Abstract
Hepatitis C virus (HCV) replicates its genome in a membrane-associated replication complex (RC). Specific membrane alterations, designated membranous webs, represent predominant sites of HCV RNA replication. The principles governing HCV RC and membranous web formation are poorly understood. Here, we used replicons harboring a green fluorescent protein (GFP) insertion in nonstructural protein 5A (NS5A) to study HCV RCs in live cells. Two distinct patterns of NS5A-GFP were observed. (i) Large structures, representing membranous webs, showed restricted motility, were stable over many hours, were partitioned among daughter cells during cell division, and displayed a static internal architecture without detectable exchange of NS5A-GFP. (ii) In contrast, small structures, presumably representing small RCs, showed fast, saltatory movements over long distances. Both populations were associated with endoplasmic reticulum (ER) tubules, but only small RCs showed ER-independent, microtubule (MT)-dependent transport. We suggest that this MT-dependent transport sustains two distinct RC populations, which are both required during the HCV life cycle.
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide (50). Current therapeutic options are limited, which motivates intensive antiviral drug development efforts (8). HCV is a positive-strand RNA virus that belongs to the family Flaviviridae (27, 36, 55). Its genome encodes a polyprotein precursor of about 3,000 amino acids which is co- and posttranslationally processed by cellular and viral proteases to yield the mature structural proteins core, E1, and E2, as well as the nonstructural proteins p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B. NS3 to NS5B form a replicase that is sufficient for autonomous HCV RNA replication (3, 30).
Formation of a membrane-associated replication complex (RC), composed of viral proteins, replicating RNA, altered cellular membranes, and other host factors, is a hallmark of all positive-strand RNA viruses (reviewed in references 32 and 44). Depending on the virus, replication may occur on altered membranes derived from the endoplasmic reticulum (ER), Golgi apparatus, mitochondria, or even lysosomes. The role of membranes in viral RNA synthesis is not well understood. It may include (i) the physical support and organization of the RC (31), (ii) the compartmentalization and local concentration of viral products (24, 48), (iii) tethering of the viral RNA during unwinding, (iv) provision of lipid constituents important for replication, and (v) protection of the viral RNA from 5′-triphosphate- and double-strand RNA-mediated host defenses or RNA interference.
A specific membrane alteration, designated as membranous web, was identified by electron microscopy (EM) as the predominant site of RNA replication in Huh-7 cells harboring subgenomic HCV replicons (13). The membranous web can be induced by NS4B alone and closely resembles the “sponge-like inclusions” previously found by EM in the livers of HCV-infected chimpanzees (11). Membranous webs are composed of vesicles 80 to 180 nm in diameter embedded in a membrane matrix. By immunofluorescence microscopy, membranous webs appear as dotlike structures with diameters of up to several microns (13).
The cellular processes involved in HCV RC and membranous web formation are poorly understood. The NS3-4A complex, NS4B, NS5A, and NS5B associate predominantly with ER membranes when expressed individually (5, 19, 46, 59; reviewed in reference 36). Membrane association of these proteins and formation of the RC most likely occur cotranslationally at the ER. Therefore, and based on additional biochemical and ultrastructural evidence (reviewed in reference 38), it is believed that the membranous web is derived from ER membranes.
We previously described HCV replicons harboring a green fluorescent protein (GFP) insertion in the C-terminal domain of NS5A, and we have shown that the NS5A-GFP fusion protein is incorporated into functional HCV RCs (37). In the present study, we exploited this system to investigate HCV RCs in live cells. Time-lapse microscopy revealed two distinct patterns of NS5A-GFP fluorescence. Large structures, representing membranous webs, showed only limited movement, were stable over many hours, segregated among daughter cells during cell division, and displayed a static internal architecture without detectable exchange of NS5A-GFP. In contrast, small structures, presumably representing small HCV RCs, showed fast, saltatory movement over long distances. Both populations were associated with the ER, but only the small RCs showed microtubule (MT)-dependent active transport. Our results show that HCV RCs are subject to intracellular transport processes and that their fate is tightly linked to the dynamic organization of the ER and MT network. The two populations of HCV RCs identified in live cells may have distinct functions during the viral life cycle.
MATERIALS AND METHODS
Antibodies and constructs.
Mouse monoclonal antibody (MAb) 9E10 against HCV NS5A has been described previously (28). HCV NS3 was detected with MAb 7019P (Maine Biotechnology Services, Portland, ME). Rabbit polyclonal antisera against Sec13 and Sec31 (52, 53) were kindly provided by Wanjin Hong (University of Singapore). Sec31A-specific MAb (clone 32) and calnexin-specific MAb (clone 37) were obtained from BD Transduction Laboratories (Lexington, KY), anti-human golgin-97 MAb CDF4 was from Molecular Probes (Eugene, OR), and anti-alpha-tubulin MAb DM1A was from Sigma (St. Louis, MO). Alexa 546 goat F(ab′)2 anti-mouse immunoglobulin G (IgG) and anti-rabbit IgG (Molecular Probes) were used as secondary antibodies. Plasmid pRSET-B/mRFP1 (6), encoding monomeric red fluorescent protein mRFP1, was kindly provided by Roger Y. Tsien (University of California, San Diego). An ER-targeted mRFP1 version, termed IgLdR1kdel, was generated as follows. The mRFP1 sequence was amplified from pRSET-B/mRFP1 with the primer pair mRFP1/PstI-fwd (5′-TCC CAG GTC CAA CTG CAG GCC TCC TCC GAG GAC GTC ATC AAG-3′) and mRFP1/KDEL-rev1 (5′-CAG CTC GTC CTT TTA GGC GCC GGT GGA GTG GCG-3′). A mouse IgH leader sequence-derived ER targeting signal was amplified from pGFP-ER (kindly provided by Roland Nitschke, University of Freiburg, Freiburg, Germany) with the primers IgHVL-fwd (5′-TCT GAA TTC GAC CTC ACC ATG GGA TGG AGC TGT ATC-3′) and IgHVL/Pst1-rev (5′-CTG CAG TTG GAC CTG GGA GTG GAC ACC-3′). PCR products were fused by overlap extension PCR with the primers IgHVL-fwd and mRFP1/KDEL-rev2 (5′-GAT CTA GAG TCG CGG CCG CTT TAC AGC TCG TCC TTT TAG GCG CCG GTG-3′), followed by digestion with EcoRI and XbaI and ligation into the corresponding restriction sites of pcDNA3.1 (Invitrogen, Carlsbad, CA).
Cell culture.
Stable Huh-7.5 cells autonomously replicating the I/NS5A-GFP-6 subgenomic replicon, Huh-7.5-I/5A-GFP-6, have been described (37). For live cell imaging, cells were grown in no. 1.5 glass-bottom dishes (MatTek, Ashland, MA). Cells were maintained at 37°C in CO2-independent imaging media (Dulbecco modified Eagle medium without phenol red supplemented with 10% fetal calf serum, 100 μg of streptomycin/ml, and 100 U of penicillin/ml and buffered with 20 mM HEPES). Imaging medium was exchanged at least 1 h prior to image acquisition, and from that point on cells were maintained in ambient air at 37°C.
Live cell microscopy.
Wide-field imaging was performed with an Axiovert 100M inverted microscope utilizing a Plan-Apochromat 100× NA 1.4 oil immersion objective (Carl Zeiss, Jena, Germany). The optical configuration used to excite NS5A-GFP included a 488-nm peak from a monochromator (TILL-Photonics, Pleasanton, CA) that was filtered through an HQ470/40 emission filter and reflected off a dichroic Q 495 LP filter. Emitted light was collected through a HQ525/40 filter and collected with a 12-bit cooled charge-coupled device camera with a maximum resolution of 1,392 × 1,040 pixels (Coolsnap HQ; Photometrics, Tucson, AZ). To increase frame rates, 2×2 binning was used, resulting in an effective resolution of 696 × 520 pixels.
Spinning disk confocal imaging was performed with an Axiovert 200 inverted microscope (Carl Zeiss) equipped with a spinning disk confocal head (UltraView; Perkin-Elmer, Boston, MA) Plan-Apochromat 100× NA 1.4 oil immersion objective. For single-color imaging of NS5A-GFP, the 488-nm laser line of an argon/krypton multiline laser (Melles Griot, Carlsbad, CA) was utilized. A 488-nm dichroic mirror was used to reflect the excitation light and to serve as a long-pass filter for the emission light. For dual-color imaging of NS5A-GFP and IgLdR1kdel, monomeric red fluorescent protein was excited along with GFP with the 488-nm laser line. In these experiments, a 488/568/647-nm tricolor dichroic mirror was used as the primary beam splitter. Images of green and red fluorescence were then separated with a dichroic mirror (565dcxr) and filtered with HQ510/30m and HQ650/75, respectively (filter set OI-30-EM in a Dual-View system; Optical Insights LLC, Tucson, AZ). Images were collected with a 14-bit back-thinned cooled charge-coupled device with a resolution of 512 × 512 pixels (C9100-12; Hamamatsu Photonics, Hamamatsu, Japan) without binning.
Time-lapse experiments over several hours were performed on a LSM 510 inverted confocal laser scanning microscope with a Plan-Neofluar 63× NA 1.25 oil immersion objective (Carl Zeiss). For single-color imaging, GFP was excited with a 488-nm laser line, and emission light was collected through a 488-nm dichroic mirror and a 505- to 550-nm band-pass filter. For dual-color imaging, the 488-nm laser line was sufficient to excite monomeric red fluorescent protein. In addition to a primary 488-nm dichroic mirror, a secondary 545-nm beam splitter was used to separate green and red emission signals. Those were further filtered with a 505- to 530-nm band-pass and 560-nm-long pass filter, respectively.
Wide-field and spinning disk confocal setups were controlled with MetaMorph (Universal Imaging, Downingtown, PA) and the laser scanning microscope with the LSM510 software (Carl Zeiss), respectively. For all experimental setups, naive Huh-7.5 cells were used as controls. Similarly, cells expressing either NS5A-GFP or IgLdR1kdel were used to control for cross talk between both fluorophores and the complementary detection channels.
Indirect immunofluorescence staining and colocalization analyses of fixed samples.
Indirect immunofluorescence staining was performed as described previously (59) with the following modifications: cells were fixed with 1% paraformaldehyde in phosphate-buffered saline at 37°C for 40 min, and all steps after permeabilization with 0.05% saponin were performed without detergent. Colocalization analyses of fixed samples were performed on a LSM 510 confocal laser scanning microscope setup with a 100× NA 1.4 Plan-Apochromat oil immersion objective (Carl Zeiss). Acquisition settings were chosen with respect to the Rayleigh criterion and the Nyquist rate. In all colocalization experiments samples with only GFP, mRFP1, or Alexa 543 staining was used to ensure that no cross talk occurred between fluorophores and complementary color channels. Pearson's correlation coefficient (33) was calculated based on raw image data to quantify colocalization. To reduce image noise, a Gaussian filter with a radius of 2.0 pixels was used on raw image data before generation of dot plots.
Data analyses and image presentation.
Selected confocal images were deconvoluted with Huygens Essential (Scientific Volume Imaging, Hilversum, The Netherlands) as indicated. Raw or deconvoluted image data were further processed with ImageJ version 1.37g (1) as specified in the legends, subsequently processed with a gamma function where specified in the legend, and finally linearly scaled to an 8-bit range. In Fig. 1 and 6, the images were mapped to the “Fire” look-up table to further enhance perception of dim structures. Intensity scale bars, which represents a linear intensity ramp covering the entire bit-depth of the acquisition system, were added in several figures to demonstrate the effect of these adjustments. Actual minimum and maximum intensity values of the acquired raw data are indicated by the position of two triangles on the ramp. All intensity adjustments made are reflected by the intensity scale bar which was processed together with the original image.
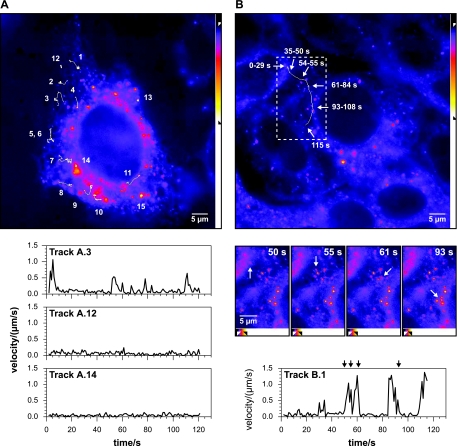
FIG. 1.
Movement patterns of HCV RCs. Live Huh-7.5-I/5A-GFP-6 cells were imaged with an Axiovert 100M inverted microscope with a Pan-Apochromat 100× NA 1.4 objective. Fluorescence was recorded with an exposure time of 750 ms at a rate of 1 Hz over a period of 120 s. Images at the top of panels A and B represent the first images of the recordings included as Videos S1 and S2 in the supplemental material. (A) Time-lapse recording of a single Huh-7.5-I/5A-GFP-6 cell. Representative structures were selected and their movement tracked as specified in Materials and Methods. Tracks 1 to 11 depict small structures that show one or more saltatory movement events. Tracks 12 and 13 are examples of stationary small structures. Tracks 14 and 15 are examples of large structures that show only confined movement patterns. Velocity versus time graphs for three selected structures are shown at the bottom. The small structure of track A.3 (dFWHM = 0.48 μm) shows three saltatory movement events starting at 0, 51, and 110 s. The small structure in track A.12 (dFWHM = 0.39 μm) and the large structure in track A.14 (FWHM dimensions: 2.1 by 1.2 μm) display peak velocities (vpeak) < 0.3 μm/s. (B) Saltatory long distance movement of a small structure. Tracking of a small structure (dFWHM = 0.62 μm) revealed five saltatory movement events which displaced the structure by a distance of 19.1 μm. The locations at various times during the 120-s recording are indicated by white arrows. The durations given at the white arrows indicate the period of time the structure remained stationary. The dashed box indicates the region for which images from various time points are shown in the middle of the panel. Five saltatory movement events can be identified in the velocity versus time graph shown at the bottom. The four images in the middle show the position of the structure immediately before the second movement event at time point t = 50 s, during a short stop before the third event (t = 55 s), immediately after the third event (t = 61 s), and after the fourth event (t = 93 s). The black arrows at the top of the graph reflect these time points. Note that the structure leaves the focal plane during the last frames. All images were processed as reflected by the intensity scale bars on the right or at the bottom of the images, respectively (see Materials and Methods for further details; γ = 0.75). Scale bars, 5 μm.
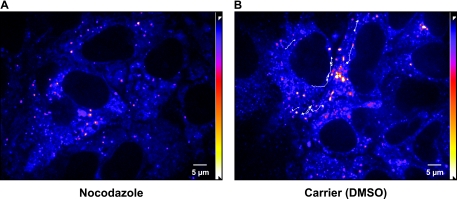
FIG. 6.
Saltatory movement of small HCV RCs is MT dependent. Huh-7.5-I/5A-GFP-6 cells were cultured in the presence of 20 μM nocodazole (A) or carrier alone (dimethyl sulfoxide, 0.1% final concentration) (B). Images were recorded with an acquisition rate of 3.5 Hz over a period of 4.5 min by spinning disk confocal microscopy, and pixel intensities were adjusted as specified in Materials and Methods (γ = 0.9). Long-distance saltatory movement events were tracked as specified in Materials and Methods. Tracks for representative structures, which display saltatory movement events, are shown as a white overlay on top of the images. Images shown in each panel are medium intensity t-projections of the first four images of the recordings to reduce noise. The entire recordings can be found as Videos S4 and S5 in the supplemental material. Cells treated with nocodazole did not display saltatory movement events. Scale bar, 5 μm.
Manuel tracking of RCs was performed as follows: the location of a RC was defined to be its local intensity maximum in images that were processed with a Gaussian blur filter. The approximate location was initially defined with the Manual Tracking plugin version 2.0 for ImageJ from Fabrice P. Cordelières. A self-written ImageJ plugin was used to refine subresolution coordinates from the approximate locations as follows. Images were threefold enlarged with pixel interpolation and processed with a Gaussian blur filter with a radius of 1.5 pixels. Structure positions were then adjusted to represent the closest local intensity maximum, and final positions were marked in the enlarged image and verified by eye.
Inhibition experiments.
The following stock solutions were prepared in dimethyl sulfoxide and stored at −20°C: 30 mM nocodazole, 100 mM nordihydroguaiaretic acid (NDGA), 20 mM brefeldin A, and 1 mg of cytochalasin D/ml. Colchicine was dissolved in water, and stock solutions had a concentration of 20 mM and were stored at 4°C. Stock solutions were diluted in imaging media to obtain the following final concentrations: 20 to 33 μM nocodazole, 30 μM NDGA, 10 μM colchicine, 5 mg of BFA/ml, and 1 μg of cytochalasin D/ml.
RESULTS
Identification of HCV RC populations with distinct dynamic features.
The general distribution and dynamics of NS5A-GFP in live Huh-7.5-I/5A-GFP-6 cells was assessed initially by wide-field microscopy. Typical recordings are shown in Fig. 1 and in Videos S1 and S2 in the supplemental material. Time-lapse recordings revealed two patterns of NS5A-GFP fluorescence. The first pattern consisted of large, polymorphic, brightly fluorescent dot-like structures with a full-width half-maximum (FWHM) diameter up to several microns located predominantly in the perinuclear region (Fig. 1A, structures 14 and 15). As shown previously, these large structures correspond to membranous webs, and the remaining nonstructural proteins are also found in these large structures (13, 37), which was confirmed by the colocalization of NS5A-GFP and NS3 in the present study (Fig. 2). Tracking of these structures revealed a confined movement pattern, which resulted in practically no displacement during the 2-min observation period. Membranous webs, therefore, are of largely stationary character.
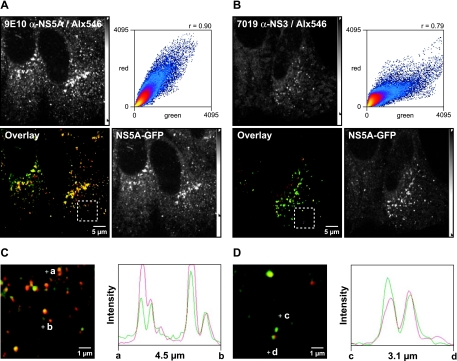
FIG. 2.
Small NS5A-GFP structures contain further HCV replicase components. Huh-7.5-I/5A-GFP-6 cells were fixed, processed for indirect immunofluorescence staining, and analyzed by confocal laser scanning microscopy as described in Materials and Methods. Viral proteins were labeled with the primary mouse MAb 9E10 anti-NS5A or 7019 anti-NS3, which were detected with an Alexa Fluor 546-labeled goat anti-mouse antibody (Alx546). (A) Colocalization between NS5A-GFP autofluorescence (green) and 9E10 anti-NS5A staining (red). Both NS5A specific signals colocalize well, which is also indicated by the high Pearson correlation coefficient (Rc = 0.90). (B) Colocalization between NS5A-GFP (green) and NS3 (red). NS5A and NS3 also colocalize well (Rc = 0.79). (C and D) Enlargement of the dashed box region in panel A (C) and enlargement of the dashed box region in panel C (D) focus on several small structures. Points a and b, as well as points c and d, define two line segments that each cross several structures. Intensity profiles along the line segments, shown on the right of the images, demonstrate that NS5-GFP and NS3 also colocalize in small structures. Scale bars indicate reference distances of 5 and 1 μm, respectively; the length of the line segments between points a and b, as well as points c and d, are depicted underneath the intensity profiles. Intensity adjustments are reflected by the intensity scale bars as detailed in Materials and Methods.
The second pattern consisted of small structures dispersed throughout the cytoplasm. Most small structures appeared as small dots, but irregular shapes and elongated forms were also observed. By confocal microscopy the measured FWHM diameter (dFWHM) of these structures was between 0.3 and 0.4 μm (Fig. 2) and above the theoretical value of a point source (dFWHM = 0.5 λ/√2 NA = 0.13 μm); yet the true size may be significantly smaller, as was shown for the dFWHM of fluorescent beads (7). The small structures displayed two types of motility: a basic type that was similar to the confined movement observed for the large structures and a second type represented by saltatory long distance movement throughout the cytoplasm.
Based on these observations, on the colocalization of NS5A-GFP with other HCV replicase components in both large and small structures (37) (Fig. 2), and on recent data obtained in an infection setting using cell culture-derived HCV (HCVcc) (see Discussion), we propose that there are two distinct forms of HCV RCs that can be distinguished by size and dynamic properties. Quantitative analyses of NS5A-GFP structure size and fluorescence intensity distribution did not allow the definition of two discretely separable populations, since structures in the intermediate parameter range were found as well (data not illustrated). Therefore, an arbitrary cutoff size, measured as the longest dFWHM, was chosen to define the large structures to have a dFWHM ≥ 1 μm and the small structures of having a dFWHM < 0.7 μm. This rigorous cutoff allowed to conclusively distinguish large and small structures also in wide field recordings. Cells with robust HCV replicon expression levels typically contained between 0 to 5 large structures defined in this way. To estimate the total number of RCs per cell, all fluorescence intensity maxima within a cell were counted. The resulting number ranged from 44 to 327 structures per cell (mean = 119, standard deviation = 76, n = 17 cells) and represents the lower limit of the true number of structures, since only structures close to the focal plane yielded sufficient fluorescence signals to be counted.
Large and small structures were tracked to quantify the differences in the movement patterns observed. Both structures showed nondirected, confined movement with peak velocities (vpeak) of <0.3 μm/s. However, for small structures the peak velocities of saltatory long-distance movements were significantly higher. The different movement patterns and velocities are illustrated in Fig. 1. Tracks of selected structures over the entire length of the recording are drawn as white lines. A single cell harboring several large and small structures is depicted in Fig. 1A and Video S1 in the supplemental material. Tracks 14 and 15 represent typical confined movement patterns of large structures. Their peak velocities were 0.13 and 0.22 μm/s, respectively. As stated above, confined movement was also found for the small structures (Fig. 1A, tracks 12 and 13). However, during the 2-min observation period a subset of these structures showed saltatory movement events, with peak velocities ranging from 0.56 to 1.71 μm/s, most commonly 0.8 to 0.9 μm/s. (Fig. 1A, tracks 1 to 11). Figure 1B and Video S2 in the supplemental material show an example of a small structure traversing the cytoplasm in a saltatory fashion over a distance of 19.1 μm, including at least four movement events with a peak velocity of 1.38 μm/s. Similar directional long distance saltatory movements were found in numerous recordings.
Taken together, time-lapse microscopy of live Huh-7.5-I/5A-GFP-6 cells revealed two HCV RC populations with distinct dynamic features.
Membranous webs are stable over several hours and are partitioned among daughter cells during cell division.
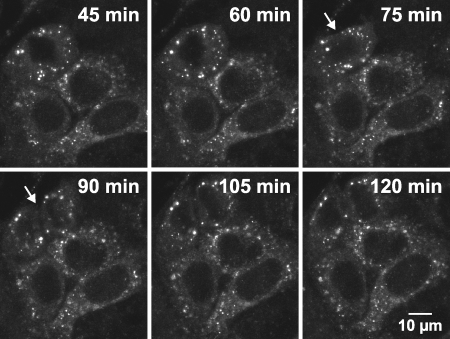
The dynamic behavior of large dot-like structures, representing membranous webs, was examined over 12 to 18 h by confocal laser scanning microscopy. Image stacks comprising the entire volume of Huh-7.5-I/5A-GFP-6 cells were acquired every 6 to 15 min and at reduced spatial resolution to avoid significant bleaching. As shown in Fig. 3 and 4, as well as in Video S3 in the supplemental material, numerous large structures could be monitored over the entire length of the recordings, indicating that membranous webs are stable over at least 12 to 18 h without significant turnover. The motility of membranous webs was generally very limited. Only little and slow displacement was observed and often reflected changes of cell shape and movement of the entire cell. However, for a minor proportion of large structures this slow intracellular displacement eventually resulted in a relocation from one side of the nucleus to the other. Importantly, directed movement patterns or specific sites of membranous web formation or disintegration could not be observed. Time-lapse recordings of several hours also revealed that membranous webs were sustained and partitioned among daughter cells during cell division (Fig. 4).
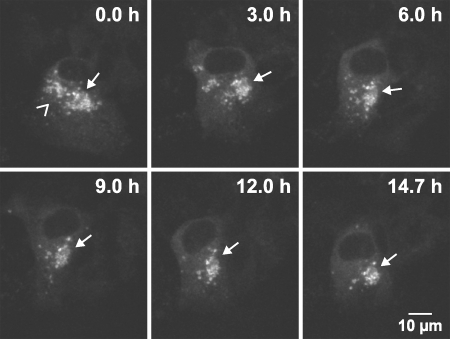
FIG. 3.
Large HCV RCs are stable for many hours. Live Huh-7.5-I/5A-GFP-6 cells were analyzed with an inverted Zeiss LSM 510 confocal laser scanning microscope with a Plan-Apochromat 100× NA 1.4 objective for a time period of t = 14.7 h. An image stack comprising the full cell dimension in the z-direction was recorded every 6 min. Images were processed with a 3 × 3 median filter to reduce noise, for each time point a mean intensity z-projection of the image stack was generated, and pixel intensities were adjusted as specified in Materials and Methods (γ = 0.7). Projections of selected time points are shown as indicated in the captions. The recording can be found as Video S3 in the supplemental material. Two clusters of large structures can be identified in the perinuclear region. The size of the left cluster (angel bracket symbol) is about 7 by 10 μm, and about 15 individual large structures 1 to 2 μm in diameter can be counted in the projection image. The right cluster (arrow) has a size of 11 by 15 μm and contains about 20 large structures. While the left cluster disintegrates throughout the first 6 h of the time lapse, the right cluster can be followed throughout the 14.7-h recording. Of note, the settings used during acquisition and data processing do not allow the detection of the small structures shown in Fig. 1, 2, 6, 7, 8, and 9. Scale bar, 10 μm.
FIG. 4.
Large HCV RCs persist during cell division. Live Huh-7.5-I/5A-GFP-6 cells were analyzed with an inverted Zeiss LSM 510 confocal laser scanning microscope with a Plan-Neofluar 63× NA 1.25 objective for a time period of 10 h. An image stack comprising the full cell dimension in the z-direction was recorded every 15 min. Images were processed with a 3 × 3 median filter to reduce noise, and for each time point mean intensity z-projections were generated. Cell division occurred between 75 and 90 min of the recording. Projections of time points encompassing the cell division (white arrow) are shown, as indicated by the captions. Scale bar, 10 μm.
Taken together, these observations indicate that membranous webs are relatively stable over time and are embedded in a conserved spatial organization within the cell.
Static internal architecture of membranous webs.
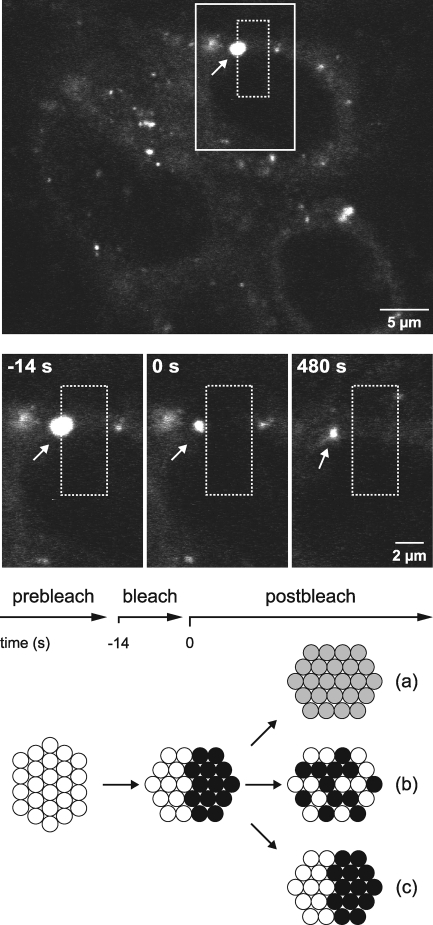
The results presented above suggested that membranous webs are stable structures that persist over at least several hours and do not exhibit significant motility. However, it is conceivable that viral replication sites require a dynamic internal structure that is constantly reorganized to facilitate translation, replication, and packaging of the viral genome. Therefore, fluorescence recovery after photobleaching (FRAP) experiments were performed to investigate the dynamics within membranous webs. In a representative experiment shown in Fig. 5, the right half of a large NS5A-GFP structure was bleached with a laser pulse and, subsequently, bleached and nonbleached halves were monitored for fluorescence recovery and redistribution. There are at least three scenarios which could dictate the outcome of this experiment (see also the diagram in Fig. 5): first, NS5A-GFP could migrate from one vesicle to another within a membranous web; second, NS5A-GFP may be tightly associated with individual vesicles but the vesicles might undergo constant reshuffling within a membranous web; and third, NS5A-GFP and individual vesicles might be tightly fixed over time. As shown in Fig. 5, the latter scenario was observed, since redistribution and recovery of NS5A-GFP did not occur. Of note, a control GFP-tagged ER protein showed fluorescence recovery within a few seconds (data not illustrated), which matches previously described recovery times of NS4B-GFP expressed alone (15). In similar experiments, we also bleached entire membranous webs and did not detect any import of peripheral NS5A-GFP within a 15-min observation period (data not illustrated). Extended observations were precluded by the overall intracellular reorganization of the cells, which made it impossible to precisely track the bleached area.
FIG. 5.
Static internal architecture of large HCV RCs. Live Huh-7.5-I/5A-GFP-6 cells were analyzed with an inverted Zeiss LSM 510 confocal laser scanning microscope with a Plan-Neofluar 63× NA 1.25 objective for a specified time period. Images of cells were recorded every 7 s. At the top, an overview image from the beginning of the experiment is shown. A rectangular region (dashed box) was bleached with a 488-nm laser pulse at time point t = −14 s for a duration of 14 s, as indicated in the time line. This area overlapped with one-half of a large structure representing a membranous web (dFWHM = 1.7 μm; arrow). Enlargements of the continuous line box area for the time point before bleaching (t = −14 s), immediately after bleaching (t = 0 s), and 8 min after bleaching (t = 480 s) are shown in the middle panels. Directly after bleaching, no fluorescence could be detected in the right half of the large structure. The structure slowly migrated to the left during a time period of 8 min, as indicated by the distance to the dashed box, which was drawn at the original coordinates. However, no fluorescence recovery was detected in the bleached half. The nonbleached half, which was the only portion of the large structure that remained visible, sustained its fluorescence intensity and size, which indicates that NS5A-GFP was not redistributed between the bleached and nonbleached halves. Three potential outcomes of this experiment are illustrated at the bottom, as discussed in Results. Note that the outcome illustrated in diagram c was observed in this experiment. Scale bars indicate reference distances of 5 and 2 μm, respectively.
In conclusion, the internal architecture of membranous webs appears relatively static, with limited exchange of viral proteins within and between neighboring RCs.
MT-dependent transport of small HCV RCs.
Saltatory movement of small NS5A-GFP structures suggested a cytoskeleton-dependent transport mechanism. This notion was supported by the observation that independent small structures often traveled along an identical yet invisible track within the cell. In addition, the measured peak velocities would be compatible with MT-dependent transport. To substantiate this idea, Huh-7.5-I/5A-GFP-6 cells were treated with nocodazole, which depolymerizes MTs (9). Preliminary experiments using α-tubulin staining as a control indicated that incubation of Huh-7.5 cells with 20 μM nocodazole for 2 h at 37°C disrupts >95% MTs (data not illustrated). As shown in a representative recording of Huh-7.5-I/5A-GFP-6 cells in Fig. 6 and in Videos S4 and S5 in the supplemental material, no saltatory NS5A-GFP movements could be observed in the presence of 20 μM nocodazole, whereas numerous medium- and long-distance saltatory movement events were observed in cells incubated with carrier alone. Saltatory movements gradually disappeared 30 min after nocodazole addition and reappeared 90 min after nocodazole wash out (data not illustrated). Colchicine, another MT-disrupting agent (21), also abrogated saltatory movement, while cytochalasin D, which blocks actin-dependent transport (17), as well as nordihydroguaiaretic acid and brefeldin A, which among other things interfere with ER-to-Golgi transport (12, 39, 42), had no effect (data not illustrated).
Taken together, these inhibition experiments demonstrate that the saltatory movement of HCV RCs is MT dependent.
HCV RCs are associated with MTs and follow ER tubule movement.
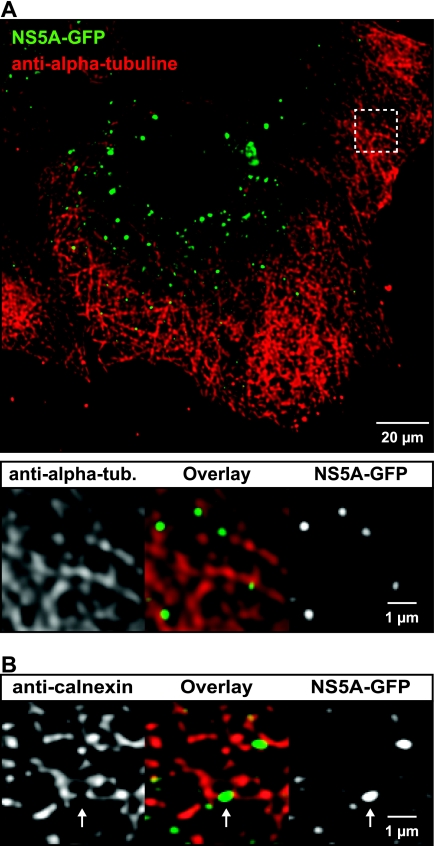
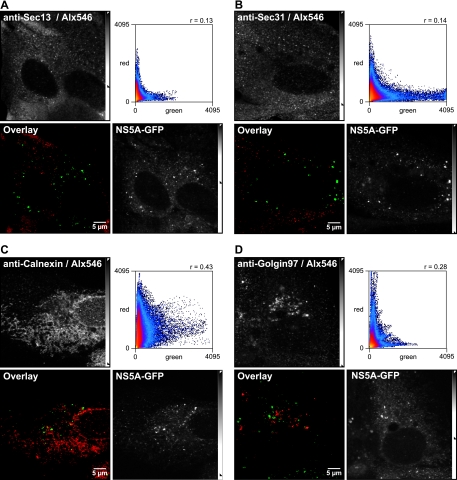
To further investigate the association of HCV RCs with MTs, Huh-7.5-I/5A-GFP-6 cells were processed for indirect immunofluorescence staining of α-tubulin, followed by confocal laser scanning microscopy and image deconvolution. As shown in Fig. 7, NS5A-GFP was found in close association with MTs. Large structures were found entangled with MTs, while the small structures were found almost exclusively side by side with or on top of MTs. Since MTs and the ER are known to associate (56), it is conceivable that the association of small NS5A-GFP structures with MTs is mediated by the ER. Indeed, colocalization studies using calnexin as an ER marker (Fig. 7B) revealed that small NS5A-GFP structures are located at ER tubules. Interestingly, the ER marker was excluded from these structures, resulting in imperfect colocalization at the pixel level. Thus, the ER marker appears to be excluded during formation of the HCV RC, as has been noted previously in the case of other positive-strand RNA viruses (43). Of note, NS5A-GFP did not colocalize with Sec13 and Sec31 (Fig. 8), indicating that small HCV RCs are not associated with ER exit sites (ERES), which are distinct locations at the ER, where COPII-mediated ER-to-Golgi transport is initiated (54).
FIG. 7.
HCV RCs are associated with MTs and the ER. Huh-7.5-I/5A-GFP-6 cells were fixed and processed for indirect immunofluorescence staining as described in Materials and Methods. Primary mouse MAbs against α-tubulin or calnexin were used to detect MTs and the ER, respectively. Stacks of images were acquired by confocal laser scanning microscopy and deconvoluted. (A) Association of HCV RCs with MTs. An overview image is shown at the top. MTs (red) form a dense network in Huh-7.5 cells. NS5A-GFP (green) is found in large and small structures. The area marked by the dashed box is enlarged below and shows that the structures are located in close proximity to MTs. Green intensity settings were adjusted in the enlargement to match the ER signal in this area. (B) Association of HCV RCs with the ER. An enlarged region from a deconvoluted image stack is shown (see Fig. 8C for an overview image). Small NS5A-GFP structures (green) are closely associated with the ER (red) and are found either on top or directly adjacent to ER tubules; however, colocalization on a per pixel basis is hardly detectable. NS5A-GFP structures often bridge a gap in the ER marker signal (arrow), which suggests that they may exclude the ER marker protein. Scale bars indicate reference distances of 20 and 1 μm, respectively.
FIG. 8.
HCV RCs are not associated with ERES. Huh-7.5-I/5A-GFP-6 cells were fixed, processed for indirect immunofluorescence staining, and analyzed by confocal laser scanning microscopy as described in Materials and Methods. Antibodies against (A) Sec13 and Sec31 (B) were used as markers for ERES, and antibodies specific for calnexin (C) and golgin-97 (D) were used as markers for ER and the Golgi apparatus, respectively. Primary antibodies were detected with Alexa Fluor 546-labeled secondary antibodies (Alx546). Interestingly, no colocalization was found between NS5A-GFP and ERES. The dot plots show that image pixels were only positive for either NS5A-GFP or the ER exit marker (Rc = 0.13 or Rc = 0.17). Slightly better colocalization was found with the ER marker calnexin (Rc = 0.43). No colocalization was found with golgin-97 (Rc = 0.28). Scale bars, 5 μm.
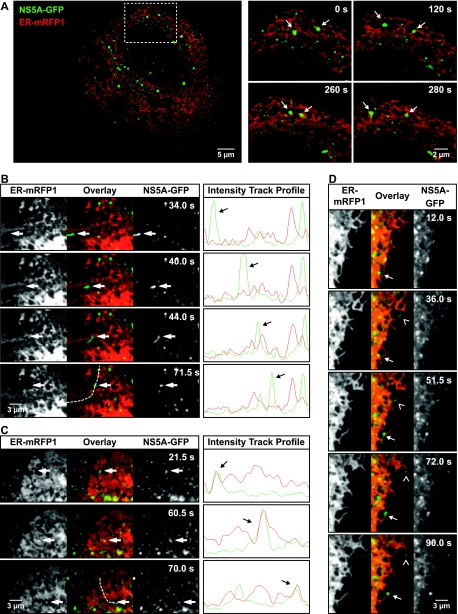
To analyze the relationship between ER tubules and small NS5A-GFP structures, we used a monomeric red fluorescent protein targeted to the ER via an immunoglobulin leader sequence and retained in the ER lumen by a KDEL retention signal (IgLdR1kdel). Live Huh-7.5-I/5A-GFP-6 cells transfected with IgLdR1kdel were analyzed by confocal laser scanning microscopy. As shown in Fig. 9A and Video S6 in the supplemental material, both large and small NS5A-GFP structures comigrated with the ER, reflecting the basic confined movement described above.
FIG. 9.
Association of HCV RCs with the ER in live cells. Huh-7.5-I/5A-GFP-6 cells were examined 36 h after transfection with the red fluorescent ER marker IgLdR1kdel. (A) Confocal laser scanning microscopy was used to acquire green NS5A-GFP and red ER fluorescence simultaneously with a rate of 0.1 Hz. An overview image is shown on the left. The area in the dashed box is shown on the right for four different time points, as indicated by the captions. The entire recording is shown in Video S6 in the supplemental material. Arrows indicate two NS5A-GFP structures that were monitored over time. Both large and small structures follow the highly dynamic ER movement. Of note, fast saltatory movements could not be recorded due to the slow frame rate. (B to D) Spinning disk confocal microscopy in combination with a beam splitter was used to simultaneously detect NS5A-GFP and the red ER marker at high acquisition rates of 2 Hz. Images of the time points indicated are shown. For panels B and C, intensity profiles along the indicated dashed lines are shown on the right for all time points. (B) Fast saltatory movement of small structures was found to occur independent of ER movement. The highlighted structure moves along ER tubules (vpeak = 1.8 μm/s), but no ER movement can be detected in parallel. The entire recording is shown in Video S7 in the supplemental material. (C) Comigration of ER (vpeak = 0.8 μm/s) was observed only in some instances. (D) Small NS5A-GFP structures were also found to follow reorganization or growth of ER tubules either directly at the tip (angel bracket symbol) or at some distance from the tip (arrow). All images were processes as specified in Materials and Methods. For noise reduction, a 3 × 3 median filter was applied to images in panel A. In the images in panels B to D, a Kalman filter was applied. This better preserved spatial separation perpendicular to the direction of movement but resulted in artificial trail formation along the axis of migration. Therefore, the intensity track profiles shown are based on images that were filtered by a spatial Gaussian blur filter only.
Saltatory movement of small HCV RCs does not depend on ER movement.
The findings presented above suggested that HCV RCs are generally linked to the ER. However, low acquisition rates of 0.1 Hz in the confocal laser scanning microscopy experiments prohibited tracking of the fast saltatory movements of small NS5A-GFP structures. Therefore, NS5A-GFP and IgLdR1kdel were recorded simultaneously with a rate of 2 Hz on a spinning disk confocal microscope equipped with a beam splitter to investigate the relationship between HCV RCs and ER motility in greater detail. As shown in Fig. 9B and Video S7 in the supplemental material, fast saltatory long-distance movement of NS5A-GFP was generally not accompanied by movement of the red fluorescent ER marker. Only in rare instances was comigration observed (Fig. 9C). As shown in Fig. 9D, some small NS5A-GFP structure movement accompanied ER tubule growth.
Taken together, our data indicate that movement of small HCV RCs is closely linked to ER motility. However, fast saltatory movements occur in an ER-independent, MT-dependent fashion.
DISCUSSION
Previous EM studies have identified the membranous web as a predominant site of HCV RNA replication (11, 13). Membranous webs are composed of vesicles embedded in a membrane matrix. By light microscopy, they appear as cytoplasmic dotlike structures with a diameter of up to a few microns (13, 37). We have identified here by time-lapse microscopy of Huh-7 cells harboring a functional HCV replicon with a GFP insertion in NS5A at least two populations of putative HCV RCs with different dynamic features: large structures (dFWHM ≥ 1.0 μm), corresponding to membranous webs, with only limited movement and a static internal architecture and small structures (dFWHM < 0.7 μm), which were transported over long distances by MT-dependent active cellular transport processes.
Large structures were found to be relatively stable over time. They did not move significantly within minutes and displayed only a confined movement pattern with vpeak values of <0.3 μm/s. When monitored over 12 to 18 h, these structures displayed only very limited motility, which mostly paralleled physiologic cell movement, and were partitioned among daughter cells during cell division. This idea is supported by our colocalization analyses, which revealed a tight entanglement of these structures with the MT and ER network, and by the characteristic confined movement, which was previously termed “corralled” movement and is indicative of the presence of barriers hindering the movement out of a so called “corralled radius” (49). Small structures were associated with the ER network in a similar fashion but, additionally, they could be rapidly transported throughout the cytoplasm by an ER-independent, MT-dependent process (vpeak = 0.6 to 1.7 μm/s).
We propose that the small NS5A-GFP structures represent small or early HCV RCs based on a number of arguments. First, colocalization studies revealed that these structures contain replicase components. Second, previous fluorescence in situ hybridization analyses revealed viral positive-strand RNA in both large and small structures (13). Third, metabolic labeling of nascent viral RNA by bromouridine also identified small and large structures (membranous webs) as sites of HCV RNA replication (13, 25, 37). Fourth, immunofluorescence staining of Huh-7.5 cells infected with HCVcc revealed that small structures are the morphological correlate of HCV RCs during the initial 12 to 24 h postinfection and that large structures appear only 24 to 48 h postinfection (28; B. Wölk and C. M. Rice, unpublished data). The HCVcc system and GFP-tagged infectious HCV genomes (45) should allow us to investigate whether large and small RCs have distinct functions during different phases of the viral life cycle, including the involvement of lipid droplets in assembly of infectious virus (35; reviewed in reference 58).
The ultrastructural correlate of small structures is unknown. It is likely that they consist of a single vesicle carrying HCV replicase proteins and viral RNA (41). These structures may resemble cellular cargo vesicles, and their interaction with cellular motor proteins may be mediated either directly by one or more HCV nonstructural proteins or by additional cellular trafficking proteins present in the vesicles. Whether the lumina of these vesicles are still connected with the cytoplasm, as was shown for the spherules induced by flock house virus RCs (24), remains unknown. Future EM and cryo-EM studies will be aimed at elucidating the ultrastructure of the small HCV RCs.
FRAP experiments revealed a static internal architecture of membranous webs and, consistent with a recent study (22), limited exchange of NS5A-GFP between RCs and their periphery. This is remarkable, since one might expect that sites of active replication would require a continuous exchange and reorganization of viral material. Indeed, continuous reorganization has recently been shown for replication foci during the S phase of fission yeast nuclei (34). Interestingly, it has been previously reported that impaired NS5A mutants can be rescued with wild-type NS5A in colony formation and transient-transcomplementation assays (2). Still, in these assays rescue may occur during de novo formation of RCs. In contrast, our FRAP experiments measured the exchange of NS5A within large RCs that were already established. Thus, the relatively static internal architecture of HCV RCs we found may actually explain, at least in part, the overall limited capacity to transcomplement HCV nonstructural proteins (2) and the rare occurrence of HCV recombinants (23, 40).
Both large and small RCs were found to be associated with the ER and to follow basic ER movement. However, long-distance saltatory movement was a specific feature of small structures, resulting in displacement over as much as 50% of the cell diameter within a period of 2 min. In this context, it is interesting that a previous study reported a requirement of HCV RNA replication for intact MT and actin polymerization (4). Interestingly, MT-dependent transport of small structures did not depend on parallel ER movement, which suggests that small HCV RCs interact with cellular motor proteins in an ER-independent fashion either directly or via a cellular factor tethered to the RC compartment. This observation also suggests that HCV RCs are not intrinsically tied to native, rapidly moving ER tubules but to an as-yet-undefined ER-derived subcompartment. This is consistent with the lack of FRAP of NS4B-induced membrane-associated foci (15) and a recent motility analysis of NS5A-GFP expressed alone versus NS5A-GFP expressed in the context of a functional HCV replicon (22). Interestingly, while the present report was under review it was shown that NS3-4A and NS5A coprecipitated with and are associated with tubulin and actin, and it was suggested that this might facilitate RC movement along the cytoskeleton (25). However, whether both viral factors are sufficient for RC motility or if further viral proteins or additional cellular factors, such as ER-resident proteins known to mediate MT-dependent motility of the ER (26, 57), are required for HCV RC motility still needs to be defined.
Anterograde transport of cellular proteins from the ER to the Golgi apparatus is MT-dependent and mediated through COPII-coated vesicles which leave the ER through distinct ERES (54). Interestingly, the morphology described for ERES (52, 53) is similar to the scattered appearance of small HCV RCs, and our colocalization analyses looked very similar to those of the ERES protein Sec13 and the ER (16). Thus, it is tempting to speculate that HCV, similar to poliovirus (43), may hijack COPII-coated vesicles to leave the ER through ERES and form RCs. However, our data indicate that the process of HCV RC formation is independent from the classical COPII-dependent anterograde pathway. First, no association of HCV RCs with various ERES marker proteins was found. Second, nocodazole treatment did not lead to the typical redistribution to regions close to the Golgi apparatus (16). Third, no colocalization of Golgi markers and the HCV RC was detected. Moreover, persistence of the large structures during cell division also indicates that HCV RC trafficking is independent of ER-to-Golgi transport, since it has been shown that mitosis induces Golgi apparatus break down and alterations in ER-to-Golgi trafficking (29). Finally, live colocalization analyses showed that after ER-independent long-distance saltatory movement small RCs were always targeted back to ER tubules. Taken together, these observations suggest that HCV RCs do not enter the secretory pathway and maintain a high affinity for the ER throughout their lifetime. This view is supported by our previous findings indicating that HCV RNA replication takes place in a compartment that sustains endoglycosidase H-sensitive glycosylation (20).
Numerous examples of MT-mediated transport of viral components have been reported. However, studies of viral trafficking have thus far focused on the entry, as well as the assembly and release, of viral particles (14). Cellular transport processes involved in viral replication have not been studied extensively. Tobacco mosaic virus encodes the P30 movement protein, which is essential for distribution of viral replication sites within infected cells and mediates cell-to-cell spread through plasmodesmata. Interestingly, this MT-dependent transport is also closely coupled with the viral replicase and the ER (18). Thus, our findings might reflect a general feature of positive-strand RNA viruses.
We initially assumed that small structures would be transported to distinct locations, where they would accumulate to large structures. However, at least with currently available experimental techniques and possibly explained by the expected low frequency of such putative events, transport of small structures into large structures could not be observed on a regular basis. Interestingly, during early poliovirus infection emerging RCs are transported to the perinuclear region in an MT-dependent fashion (10). However, preliminary studies in HCV replicons suggests that MTs are at least not required to maintain morphology and perinuclear location of already existing membranous webs (Wölk and Rice, unpublished).
Membranous web formation may occur independently of MT-dependent transport of small RCs. Indeed, formation of clusters of large structures suggests that HCV RCs may intrinsically coalesce. Thus, formation of large RCs may be favored by an increased local density of small RC structures, and this process may be promoted by random ER movement. In this scenario, one might speculate that saltatory long-distance transport of small RCs represents a mechanism by which HCV ensures the spread of replication sites over the cytoplasm. This could be beneficial for HCV in several ways. Distribution of small viral replication factories might preserve physiologic cell structure and function, consistent with the generally noncytolytic nature of HCV infection. Moreover, independent replication sites within a given cell may increase the chances of highly adapted genomes to establish productive infection and at the same time limit the detrimental effects of deleterious mutations. In this context, a wealth of discrete replication sites has also been reported for other positive-strand RNA viruses, e.g., rotavirus (51). In contrast, during infection with vaccinia virus, whose DNA genome is not prone to a high mutation rate, the formation of large perinuclear replication sites goes along with the loss of small peripheral dots (47). Thus, it may be important for HCV to maintain small RCs. It will be interesting to study viral replication in the light of these hypotheses once the viral and cellular factors required for saltatory movements are identified and can be subjected to selective interference.
Taken together, our results suggest the presence of two populations of HCV RCs with distinct dynamic features and with potentially distinct functions during the viral life cycle. An improved understanding of the molecular and cellular mechanisms involved in HCV RC formation may reveal novel targets for antiviral intervention in the future.
Supplementary Material
Acknowledgments
We gratefully acknowledge Alison North of the Rockefeller University Bio-Imaging Resource Center and Roland Nitschke of the University of Freiburg Live Imaging Center for their extensive support; Kurt Bienz, Denise Egger, Rainer Gosert, Joshua Z. Rappoport, and Jyoti K. Jaiswal for fruitful discussions and helpful suggestions; and Hubert E. Blum and Thomas F. Schulz for generous support and constant encouragement.
This study was supported by the Deutsche Forschungsgemeinschaft (MO 799/1-3 and WO 900/2-1), the Swiss National Science Foundation (3100A0-107831/1), the Swiss Cancer League/Oncosuisse (OCS-01762-08-2005), the Leenaards Foundation, the Greenberg Medical Research Institute, the Starr Foundation, the Ellison Medical Foundation, and the U.S. Public Health Service (CA57973).
Footnotes
Published ahead of print on 20 August 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Bost, A. G., D. Venable, L. Liu, and B. A. Heinz. 2003. Cytoskeletal requirements for hepatitis C virus (HCV) RNA synthesis in the HCV replicon cell culture system. J. Virol. 774401-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass, V., E. Bieck, R. Montserret, B. Wölk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2778130-8139. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 997877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, G., and C. J. R. Sheppard. 2004. Practical limits of resolution in confocal and non-linear microscopy. Microsc. Res. Tech. 6318-22. [DOI] [PubMed] [Google Scholar]
- 8.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436953-960. [DOI] [PubMed] [Google Scholar]
- 9.Döhner, K., and B. Sodeik. 2005. The role of the cytoskeleton during viral infection. Curr. Top. Microbiol. Immunol. 28567-108. [DOI] [PubMed] [Google Scholar]
- 10.Egger, D., and K. Bienz. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86707-718. [DOI] [PubMed] [Google Scholar]
- 11.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 765974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara, T., N. Takami, Y. Misumi, and Y. Ikehara. 1998. Nordihydroguaiaretic acid blocks protein transport in the secretory pathway causing redistribution of Golgi proteins into the endoplasmic reticulum. J. Biol. Chem. 2733068-3075. [DOI] [PubMed] [Google Scholar]
- 13.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124741-754. [DOI] [PubMed] [Google Scholar]
- 15.Gretton, S. N., A. I. Taylor, and J. McLauchlan. 2005. Mobility of the hepatitis C virus NS4B protein on the endoplasmic reticulum membrane and membrane-associated foci. J. Gen. Virol. 861415-1421. [DOI] [PubMed] [Google Scholar]
- 16.Hammond, A. T., and B. S. Glick. 2000. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 113013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinlein, M., B. L. Epel, H. S. Padgett, and R. N. Beachy. 1995. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 2701983-1985. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann, C., A. Sambade, and M. Heinlein. 2007. Plasmodesmata and intercellular transport of viral RNA. Biochem. Soc. Trans. 35142-145. [DOI] [PubMed] [Google Scholar]
- 19.Hügle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 28470-81. [DOI] [PubMed] [Google Scholar]
- 20.Ivashkina, N., B. Wölk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 7613088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob, R., M. Heine, A. Marwan, and H. Y. Naim. 2003. Distinct cytoskeletal tracks direct individual vesicle populations to the apical membrane of epithelial cells. Curr. Biol. 13607-612. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. M., S. N. Gretton, J. McLauchlan, and P. Targett-Adams. 2007. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J. Gen. Virol. 88470-475. [DOI] [PubMed] [Google Scholar]
- 23.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 764034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, C., K. Jeng, K. Machida, and M. M. C. Lai. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 828838-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane, J. D., and V. J. Allan. 1999. Microtubule-based endoplasmic reticulum motility in Xenopus laevis: activation of membrane-associated kinesin during development. Mol. Biol. Cell 101909-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., H. J. Thiel, and C. M. Rices. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 28.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wölk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 29.Lippincott-Schwartz, J., T. H. Roberts, and K. Hirschberg. 2000. Secretory protein trafficking and organelle dynamics in living cells. Annu. Rev. Cell Dev. Biol. 16557-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 31.Lyle, J. M., A. Clewell, K. Richmond, O. C. Richards, D. A. Hope, S. C. Schultz, and K. Kirkegaard. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J. Biol. Chem. 27716324-16331. [DOI] [PubMed] [Google Scholar]
- 32.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manders, E. M., J. Stap, G. J. Brakenhoff, R. van Driel, and J. A. Aten. 1992. Dynamics of three-dimensional replication patterns during the S-phase, analyzed by double labeling of DNA and confocal microscopy. J. Cell Sci. 103(Pt. 3)857-862. [DOI] [PubMed] [Google Scholar]
- 34.Meister, P., A. Taddei, A. Ponti, G. Baldacci, and S. M. Gasser. 2007. Replication foci dynamics: replication patterns are modulated by S-phase checkpoint kinases in fission yeast. EMBO J. 261315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 36.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5453-463. [DOI] [PubMed] [Google Scholar]
- 37.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 787400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moradpour, D., R. Gosert, D. Egger, F. Penin, H. E. Blum, and K. Bienz. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antivir. Res. 60103-109. [DOI] [PubMed] [Google Scholar]
- 39.Nehls, S., E. L. Snapp, N. B. Cole, K. J. Zaal, A. K. Kenworthy, T. H. Roberts, J. Ellenberg, J. F. Presley, E. Siggia, and J. Lippincott-Schwartz. 2000. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2288-295. [DOI] [PubMed] [Google Scholar]
- 40.Noppornpanth, S., T. X. Lien, Y. Poovorawan, S. L. Smits, A. D. M. E. Osterhaus, and B. L. Haagmans. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 807569-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 7913594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richards, A. A., E. Stang, R. Pepperkok, and R. G. Parton. 2002. Inhibitors of COP-mediated transport and cholera toxin action inhibit simian virus 40 infection. Mol. Biol. Cell 131750-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 759808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 814591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt-Mende, J., E. Bieck, T. Hügle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 27644052-44063. [DOI] [PubMed] [Google Scholar]
- 47.Schramm, B., C. A. M. de Haan, J. Young, L. Doglio, S. Schleich, C. Reese, A. V. Popov, W. Steffen, T. Schroer, and J. K. Locker. 2006. Vaccinia-virus-induced cellular contractility facilitates the subcellular localization of the viral replication sites. Traffic 71352-1367. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9505-514. [DOI] [PubMed] [Google Scholar]
- 49.Shav-Tal, Y., X. Darzacq, S. M. Shenoy, D. Fusco, S. M. Janicki, D. L. Spector, and R. H. Singer. 2004. Dynamics of single mRNPs in nuclei of living cells. Science 3041797-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5558-567. [DOI] [PubMed] [Google Scholar]
- 51.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 787763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang, B. L., F. Peter, J. Krijnse-Locker, S. H. Low, G. Griffiths, and W. Hong. 1997. The mammalian homolog of yeast Sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol. Cell. Biol. 17256-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang, B. L., T. Zhang, D. Y. Low, E. T. Wong, H. Horstmann, and W. Hong. 2000. Mammalian homologues of yeast sec31p. An ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J. Biol. Chem. 27513597-13604. [DOI] [PubMed] [Google Scholar]
- 54.Tang, B. L., Y. Wang, Y. S. Ong, and W. Hong. 2005. COPII and exit from the endoplasmic reticulum. Biochim. Biophys. Acta 1744293-303. [DOI] [PubMed] [Google Scholar]
- 55.Tellinghuisen, T. L., M. J. Evans, T. von Hahn, S. You, and C. M. Rice. 2007. Studying hepatitis C virus: making the best of a bad virus. J. Virol. 818853-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terasaki, M., L. B. Chen, and K. Fujiwara. 1986. Microtubules and the endoplasmic reticulum are highly interdependent structures. J. Cell Biol. 1031557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vedrenne, C., and H. Hauri. 2006. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic 7639-646. [DOI] [PubMed] [Google Scholar]
- 58.Wölk, B., and H. Wedemeyer. 2008. Hepatitis C virus living off the fat of the land. Hepatology 47343-345. [DOI] [PubMed] [Google Scholar]
- 59.Wölk, B., D. Sansonno, H. G. Kräusslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 742293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.