Abstract
Intrahepatic virus-specific CD8+ T cells are thought to be important for the control of hepatitis C virus (HCV) infection, yet the precise kinetics for the expansion of epitope-specific T cells over the course of infection are difficult to determine with currently available methods. We used a real-time PCR assay to measure the frequency of clonotypic HCV-specific CD8+ T cells in peripheral blood and snap-frozen liver biopsy specimens of two chimpanzees (Pan troglodytes) with previously resolved HCV infection who were rechallenged with HCV. In response to rechallenge, the magnitude of each clonotypic response was 10-fold higher in the liver than in the blood, and the peak clonotype frequency was concurrent with the peak viral load. The higher frequency of HCV-specific clonotypes in the liver than in peripheral blood was maintained for at least 3 months after the clearance of viremia. After antibody-mediated CD8+ T-cell depletion and another viral challenge, the rebound of these clonotypes was seen prior to an appreciable reconstitution of CD8+ T-cell values and, again, at higher frequencies in the liver than in peripheral blood. These data demonstrate the importance of intrahepatic virus-specific CD8+ T cells for the clearance of infection and the rapid kinetics of expansion after virus challenge.
The results of several studies have demonstrated the importance of cellular immune responses for the clearance of hepatitis C virus (HCV) infection (3, 5, 18, 19, 20, 22, 24, 25). The chimpanzee (Pan troglodytes) is the only available animal model for hepatitis C infection, and the results of a previous study demonstrated that in animals with resolved infection, the depletion of CD8+ T cells caused delayed clearance of the virus after rechallenge with homologous virus. The clearance of infection coincided with the return of small numbers of CD8+ T cells in the peripheral blood and liver (18) and, more importantly, with the appearance of epitope-specific CD8+ T cells in the liver. Functional HCV antigen-specific CD8+ T-cell responses were detected after mitogenic (anti-CD3 antibody) expansion of mononuclear cells recovered from the liver. Without this in vitro expansion step, too few mononuclear cells (approximately 100,000) are recovered from a 1- to 2-cm-long needle biopsy specimen to directly assess CD8+ or CD4+ T-cell frequencies by enzyme-linked immunospot assay (10). That study was performed in real-time on freshly isolated liver mononuclear cells from animals for whom the dominant immune responses were previously characterized (18). From the results of the experiments presented here, we demonstrate the ability to simultaneously track the frequencies of individual T cells from snap-frozen liver biopsy specimens and peripheral blood mononuclear cells (PBMC) and provide a detailed kinetic study of the return of HCV-specific CD8+ T cells after antibody-mediated T-cell depletion.
The construction of major histocompatibility complex (MHC) class I tetramers has facilitated the direct visualization of HCV epitope-specific T-cell responses in liver tissue of animals and humans (1). The chief advantage of tetramers is the ability to directly stain peripheral blood lymphocytes or, in the case of HCV infection, lymphocytes directly isolated from the liver. However, with the current technology, the cognate epitope must be identified, and the reagent must be available at the time the cells are obtained. Limited liver sample availability has made it difficult to track immune response in the liver, especially in longitudinal studies. Our goal was to use a combination of immunological and molecular techniques to identify HCV-specific CD8 T-cell responses and to quantify and track the frequency of individual-epitope-specific T cells over the course of CD8+ T-cell depletion and HCV infection of chimpanzees.
The specificity of T cells for their cognate peptides is conferred by the CDR3 region of the T-cell receptor (TCR), which in the beta chain is encoded by recombination of the variable, diversity, and joining regions of TCR genes. Additional diversity is conferred by random insertion of nucleotides between these regions. PCR primers (6, 11) or labeled oligonucleotide probes (9) corresponding to the TCR beta chain CDR3 region confer exquisite sensitivity for the detection of individual TCR clonotypes. In a previous study, we identified CD8+ HCV-specific TCR clonotypes in animals that resolved HCV infection (13). From the results of the experiments presented here, we demonstrate the ability to simultaneously track the frequencies of individual T cells from snap-frozen liver biopsy specimens and PBMC and provide a detailed kinetic study of the return of HCV-specific CD8+ T cells after antibody-mediated T-cell depletion. We designed PCR primers specific for dominant clonotypes detected in chimpanzees with resolved HCV infection. Using real-time PCR, we were able to retrospectively track the frequencies of these clonotypes in the liver and PBMC from RNA samples extracted at several time points. This provided a detailed kinetic analysis of TCR clonotype frequency after HCV rechallenge in animals with previously resolved infection and after CD8+ T-cell depletion and subsequent HCV challenge in these same animals. It also provided the relative frequencies of virus-specific T cells in different anatomic compartments.
MATERIALS AND METHODS
HCV infections.
The chimpanzees were maintained under standard conditions for humane care and in compliance with NIH guidelines at the New Iberia Research Center, Lafayette, LA. They were infected intravenously with 100 chimpanzee infectious doses of HCV-1/910 stock (2) for the first time in 1994 and then rechallenged with the same inoculum size and strain 7 years later in 2001. Six months after clearance of the second infection, three doses of the anti-CD8 antibody cM-T807 (17, 23) were administered intravenously at days −14, −11, and −7 prior to the third HCV challenge, each at a dose of 2.5 mg/kg of body weight. HCV challenge was with 100 chimpanzee infectious doses of HCV-1/910.
Isolation of lymphocytes from blood and liver.
Human and chimp blood samples were collected in EDTA tubes, and PBMCs were isolated by using a Ficoll density gradient according to the standard protocol. Needle biopsy liver samples were stored in 1.0 ml of RNAlater reagent (Ambion, Austin, TX) at −80°C until RNA was isolated with RNA Stat-60 (Tel Test, Inc., TX) according to the manufacturer's instructions.
Identification of optimal peptide epitopes.
T-cell clones were screened against nine pools containing 30 to 40 peptides which were prepared from a set of 301 overlapping peptides (Mimotypes Pty.) encompassing the entire HCV-1 polyprotein according to the sequence published by Choo et al. (2). The peptides were 20 amino acids long and overlapped by 10 residues. Recognized peptides were mapped to optimal sequences that were used to synthesize Patr class I tetramers (18).
Flow cytometry.
The antibodies used included anti-human CD3, CD4, and CD8, all purchased from BD Biosciences. The Patr B*2301/E2445 (HKFNSSGCPERL) and Patr A*0701/p7756 (AASLAGTHGLVSFL) tetramers were synthesized at the NIH tetramer core facility, Emory University, Atlanta, GA.
Sorting tet+ cells.
Tetramer-positive (tet+) cells were sorted directly from cryopreserved PBMC (week 2 postinfection). Phycoerythrin-labeled tetramer diluted 1:100 was added to cells and incubated in the dark for 15 min at room temperature, followed by surface labeling with anti-CD3, anti-CD4, and anti-CD8 antibodies (BD Biosciences, San Diego, CA) for 30 min on ice. After washing with fluorescence-activated cell sorter buffer (phosphate-buffered saline, 2% fetal calf serum, 0.1% NaN3), viability dye (Viaprobe; BD Biosciences, Pharmingen, San Diego, CA) was added, and CD8+ tet+ cells were sorted to more than 95% purity.
TCR sequencing.
RNA was isolated from sorted tetramer-specific cells (at least 5,000 cells from each sort) and an equal number of CD8+ tetramer-negative (tet−) cells with RNA Stat-60 (Teltest, Inc., TX). Anchored reverse transcription-PCR was performed by using a modified version of the SMART (switching mechanism at 5′ end of RNA transcript) procedure (Clontech, Mountain View, CA) and a TCR beta constant region 3′ primer (5′-ATT CCT TTC TCT TGA CCA TG-3′). cDNA amplification was performed using the TCR constant region-based primer (5′-TTC ACC CAC CAG CTC AGC TC-3′) and 10× universal primer A mix (Clontech, Mountain View, CA). PCR products of 600 to 700 bp were gel purified (Qiagen, Valencia, CA), ligated into the pCR-II vector (Invitrogen, Carlsbad, CA), and used to transform chemically competent Escherichia coli TOP10 cells (Invitrogen, Carlsbad, CA). Bacterial colonies were selected and screened for the presence of the insert by using PCR with M13 primers. DNA was sequenced with a Taq dye deoxy Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and capillary electrophoresis on a Prism automated sequencer (Applied Biosystems, Foster City, CA).
Real-time PCR.
PBMC were thawed, counted, and lysed in RNA Stat-60. Liver biopsy fragments were placed directly in RNAlater (Ambion, Austin, TX) solution and stored in liquid nitrogen until used. Biopsy tissue removed from RNAlater was homogenized in RNA Stat-60 by using a syringe and fine needle. cDNA was synthesized with a 3′ constant region primer appropriately diluted in Tricine-EDTA solution. Real-time quantitative PCR was performed with clonotype-specific primers and probes. Probes were labeled with 6′-carboxyfluorescein and 6-carboxytetramethylrhodamine quencher (Applied Biosystems, Foster City, CA). Unique plasmid clones were used as standards for each clonotype and serially diluted to generate a standard curve. The frequency of a particular clonotype was calculated by dividing the clonotype copy number by the total T-cell copy number (based on total constant region amplification). Samples were run in triplicate. Negative controls for each experiment included a control without template, cDNA derived from human PBMC, and cDNA from an unrelated chimpanzee. When experiments were run at different times or in several plates, a high and low control were also included as a quality control measure. Standard curves of the results from several different experiments were compared (data not shown) to ensure that the plasmid standards did not degrade over time. Full details of all primers, fluorescent probes, components, and cycling temperatures are available upon request.
Statistical analysis.
Real-time PCRs for the clonotype analysis and constant region were done in triplicate. The standard error was calculated according to the formula SQRT{[SD(x)2/mean(x)2 + SD(y)2/mean(y)2 − 2 × correlation(x,y) × SD(x) × SD(y)/[mean(x) × mean(y)]]/n}, where x and y are quantities of clonotype and total TCR, respectively; n is the number of replicates for each sample; SQRT is square root; and SD is standard deviation.
RESULTS
TCR sequence analysis of p7756-specific CD8 T cells.
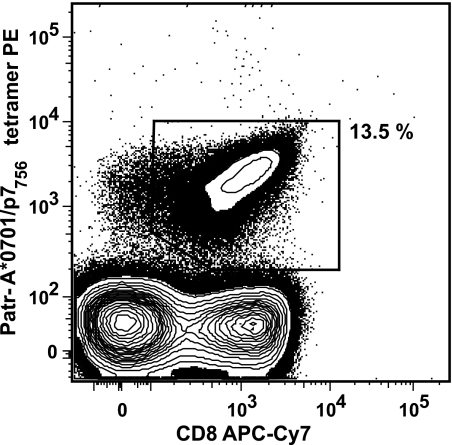
The peak magnitude of the immunodominant immune response in chimpanzee CB0556 in peripheral blood occurred 2 weeks after HCV rechallenge when the tetramer frequency was 13.5% of CD8+ T cells (Fig. 1). Cryopreserved cells from this time point were stained with A*0701/P7756-phycoerythrin tetramer and directly sorted for TCR analysis. By day 90 after infection, the tetramer frequency declined to 0.01% and was too low for direct sorting. For this time point, cells were expanded in vitro with the relevant epitopic peptide, as previously described (13, 18), and sorted. In each case, an identical number of CD8+ tet− cells were sorted as a control.
FIG. 1.
Tetramer frequency of p7756-specific T cells as the percentage of CD8+ T cells in animal CB0556 2 weeks after rechallenge infection. PE, phycoerythrin; APC, allophycocyanin.
Analysis of the TCR sequences of A*0701-specific CD8+ T cells directly sorted from PBMC of animal CB0556 sampled on day 14 postinfection (n = 58 sequences) revealed three dominant clonotypes that made up over 90% of the TCR repertoire, designated TRBV27-PEG (63%), TRBV6-3-QEST (22%), and TRBV27-ITR (6%), with four additional clonotypes appearing as single sequences (Fig. 2A). There were two dominant clonotypes in the in vitro-expanded population, representing 37% of the analyzed sequences (Fig. 2C). The two dominant clonotypes at day 14 were detectable in the expanded cells from week 13 but were present at low frequencies (Fig. 2A and C). The dominant clonotype seen in the expanded cells (TRBV27-ITR) was present in the day 14 sequences and represented 24% of the sequences of the expanded cells. These differences highlight either different frequencies of clonotypes or different in vitro expansion potentials of clonotypes at different times postinfection. Despite the presence of a dominant clonotype (TRBV4-1-TTVN) in tet− CD8+ T cells in each case, the remaining CD8+ TCR repertoire was diverse, and sequences did not match the TCR sequences from the tet+ populations (Fig. 2B). This dominant clonotypic expansion is similar to what we observed after HCV rechallenge of a second animal, CB0572, in which the E2445 epitope was recognized. In that case, 80% of the TCR repertoire was dominated by four clonotypes (13).
FIG. 2.
TCR beta repertoire of chimpanzee CB0556. TCR beta sequences of A*0701/p7756 tetramer-specific CD8+ T cells from PBMC of animal CB0556 sampled 2 weeks postinfection (A), TCR repertoire of A*0701/p7756 tet− CD8+ T cells 2 weeks postinfection (B), and TCR beta sequences of A*0701/p7756 tetramer-specific CD8+ T cells obtained after two rounds of in vitro expansion of PBMC of animal CB0556 sampled 13 weeks postinfection (C). The purity of the sorted T cells was at least 98%. Clonotypes in bold letters were selected for T-cell tracking by real-time PCR. *, clonotypes also found in direct-sort TCR repertoire. TRBV, T-cell receptor beta chain variable region; TRBJ, TCR beta chain joining region.
Real-time PCR quantitation of clonotype frequency in peripheral blood and liver.
The availability of a CD8+ HCV epitope-specific clone from animal CB0572 allowed us to evaluate the sensitivity of a TCR clonotype-specific real-time PCR assay. Primers and a probe for the variable region and CDR3 region of the TCR beta chain were used to detect E2445 epitope-specific T-cell clone 5A, a CD8+ T-cell clone derived from the liver after the primary HCV infection, which was spiked into aliquots of 100,000 normal human PBMC at frequencies ranging from 10 to 0.001% (with 100% 5A clone and 100% PBMC used as positive and negative controls, respectively). The curve generated from this experiment was linear over 6 orders of magnitude and sensitive down to a frequency of 0.001%, representing only one 5A cell spiked into 100,000 PBMC (data not shown). This is similar to the sensitivity of real-time PCR detection of T-cell clonotypes described in other studies (7, 12).
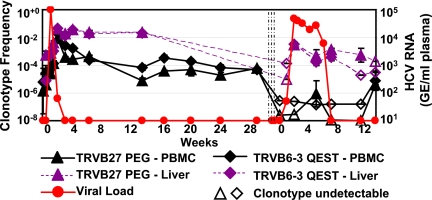
In addition to the PCR primer set specific for CB0572, we designed two primer sets for the dominant clonotypes within the tet+ population of CB0556 (Fig. 2). The baseline memory frequencies (before HCV reinfection) of TCR clonotypes in the peripheral blood for the p7756 Patr A*0701 epitope were 0.0008% (TRBV27-PEG) and 0.0002% (TRBV6-3-QEST) of total T cells. In the liver, the corresponding frequency of TRBV27-PEG was 0.07% and of TRBV6-3-QEST was 0.05%. Peak viremia reached 100,000 copies/ml by day 7, at which time the blood frequencies of these two clones were 0.005% (TRBV27-PEG) and 0.002% (TRBV6-3-QEST) and the liver frequencies were 0.1% (TRBV27-PEG) and 0.4% (TRBV6-3-QEST). The enhanced kinetics of the response in the liver also coincided with higher peak clonotype frequencies in the liver for each T-cell clone. By 2 weeks after rechallenge, virus was almost cleared (viral load of 64), and by this time the clonotype frequencies in peripheral blood had increased approximately 1,000-fold, to 2.09% for TRBV27-PEG and 0.8% for TRBV6-3-QEST. In the liver, the peak responses were 4.5% for TRBV27-PEG and 4% for TRBV6-3-QEST (Fig. 3).
FIG. 3.
Clonotype frequency of two p7756-770 peptide-specific (AASLAGTHGLVSFL; restricted by Patr A*0701) and dominant TCR clonotypes measured using real-time PCR assay of PBMC (solid lines) and liver biopsy samples (dotted lines) of HCV-infected chimpanzee (CB0556). Each point indicates the mean of triplicate values and its standard error. Open symbols indicate undetectable levels of the specific clonotype, and the minimal detectable frequency was calculated by using the lowest limit of detection on the standard curve as the nominator. The sensitivity of detection of each clonotype was at least 10 copies as measured by using molecularly cloned TCRs. The three vertical lines indicate the administration of three doses of anti-CD8 antibody. Week 0 indicates the times of HCV infection. GE, genome equivalents.
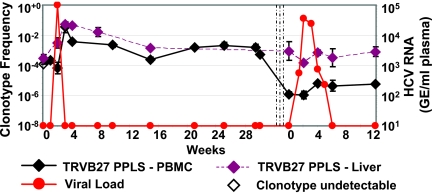
TCR tracking for clone 5A in animal CB0572 (E2445 Patr B*2401 tet+) showed similar kinetics. The clonotype frequency in the blood was 0.02% prior to rechallenge. Two weeks after reinfection, when viral load peaked at 100,000 copies/ml, the clonotype frequency increased to a maximum of 3.65% of T cells in blood, and this was followed by a slow decrease in the clonotype frequency to 0.1% from day 14 to day 94 postinfection. In the liver, the 5A frequency increased from 0.03% to 5.11% by day 14 and remained high through day 21 (4.41%) despite the clearance of viremia (Fig. 4). In each animal, the frequency of clonotypes as measured by real-time PCR tracked closely with tetramer frequencies on directly stained T cells isolated from the liver (18). T-cell clonotypes expanded more rapidly in the liver and persisted at higher frequencies than in peripheral blood after plasma virus was no longer detectable.
FIG. 4.
Clonotype frequency of E2445-457 peptide-specific (HKFNSSGCPERL; restricted by Patr B*2301) and dominant TCR clonotype measured using real-time PCR assay in PBMC (solid line) and liver biopsy samples (dotted line) of HCV-infected chimpanzee (CB0572). Each point indicates the mean of triplicate values and its standard error. Open symbols indicate undetectable levels of the specific clonotype, and the minimal detectable frequency is calculated by using the lowest limit of detection on the standard curve as the nominator. The sensitivity of detection of this clonotype is at least 10 copies as measured by using the molecularly cloned TCR. The three vertical lines indicate the administration of three doses of anti-CD8 antibody. Week 0 indicates the times of HCV infection. GE, genome equivalents.
Tracking TCR clonotypes in PBMC and liver after CD8 depletion and third infection.
In our prior study of these animals, after CD8+ T-cell depletion the clearance of virus was associated with the return of CD8+ T cells in the periphery, as well as with the ability to expand HCV-specific T cells from the liver (18). We next evaluated the frequency of these TCR clonotypes in the liver and peripheral blood after CD8+ T-cell depletion and subsequent infection. Prior to reinfection, both clonotypes in animal CB0556 were below the limits of detection in both PBMC (<0.0003%) and the liver (<0.15%) (Fig. 3). By day 7 after infection, both clonotypes were detectable in the liver at frequencies of 0.14% (TRBV27-PEG) and 0.1% (TRBV6-3-QEST), substantially lower than the frequencies observed in the absence of CD8+ T-cell depletion, yet were not detectable in peripheral blood. From day 7 through day 42 postinfection, the frequency of each clonotype declined in the liver to frequencies in the 0.01% range, and this coincided with the clearance of virus after the third challenge. Throughout this time, clonotypes were either absent or barely detectable in the peripheral blood in the case of TRBV6-3-QEST (0.01%) at a single time point (Fig. 3).
We observed a similar pattern of TCR clonotype frequency in chimpanzee CB0572. After CD8 depletion, the 5A clonotype was still detectable at very low levels in the liver (0.09%) and blood (0.00016%), with a striking 3-log difference in frequencies of the clonotype between the two compartments (Fig. 4). Perhaps reflecting the efficiency of CD8+ T-cell depletion, there were few T cells in the liver on day 14, and the clonotype frequency was <0.015%. However, the clonotype was detectable at days 28 (0.07%) and 42 (0.03%), by which time virus was cleared (Fig. 4). Throughout this time, the clonotype was present at a persistently low level in peripheral blood. Therefore, in each animal, HCV clonotypes persisted in the liver at higher frequencies than in peripheral blood even after nearly complete CD8+ T-cell depletion.
DISCUSSION
The results of immunohistochemical studies have confirmed the presence of activated CD8 T cells in the liver, and the results of flow cytometric studies have shown a higher frequency of HCV-specific CD8 T cells in the liver than in blood of chronically infected subjects (8, 10, 14). Most studies evaluating intrahepatic HCV-specific T cells rely on ex vivo expansion with either recombinant HCV proteins or HCV peptides or on nonspecific stimulation using phytohemagglutinin or anti-T-cell antibodies prior to analysis (14). An alternate way to directly analyze unexpanded cells is through the use of MHC class I tetramers (8). Although our group has used these reagents with some success (18), this method requires the use of the majority of freshly isolated cells available from a liver biopsy specimen, and the low frequencies of these cells after virus is cleared are typically below the limit of detection by flow cytometry. Compounding these limitations are the lack of availability of all MHC class I alleles, the requirement that the epitopes to be evaluated need to be precisely known in order to synthesize tetramers, and the requirement that the flow analysis take place in real time on small numbers of freshly isolated cells. In this study, we used real-time PCR to perform a longitudinal analysis of T-cell clonotype expansion on frozen needle biopsy liver samples.
In both animals, the rapid resolution of viremia after rechallenge temporally coincided with massive expansion of the dominant memory T-cell clonotype, highlighting the importance of memory CD8+ T cells to the outcome of infection. While immunological memory conferred by the spontaneous resolution of acute hepatitis C does not protect against reinfection, it does significantly reduce the time of viremia upon reexposure. Here we show that despite the effective depletion of CD8+ T cells, each animal was able to clear virus, albeit at a lower rate. In a previous study (18), we documented that viral clearance was associated with the return of a detectable number of CD8+ T cells in the periphery and with the ability to expand HCV-specific T cells from the liver and peripheral blood. Here we show that at the clonotype level, the number of HCV-specific T cells remaining after depletion was several logs lower in peripheral blood than in the liver. These cells persisted in the liver after rechallenge, and in the case of animal CB0556, the slower clearance of virus was associated with a smaller peak frequency of these clonotypes, followed by a gradual decay in frequency.
The inability of these clonotypes to expand robustly after HCV challenge in the setting of CD8+ depletion may be due to the persisting effect of the depleting antibody in these animals. In animal CB0556, the predepletion CD8+ T-cell number was 1,328 cells/mm3. Even though this animal cleared virus by day 42, the absolute number of CD8+ T cells was only 5 cells/mm3 by day 42, and 12 months after the depletion, CD8+ T cells in the periphery had only recovered to 634 cells/mm3. For animal CB0572, the predepletion CD8+ T-cell number was 1,073 cells/mm3. This animal had a CD8+ T-cell nadir of 1 CD8+ T cell/mm3 after antibody-mediated CD8+ T-cell depletion and also cleared virus completely by day 42, at which time the absolute CD8+ T-cell number was 146 cells/mm3. Twelve months after the depletion, CD8+ T cells had only recovered to 256 cells/mm3. We did not track the frequencies of total CD8+ T cells in the liver of these animals, but in other animals given the cM-T807 antibody with the same infusion protocol, there was a virtual absence of CD8 alpha and beta chain transcripts in the liver through day 56 after the first infusion (data not shown). Our continued ability to detect these epitope-specific T-cell clonotype transcripts in the liver suggests that they made up a significant fraction of the CD8+ T cells during peak viremia.
Despite the potential advantages of real-time PCR for tracking T-cell clonotypes, there are a few caveats to the interpretation of these data. Since we are measuring RNA transcripts, it is possible that activated T cells may generate more TCR transcripts per cell than resting memory cells, in which case this method would overestimate the actual T-cell frequency. However, our results tracked closely with the actual tetramer frequency in peripheral blood (1%, 0.58%, and 0.2% at week 4, 8, and 24, respectively) and the liver (4.4%, 3.4%, and 1.8% at week 4, 8, and 24, respectively) of chimpanzee CB0572 over the course of infection (18). It is also possible that primer efficiencies can differ when primer selection is based on the limited number of nucleotides present within the CDR3 region of the TCR beta chain. For the four primers described here, this did not appear to be a significant problem, and the efficiency of amplification was equal to or greater than that of our TCR beta chain constant region primers. Furthermore, the hierarchy of TCR sequence frequency obtained from direct sorting of tet+ T cells was identical to that measured by real-time PCR (Fig. 1A, 2, and 3).
Tracking of individual T-cell clonotypes has been used in studies of immune-based neurological disorders (15) and can be adapted to the study of T-cell clonotype expansion within peripheral blood or tissue from stored samples in any system where T cells are characterized at a time after the samples are obtained. Recent studies have characterized TCR clonotypes implicated in the pathogenesis of aplastic anemia (16) and have assessed the repertoire and frequency of melanoma-specific T-cell clonotypes in peripheral blood and tumors after therapeutic vaccination (4, 21). TCR transcript quantitation in these studies was performed via limiting dilution PCR; however, real-time PCR allows a rapid simultaneous quantitative assessment of clonotypes in peripheral blood and tissues and does not rely on prior knowledge of epitope specificity.
In this study, we confirmed that the rapid expansion of virus-specific T cells occurs as quickly as 7 days postinfection, the kinetics of clonotype expansion and contraction coincide with the clearance of viremia, and the frequencies of HCV-specific clonotypes are always higher in the liver than in PBMC. Even in the setting of robust CD8+ T-cell depletion, these clonotypes persist at low levels in the liver throughout challenge. The ability to perform such experiments on stored samples will greatly enhance our understanding of T-cell kinetics and homing during acute and memory immune responses.
Acknowledgments
We are grateful to Bryan Shepherd, Department of Biostatistics, Vanderbilt University, for assisting with the statistical analysis.
This study was supported by NIH grant U19 AI048231. D.M.-O. was supported by grants from the Kompetenznetz HIV/BMBF and Helmholtz Zentrum für Infektionsforschung.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Altman, J. D., P. A. Moss, P. J. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 27494-96. [DOI] [PubMed] [Google Scholar]
- 2.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244359-362. [DOI] [PubMed] [Google Scholar]
- 3.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10439-449. [DOI] [PubMed] [Google Scholar]
- 4.Derre, L., M. Bruyninx, P. Baumgaertner, E. Devevre, P. Corthesy, C. Touvrey, Y. D. Mahnke, H. Pircher, V. Voelter, P. Romero, D. E. Speiser, and N. Rufer. 2007. In vivo persistence of codominant human CD8+ T cell clonotypes is not limited by replicative senescence or functional alteration. J. Immunol. 1792368-2379. [DOI] [PubMed] [Google Scholar]
- 5.Diepolder, H. M., R. Zachoval, R. M. Hoffmann, E. A. Wierenga, T. Santantonio, M. C. Jung, D. Eichenlaub, and G. R. Pape. 1995. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 3461006-1007. [DOI] [PubMed] [Google Scholar]
- 6.Douek, D. C., M. R. Betts, J. M. Brenchley, B. J. Hill, D. R. Ambrozak, K. L. Ngai, N. J. Karandikar, J. P. Casazza, and R. A. Koup. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 1683099-3104. [DOI] [PubMed] [Google Scholar]
- 7.Gallard, A., G. Foucras, C. Coureau, and J. C. Guery. 2002. Tracking T cell clonotypes in complex T lymphocyte populations by real-time quantitative PCR using fluorogenic complementarity-determining region-3-specific probes. J. Immunol. Methods 270269-280. [DOI] [PubMed] [Google Scholar]
- 8.He, X.-S., B. Rehermann, F. X. Lopez-Labrador, J. Boisvert, R. Cheung, J. Mumm, H. Wedemeyer, M. Berenguer, T. L. Wright, M. M. Davis, and H. B. Greenberg. 1999. Quantitative analysis of hepatitis C virus-specific CD8+ T cells in peripheral blood and liver using peptide-MHC tetramers. Proc. Natl. Acad. Sci. USA 965692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islam, S. A., C. M. Hay, K. E. Hartman, S. He, A. K. Shea, A. K. Trocha, M. J. Dynan, N. Reshamwala, S. P. Buchbinder, N. O. Basgoz, and S. A. Kalams. 2001. Persistence of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte clones in a subject with rapid disease progression. J. Virol. 754907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakumu, S., K. Yoshioka, T. Wakita, T. Ishikawa, K. Murase, A. Kusakabe, and S. Kurokawa. 1990. Comparisons of peripheral blood and hepatic lymphocyte subpopulations and interferon production in chronic viral hepatitis. J. Clin. Lab. Immunol. 331-6. [PubMed] [Google Scholar]
- 11.Kalams, S. A., R. P. Johnson, A. K. Trocha, M. J. Dynan, H. S. Ngo, R. T. D'Aquila, J. T. Kurnick, and B. D. Walker. 1994. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J. Exp. Med. 1791261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim, A., V. Baron, L. Ferradini, M. Bonneville, P. Kourilsky, and C. Pannetier. 2002. Combination of MHC-peptide multimer-based T cell sorting with the Immunoscope permits sensitive ex vivo quantitation and follow-up of human CD8+ T cell immune responses. J. Immunol. Methods 261177-194. [DOI] [PubMed] [Google Scholar]
- 13.Meyer-Olson, D., N. H. Shoukry, K. W. Brady, H. Kim, D. P. Olson, K. Hartman, A. K. Shintani, C. M. Walker, and S. A. Kalams. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J. Exp. Med. 200307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minutello, M. A., P. Pileri, D. Unutmaz, S. Censini, G. Kuo, M. Houghton, M. R. Brunetto, F. Bonino, and S. Abrignani. 1993. Compartmentalization of T lymphocytes to the site of disease: intrahepatic CD4+ T cells specific for the protein NS4 of hepatitis C virus in patients with chronic hepatitis C. J. Exp. Med. 17817-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muraro, P. A., K. P. Wandinger, B. Bielekova, B. Gran, A. Marques, U. Utz, H. F. McFarland, S. Jacobson, and R. Martin. 2003. Molecular tracking of antigen-specific T cell clones in neurological immune-mediated disorders. Brain 12620-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risitano, A. M., J. P. Maciejewski, S. Green, M. Plasilova, W. Zeng, and N. S. Young. 2004. In-vivo dominant immune responses in aplastic anaemia: molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing. Lancet 364355-364. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283857-860. [DOI] [PubMed] [Google Scholar]
- 18.Shoukry, N. H., A. Grakoui, M. Houghton, D. Y. Chien, J. Ghrayeb, K. A. Reimann, and C. M. Walker. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 1971645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoukry, N. H., J. Sidney, A. Sette, and C. M. Walker. 2004. Conserved hierarchy of helper T cell responses in a chimpanzee during primary and secondary hepatitis C virus infections. J. Immunol. 172483-492. [DOI] [PubMed] [Google Scholar]
- 20.Spangenberg H. C., S. Viazov, N. Kersting, C. Neumann-Haefelin, D. McKinney, M. Roggendorf, F. von Weizsäcker, H. E. Blum, and R. Thimme. 2005. Intrahepatic CD8+ T-cell failure during chronic hepatitis C virus infection. Hepatology 42828-837. [DOI] [PubMed] [Google Scholar]
- 21.Speiser, D. E., P. Baumgaertner, C. Barbey, V. Rubio-Godoy, A. Moulin, P. Corthesy, E. Devevre, P. Y. Dietrich, D. Rimoldi, D. Lienard, J. C. Cerottini, P. Romero, and N. Rufer. 2006. A novel approach to characterize clonality and differentiation of human melanoma-specific T cell responses: spontaneous priming and efficient boosting by vaccination. J. Immunol. 1771338-1348. [DOI] [PubMed] [Google Scholar]
- 22.Thimme, R., D. Oldach, K.-M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 1941395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thimme, R., S. Wieland, C. Steiger, J. Ghrayeb, K. A. Reimann, R. H. Purcell, and F. V. Chisari. 2003. CD8+ T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 7768-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wedemeyer, H., X.-S. He, M. Nascimbeni, A. R. Davis, H. B. Greenberg, J. H. Hoofnagle, T. J. Liang, H. Alter, and B. Rehermann. 2002. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J. Immunol. 1693447-3458. [DOI] [PubMed] [Google Scholar]
- 25.Woollard, D. J., A. Grakoui, N. H. Shoukry, K. K. Murthy, K. J. Campbell, and C. M. Walker. 2003. Characterization of HCV-specific Patr class II restricted CD4+ T cell responses in an acutely infected chimpanzee. Hepatology 381297-1306. [DOI] [PubMed] [Google Scholar]