luminol. Generated signals were captured on X-ray film. As shown in lanes 1 to 12 of panels A, B, and D, the exposure time was adjusted to allow an overexposure of the signals from protein C bands. Lane 13 of panels A, B, and D represents images of fraction 4 exposed for a shorter exposure time. The two viral proteins E and C are indicated.
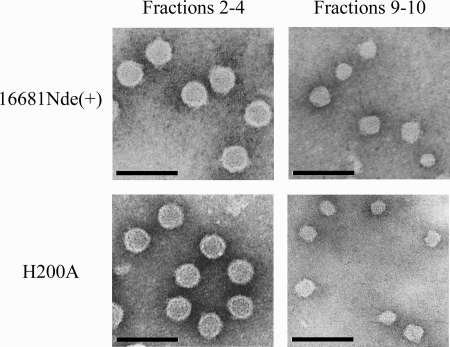
The nature of particles in the upper sucrose fractions was further examined by transmission electron microscopy. The parent virus and the H200A mutant, the virus with a nearly equal distribution of particles in the lower and upper sucrose fractions, were grown in C6/36 cells, concentrated by precipitation with PEG and pelleting of the resuspended materials through a sucrose cushion, and then subjected to rate-zonal centrifugation. When particles pooled from the lower and upper sucrose fractions were visualized with negative staining, differences in size were observed (Fig. 7). Spherical particles in the lower fractions were larger [16681Nde(+), mean diameter ± standard deviation of 45.5 ± 2.1 nm, n = 130; the H200A mutant, 45.2 ± 2.2 nm, n = 158] and homogeneous in shape, whereas those in the upper fractions were smaller (16681Nde(+), 30.8 ± 3.4 nm, n = 172; the H200A mutant, 31.4 ± 4.1 nm, n = 204) and less homogeneous. Particles in the upper sucrose fractions were similar to previously described subviral particles, which were derived from the expression of prM+E genes of dengue virus and TBEV in eukaryotic cells (2, 11, 45). Based on the results from immunoblot analysis and electron microscopy, particles in the upper sucrose fractions likely represented subviral particles that assembled without incorporating the C protein.
FIG. 7.
Transmission electron microscopic visualization of particles. The 16681Nde(+) and H200A strains were grown in C6/36 cells. Viral particles were concentrated by precipitation with PEG, pelleted through a 24 g% (wt/wt) sucrose cushion, and subjected to rate-zonal centrifugation in a 5 to 25 g% (wt/wt) sucrose gradient. Two pools of fractions 2 to 4 and 9 to 10 were concentrated using an ultrafiltration centrifugal device. Concentrated preparations were adsorbed onto the glow-discharged, Formvar/carbon-coated copper grids, fixed with 1% glutaraldehyde, and stained with 1% uranyl acetate. Pictures were taken at a magnification of ×50,000 with a defocus technique using an 80-kV transmission electron microscope. Bar represents 100 nm.
The influence of prM cleavage on the proportion of mature versus immature virions was next examined by comparing the prM/E and (prM+M)/E ratios of the fraction 4 particles following the immunoprecipitation with anti-E or anti-prM antibodies. For the parent virus, the (prM+M)/E ratio was close to 1, as expected for the equimolar relationship between prM/M and E in flaviviral particles, whereas the prM/E ratio was only 0.36 (Fig. 8A). This result indicated that if an all-or-none prM cleavage had occurred, only about two-thirds of the fraction 4 particles would have existed as mature virions and the other third as immature virions. However, the fraction 4 particles were precipitated by an anti-prM antibody almost as efficiently as the anti-E antibody (Fig. 3A and B and Fig. 4A), and the sequential immunoprecipitation employing an anti-E antibody following the anti-prM antibody reaction revealed only a small proportion of particles that were not precipitated by the anti-prM antibody (Fig. 5). It is likely then that more than one-third of the fraction 4 particles contained the remaining prM molecules. In this case, the prM/E ratio provides only a minimal estimate for the proportion of prM-containing virions, which includes immature (180 prM/particle) and partially mature (1 to 179 prM/particle) virions, among all virions present in fraction 4. For the E199A and E203A mutants with enhanced prM cleavage, the prM/E ratios were 0.29 and 0.14, respectively, indicating a decrease in the proportion of prM-containing virions compared to the parent virus level (Fig. 8A). A very low prM/E ratio was also observed for JEVpr/16681(Fig. 8A). On the contrary, the prM/E ratio was elevated in the three mutants with reduced prM cleavage (1.02, 0.99 and 0.94, respectively, for the H200A, R201A, and R202A* mutants), indicating an increase in the proportion of prM-containing virions in these mutants (Fig. 8A). The prM/E ratio of the fraction 4 particles of the S206A mutant was similar to that of the parent virus. In contrast to the wide variations of the prM/E ratio, the (prM+M)/E ratio distributed in a more restricted range of 1.05 to 1.39 in these mutants (Fig. 8A). It should be noted that the (prM+M)/E ratio of the fraction 4 particles in some mutants was higher than that of the parent virus, likely because of the difficulties in measuring signals of the M protein in these mutants. Nevertheless, when the E signal was replaced by the combined (prM+M) signals, the prM/(prM+M) ratio remained higher than that of the parent virus. For example, the prM/(prM+M) ratio of virions in fraction 4 was 0.75 for the R202A* mutant compared with 0.33 for the parent virus.
FIG. 8.
prM/E and (prM+M)/E ratios and specific infectivity. (A) prM/E and (prM+M)/E ratios. Radiolabeled particles in the PEG-precipitated, unfractionated preparations and the peak virion fractions derived from rate-zonal centrifugation were immunoprecipitated with 3H5 or prM-6.1 and subjected to SDS-polyacrylamide gel electrophoresis and phosphorimager analysis. The proportion of prM-containing particles was determined by dividing the adjusted prM signal by the adjusted E signal. For comparison, the adjusted (prM+M) signal was divided by the adjusted E signal to yield the (prM+M)/E ratio. With the exception of an unfractionated preparation of S206A virus, data represent the means and standard errors of the means derived from two to four determinations. *, P values of ≤0.02 in the pair-wise comparison of each mutant with the parent virus. (B) Specific infectivity. Specific infectivity was determined following the quantitative RT-PCR and focus immunoassay titration and shown separately in the PS-PS and C6/36-PS categories according to the cellular sources employed for the virus propagation and titration. Data represent the means and standard deviations derived from two to eight determinations. *, P values of ≤0.01 in the pair-wise comparison of each mutant with the parent virus.
Thus, analyses of the alanine-scanning pr-M junction mutants revealed that the reduction of prM cleavage was associated with higher proportions of subviral particles and prM-containing virions present in the extracellular compartment, whereas an enhancement of cleavage brought about the opposite effect. These results extend a finding from the alteration of the proportion of extracellular particles caused by various efficiencies of prM cleavage previously observed for prM+E transfected cells (2).
Alterations of infectivity and virus replication.
Our analysis of the extracellular particles by rate-zonal centrifugation and immunoprecipitation indicated the presence of prM-containing particles that cosedimented with infectious virions in the lower sucrose fractions. These findings raised the possibility that, by modifying the proportions between mature virions and prM-containing virions or virions and subviral particles, alterations of prM cleavage efficiency might affect the infectivity of dengue virus preparations in more than one direction. To assess this possibility, we determined the infectivity of the virus preparations and monitored their multiplications, employing in vitro single-step and multistep kinetics assays.
For a comparison of the infectivity of the parent with that of mutant viruses, specific infectivity was determined by quantitating the level of viral genomic RNA in virus preparations obtained from C6/36 or PS cells, employing a quantitative real-time RT-PCR method, and comparing this level with the infectious virus level that was determined by the focus immunoassay titration. The specific infectivity was expressed in two categories, C6/36-PS and PS-PS, according to the cells employed for virus multiplication and titration, respectively. As shown in Fig. 8B, there was less than a 10-fold difference between the specific infectivity levels of the parent virus following its multiplication in these two cell lines. This was also observed for mutant viruses, with the exception of the R201A and the R202A* mutants; the basis for this disparity has not been established. There was a reduction (a difference of more than 10-fold in at least one category) of specific infectivity in the R201A mutant (32.1- and 7.6-fold in the PS-PS and C6/36-PS categories, respectively; n = 4) and the R202A* mutant (31.4- and 4.1-fold, respectively; n = 2 to 4) compared with that of the parent virus. A decrease in the specific infectivity of the H200A mutant was modest but consistently observed (4.8- and 2.7-fold, respectively; n = 3 to 4). These data indicated that reduction of prM cleavage to the extent observed for these mutants, most notably the R201A and R202A* mutants, resulted in a down-modulation of the virus infectivity. This is consistent with previous findings in a TBEV mutant in which the failure to cleave prM resulted in a loss of infectivity (9) and confirms similarly low levels of the reduction in the specific infectivity of dengue virus following treatment of infected cells with acidotropic agents (36). In contrast, the increases in the specific infectivity of the E203A mutant were detected at 8.5- and 2.0-fold in the PS-PS and C6/36-PS categories, respectively (n = 4 or 5). As in the case of the H200A mutant, changes of less than 10-fold may represent a modest increase of the specific infectivity of the E203A mutant or, less likely, reflect the variations that might result from virus titration as viral samples were serially diluted in 10-fold steps during the quantitation of infectious viruses. The lack of a strong up-modulatory effect of an enhanced prM cleavage on the specific infectivity as detected in the E203A mutant reiterated a similar finding previously observed for JEVpr/16681 (Fig. 8B) (22). Changes in the specific infectivity of the E199A and S206A mutants with only slight alterations of prM cleavage were minimal (Fig. 8B).
The influence of altered prM cleavage on virus replication was next assessed with single-step and multistep kinetics studies. In the multistep kinetics study, PS cells were infected at an MOI of 0.01 for 2 h. Cells were washed extensively to remove unbound particles, and the culture medium was replaced and then collected daily for the quantitation of infectious viruses. Comparison of virus titers revealed that the H200A, R201A, and R202A* mutants, which had reduced prM cleavage, exhibited reduced multiplication over the first 5 days after the infection of PS cells (Fig. 9). On day 7 after infection, there were no differences among these and other viruses. The parent virus multiplied rapidly over the first 3 days, reaching the plateau level at day 4, and then declined afterward; the late decrease in virus titer correlated with extensive detachment of cells. The E199A, E203A, and S206A mutants were indistinguishable from the parent virus in this experiment (Fig. 9).
FIG. 9.
Multistep kinetics of virus replication. PS cells were infected with the parent virus and the pr-M junction mutants at an MOI of 0.01 for 2 h and washed extensively, and the culture medium was removed daily for 7 days for the determination of infectious virus titers, employing focus immunoassay titration. Data represent the means and standard errors of the means derived from two separate experiments. *, P values of ≤0.05 in the pair-wise comparison of each mutant with the parent virus.
The multiplication of viruses was assessed in more detail with the single-step kinetics study. PS cells were infected at an MOI of 1 for 2 h and washed, and the culture medium was collected at various hourly time points over 2 days. Following removal of the medium, infected cells were washed extensively and subjected to three freeze-thaw cycles to liberate cell-associated viruses. Comparisons were then made with both extracellular viruses and cell-associated viruses by employing the focus immunoassay titration. As reported previously (22), the parent virus rapidly generated infectious, cell-associated particles beginning at 14 h after adsorption (Fig. 10A). At 10 h later, an approximately 1,000-fold increase in the infectious virus titer was detected. Following this initial amplification, the subsequent rise to the plateau level at 36 to 48 h after adsorption was not as large. In the extracellular compartment, the rise in the infectious virus titer of 16681Nde(+) was also detected at 14 h after adsorption, with further changes following the same time course as that of the cell-associated viruses. For JEVpr/16681, a marked reduction of extracellular virus titers was again observed, despite a rise of cell-associated virus titers that was similar to that in 16681Nde(+) (Fig. 10F). Compared with the parent virus, the H200A, R201A and R202A* mutants with reduced prM cleavage displayed longer latency periods of 20 to 24 h, as well as reduced levels of infectious virus titer (Fig. 10B to D). Unlike JEVpr/16681, the reductions in titers were detected concomitantly in both compartments. At the plateau levels detected at 36 to 48 h after adsorption, the reduction was greater in magnitude in the R201A and R202A* mutants (more than 100-fold) than in the H200A mutant (slightly more than 10-fold), which correlated with the changes in their specific infectivities. As expected from the multistep kinetics study, the replication of the E203A mutant was similar to that of the parent virus in the single-step study (Fig. 10E). Despite a high level of prM cleavage in that mutant, the changes in its extracellular infectious virus titer were not affected as they were in the case of JEVpr/16681 (22). This result suggested that enhanced prM cleavage by itself did not adversely influence the accumulation of infectious viruses in the extracellular compartment. The reductions of extracellular infectious virus titer and focus size previously observed with JEVpr/16681 are likely due to another mechanism(s).
FIG. 10.
Single-step kinetics of virus replication. PS cells were infected with designated viruses at an MOI of 1 for 2 h, washed extensively, and further cultured in 1 ml of the culture medium. At the indicated time points after the infection, the culture medium was removed for the determination of infectious virus titer, and the cell monolayers were washed extensively, overlaid with the culture medium containing 20% FBS, and subjected to three freeze-thaw cycles. Cell lysates were clarified by centrifugation, and the levels of cell-associated infectious viruses were determined by focus immunoassay titration. Data represent the means and standard errors of the means derived from three to six separate experiments. *, P values of ≤0.05 in the pair-wise comparison of each mutant with the parent virus.
Among the alanine-scanning mutants employed in the multistep kinetics study, alterations of virus multiplication were detected only in the three viruses with reduced prM cleavage. Further assessment using the single-step model revealed a greater reduction of virus multiplication in the R201A and R202A* mutants than in the H200A mutant. The differences correlated with the changes in their specific infectivities but not with the levels of prM cleavage.
DISCUSSION
During the replication of flaviviruses, cleavage of prM is essential for the acquisition of infectivity (27). Previous observations for the partial prM cleavage in dengue virus raise questions regarding the underlying structural basis, as well as associated functional consequences. Our analysis of the pr-M junction mutants reveals a suppressive effect of the conserved P3 Glu residue, indicating that a mechanism responsible for down-modulating prM cleavage involves the acidic residue strategically located within the furin consensus sequence. The effect of P3 Glu, however, may be counteracted by the augmenting influences of the P5 Arg and P6 His residues. A basic P5 residue is conserved in mosquito-borne flaviviruses, while the P6 His (or Arg in serotype 4) residue is unique to dengue virus (22). The combination of up- and down-modulatory influences provided by these residues likely presents a mechanism for fine-tuning prM cleavage during virus evolution. Phylogenetic analysis revealed initial circulation of dengue viruses in monkeys and subsequent transmission into humans (17, 48). It would be interesting to determine whether the prM cleavage-modifying residues exist in sylvatic dengue viruses and to correlate the emergence of these residues with diversification and host range expansion.
Crystallographic studies of furin from micse and yeast provide an explanation for the cleavage-suppressive activity of a P3 acidic residue. In the catalytic domain of furin, three pockets accommodate each of the substrate P1, P2, and P4 basic residues through charge-charge interactions (16, 18). On the enzyme surface, residues surrounding the pockets are enriched with acidic side chains (16), which favor basic residues at positions P3, P5, and P6 of the substrate (16, 30, 37) and, in the case of the yeast furin homolog Kex2, select against Asp and Glu at position P3 (18, 38). Based on this model, P5 Arg and P6 His/Arg at the pr-M junction may facilitate cleavage by providing additional favorable interactions, whereas the P3 acidic side chain may weaken the enzyme-substrate interaction or impede docking of P1, P2, and P4 residues into the substrate binding pockets. Whether a P3 discriminatory residue(s) exists in the furin of dengue virus-relevant species, however, is not yet known.
Prior to this study, the consequences of altered prM cleavage were examined by treating flavivirus-infected cells with acidotropic agents. Virus-specific infectivity was reduced 62-fold in West Nile virus (50) to 6- to 8-fold in dengue virus (36). The pr-M junction mutants employed in this study provided a system in which the cleavage of prM could be either up- or down-modulated. Among viable mutants, prM cleavage ranged from 16.2% (for the R202A* mutant) to 90.9% (for the E203A mutant). With a baseline prM cleavage level of 60 to 70% in the parent virus, suppression of cleavage was achieved to a greater extent than enhancement. The discrepancy correlated with a marked reduction in the specific infectivity of the R202A* mutant compared with a modest increase in that of the E203A mutant. In TBEV and West Nile virus, both of which undergo complete prM cleavage, the high levels of suppression of the specific infectivity were associated with a wider window in which prM cleavage could be down-modulated by acidotropic agents (13, 50).
Changes in prM cleavage efficiency are known to affect the proportion of large and small flaviviral particles released from prM+E-transfected cells (2, 23-25). Alterations in the proportion of virus particles were found in this study during the replication of several pr-M junction mutants. How the pr-M junction mutations influence the proportion of extracellular particles is unclear. These mutations may affect the particles being formed in the ER if they are associated with a structural change that favors one particle type over another. Certain prM mutations are known to affect the assembly or stability of the viral particles (34, 51). Recent evidence revealed the proprotein convertase activity in the ER and early Golgi apparatus compartments (41), suggesting that the pr-M junction mutations may affect immature particle-furin interaction soon after budding. Alternatively, the pr-M junction mutations may affect viral particles during transport. In the presence of membrane, mature particles are triggered by acidic pH to alter the conformation and oligomerization of E protein, leading to envelope-membrane fusion (32). As large and small particles differ in their arrangement of E dimers, low-pH-triggered fusion with the membrane may not be equally efficient for these particles. Differential fusion during transport may represent a mechanism responsible for skewing the proportion of particles among the pr-M junction mutants.
An unexpected finding from this study is the indication of a large fraction of prM-containing virions released from dengue virus-infected cells. This was based on the observation that the rate-zonal fraction 4 particles were efficiently precipitated by a prM-specific antibody, while only a small proportion of virions failed to bind to this antibody during sequential immunoprecipitation. Together with another finding that revealed a low proportion of prM compared with E in the fraction 4 particles, these results suggest the presence of prM on a large fraction of particles, many of which should contain less prM than E (i.e., partially mature virions). These prM-containing virions are likely to be noninfectious and their presence may help explain the low specific infectivity of dengue virus preparations detected in this and previous studies (22, 47). Nevertheless, certain prM-specific antibodies are able to enhance dengue virus infection in vitro or protect mice from lethal infection (19, 21, 36), and some of the partially mature virions may be infectious or they may acquire infectivity upon entry into the cells. The existence of infectious dengue virions containing both prM and M remains to be verified in the future.
As in our previous study, extracellular virions of JEVpr/16681 accumulated at levels that were lower than those of the parent virus; this defect was thought to reflect a delay in virus export associated with an enhanced prM cleavage (22). However, the E203A mutant did not exhibit the same defect. The discrepancy indicates that enhanced prM cleavage cannot be responsible for the reduced accumulation of JEVpr/16681 extracellular virions as concluded earlier (22). Another mechanism, likely involving the P8, P10, and P13 basic residues, may instead play a role. It will be crucial to distinguish between the trapping of virions inside infected cells and the augmented binding of virions to the cell surface. JEVpr/16681 differs from the parent virus in a short segment just proximal to the pr-M junction. As this segment is present in the prM molecules, any responsible mechanism likely involves changes in the structure and functioning of prM-containing virions.
Acknowledgments
We thank Thongkham Taya, Aroonrung Jairungsri, Sineenath Santitheerakul, Thippawan Yasanga, Potchong Chotiyarnwong, Jetsada Ketkarn, Nuntaya Punyadee, and Sunisa Butphet for excellent technical assistance. We are grateful to Bunpote Siridechadilok for guidance and helpful discussions in the electron microscopic study and Sutha Sangiambut for critically reviewing the manuscript.
This investigation received financial support from the Thailand-Tropical Diseases Research (T-2) Program (99-2-DEN-03-008 and 02-2-DEN-03-003) and the Medical Biotechnology Unit Network of the National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Bangkok, Thailand. J.J. is supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. program (PHD/0225/2546). P.M. is a senior research scholar supported by the Thailand Research Fund.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 695816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., Y. J. Tao, G. O'Riordain, C. W. Mandl, S. C. Harrison, and F. X. Heinz. 2003. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J. Virol. 7711357-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R., S. Wang, C. Osiowy, and A. C. Issekutz. 1997. Activation of endothelial cells via antibody-enhanced dengue virus infection of peripheral blood monocytes. J. Virol. 714226-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergeron, E., A. Basak, E. Decroly, and N. G. Seidah. 2003. Processing of α4 integrin by the proprotein convertases: histidine at position P6 regulates cleavage. Biochem. J. 373475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blok, J., S. M. McWilliam, H. C. Butler, A. J. Gibbs, G. Weiller, B. L. Herring, A. C. Hemsley, J. G. Aaskov, S. Yoksan, and N. Bhamarapravati. 1992. Comparison of a dengue-2 virus and its candidate vaccine derivative: sequence relationships with the flaviviruses and other viruses. Virology 187573-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cammisa-Parks, H., L. A. Cisar, A. Kane, and V. Stollar. 1992. The complete nucleotide sequence of cell fusing agent (CFA): homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology 189511-524. [DOI] [PubMed] [Google Scholar]
- 7.Chang, G. J. J., A. R. Hunt, D. A. Holmes, T. Springfield, T. S. Chiueh, J. T. Roehrig, and D. J. Gubler. 2003. Enhancing biosynthesis and secretion of premembrane and envelope proteins by the chimeric plasmid of dengue virus type 2 and Japanese encephalitis virus. Virology 306170-180. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree, M. B., R. C. Sang, V. Stollar, L. M. Dunster, and B. R. Miller. 2003. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch. Virol. 1481095-1118. [DOI] [PubMed] [Google Scholar]
- 9.Elshuber, S., S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84183-191. [DOI] [PubMed] [Google Scholar]
- 10.Falconar, A. K. I. 1999. Identification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteins. Arch. Virol. 1442313-2330. [DOI] [PubMed] [Google Scholar]
- 11.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7593-602. [DOI] [PubMed] [Google Scholar]
- 12.He, R.-T., B. L. Innis, A. Nisalak, W. Usawattanakul, S. Wang, S. Kalayanarooj, and R. Anderson. 1995. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J. Med. Virol. 45451-461. [DOI] [PubMed] [Google Scholar]
- 13.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198109-117. [DOI] [PubMed] [Google Scholar]
- 14.Henchal, E. A., J. M. McCown, D. S. Burke, M. C. Seguin, and W. E. Brandt. 1985. Epitope analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. Am. J. Trop. Med. Hyg. 34162-169. [DOI] [PubMed] [Google Scholar]
- 15.Henchal, E. A., M. K. Gentry, J. M. McCown, and W. E. Brandt. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31830-836. [DOI] [PubMed] [Google Scholar]
- 16.Henrich, S., A. Cameron, G. P. Bourenkov, R. Kiefersauer, R. Huber, I. Lindberg, W. Bode, and M. E. Than. 2003. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Biol. 10520-526. [DOI] [PubMed] [Google Scholar]
- 17.Holmes, E. C. 2004. The phylogeography of human viruses. Mol. Ecol. 13745-756. [DOI] [PubMed] [Google Scholar]
- 18.Holyoak, T., M. A. Wilson, T. D. Fenn, C. A. Kettner, G. A. Petsko, R. S. Fuller, and D. Ringe. 2003. 2.4 Å resolution crystal structure of the prototypical hormone-processing protease Kex2 in complex with an Ala-Lys-Arg boronic acid inhibitor. Biochemistry 426709-6718. [DOI] [PubMed] [Google Scholar]
- 19.Huang, K. J., Y. C. Yang, Y. S. Lin, J. H. Huang, H.-S. Liu, T. M. Yeh, S. H. Chen, C. C. Liu, and H. Y. Lei. 2006. The dual-specific binding of dengue virus and target cells for the antibody-dependent enhancement of dengue virus infection. J. Immunol. 1762825-2832. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa, T., and E. Konishi. 2006. Mosquito cells infected with Japanese encephalitis virus release slowly-sedimenting hemagglutinin particles in association with intracellular formation of smooth membrane structures. Microbiol. Immunol. 50211-223. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman, B. M., P. L. Summers, D. R. Dubois, W. H. Cohen, M. K. Gentry, R. L. Timchak, D. S. Burke, and K. H. Eckels. 1989. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 41576-580. [DOI] [PubMed] [Google Scholar]
- 22.Keelapang, P., R. Sriburi, S. Supasa, N. Punyadee, A. Songjaeng, A. Jairungsri, C. Puttikunt, W. Kasinrerk, P. Malasit, and N. Sittisombut. 2004. Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J. Virol. 782367-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojima, A., A. Yasuda, H. Asanuma, T. Ishikawa, A. Takamizawa, K. Yasui, and T. Kurata. 2003. Stable high-producer cell clone expressing virus-like particles of the Japanese encephalitis virus E protein for a second-generation subunit vaccine. J. Virol. 778745-8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konishi, E., and A. Fujii. 2002. Dengue type 2 virus subviral extracellular particles produced by a stably transfected mammalian cell line and their evaluation for a subunit vaccine. Vaccine 201058-1067. [DOI] [PubMed] [Google Scholar]
- 25.Konishi, E., A. Fujii, and P. W. Mason. 2001. Generation and characterization of a mammalian cell line continuously expressing Japanese encephalitis virus subviral particles. J. Virol. 752204-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 28.Lobigs, M., and E. Lee. 2004. Inefficient signalase cleavage promotes efficient nucleocapsid incorporation into budding flavivirus membranes. J. Virol. 78178-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason, P. W., S. Pincus, M. J. Fournier, T. L. Mason, R. E. Shope, and E. Paoletti. 1991. Japanese encephalitis virus-vaccinia recombinants produce particulate forms of the structural membrane proteins and induce high levels of protection against lethal JEV infection. Virology 180294-305. [DOI] [PubMed] [Google Scholar]
- 30.Molloy, S. S., E. D. Anderson, F. Jean, and G. Thomas. 1999. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 928-35. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302248. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 313-22. [DOI] [PubMed] [Google Scholar]
- 33.Murray, J. M., J. G. Aaskov, and P. J. Wright. 1993. Processing of the dengue virus type 2 proteins prM and C-prM. J. Gen. Virol. 74175-182. [DOI] [PubMed] [Google Scholar]
- 34.Pryor, M. J., L. Azzola, P. J. Wright, and A. D. Davidson. 2004. Histidine 39 in the dengue virus type 2 M protein has an important role in virus assembly. J. Gen. Virol. 853627-3636. [DOI] [PubMed] [Google Scholar]
- 35.Purdy, D. E., and G. J. J. Chang. 2005. Secretion of noninfectious dengue virus-like particles and identification of amino acids in the stem region involved in intracellular retention of envelope protein. Virology 333239-250. [DOI] [PubMed] [Google Scholar]
- 36.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174450-458. [DOI] [PubMed] [Google Scholar]
- 37.Rockwell, N. C., D. J. Krysan, T. Komiyama, and R. S. Fuller. 2002. Precursor processing by Kex2/furin proteases. Chem. Rev. 1024525-4548. [DOI] [PubMed] [Google Scholar]
- 38.Rockwell, N. C., G. T. Wang, G. A. Krafft, and R. S. Fuller. 1997. Internally consistent libraries of fluorogenic substrates demonstrate that Kex2 protease specificity is generated by multiple mechanisms. Biochemistry 361912-1917. [DOI] [PubMed] [Google Scholar]
- 39.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246317-328. [DOI] [PubMed] [Google Scholar]
- 40.Russell, P. K., W. E. Brandt, and J. M. Dalrymple. 1980. Chemical and antigenic structure of flaviviruses, p. 503-529. In R. W. Schlesinger (ed.), The togaviruses. Academic Press, New York, NY.
- 41.Salvas, A., S. Benjannet, T. L. Reudelhuber, M. Chretien, and N. G. Seidah. 2005. Evidence for proprotein convertase activity in the endoplasmic reticulum/early Golgi. FEBS Lett. 5795621-5625. [DOI] [PubMed] [Google Scholar]
- 42.Sriburi, R., P. Keelapang, T. Duangchinda, S. Pruksakorn, N. Maneekarn, P. Malasit, and N. Sittisombut. 2001. Construction of infectious dengue 2 virus cDNA clones using high copy number plasmid. J. Virol. Methods 9271-82. [DOI] [PubMed] [Google Scholar]
- 43.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 718475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stollar, V., T. M. Stevens, and R. W. Schlesinger. 1966. Studies on the nature of dengue viruses. II. Characterization of viral RNA and effects of inhibitors of RNA synthesis. Virology 30303-312. [DOI] [PubMed] [Google Scholar]
- 45.Sugrue, R. J., J. Fu, J. Howe, and Y.-C. Chan. 1997. Expression of the dengue virus structural proteins in Pichia pastoris leads to the generation of virus-like particles J. Gen. Virol. 781861-1866. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Schaar, H. M., M. J. Rust, B.-L. Waarts, H. van der Ende-Metselaar, R. J. Kuhn, J. Wilschut, X. Zhuang, and J. M. Smit. 2007. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J. Virol. 8112019-12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, E., H. Ni, R. Xu, A. D. T. Barrett, S. J. Watowich, D. J. Gubler, and S. C. Weaver. 2000. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 743227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, S., R. He, and R. Anderson. 1999. PrM- and cell-binding domains of the dengue virus E protein. J. Virol. 732547-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wengler, G., and G. Wengler. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 632521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshii, K., A. Konno, A. Goto, J. Nio, M. Obara, T. Ueki, D. Hayasaka, T. Mizutani, H. Kariwa, and I. Takashima. 2004. Single point mutation in tick-borne encephalitis virus prM protein induces a reduction of virus particle secretion. J. Gen. Virol. 853049-3058. [DOI] [PubMed] [Google Scholar]
- 52.Yu, I.-M., W. Zhang, H. A. Holdaway, L. Li, V. A. Kostyuchenko, P. R. Chipman, R. J. Kuhn, M. G. Rossmann, and J. Chen. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 3191834-1837. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 222604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, A., G. Webb, X. Zhu, and D. F. Steiner. 1999. Proteolytic processing in the secretory pathway. J. Biol. Chem. 27420745-20748. [DOI] [PubMed] [Google Scholar]