Abstract
The UL11 and UL16 tegument proteins of herpes simplex virus are conserved throughout the herpesvirus family. Previous studies have shown that these proteins interact, perhaps to link UL16-bound nucleocapsids to UL11, which resides on the cytoplasmic face of the trans-Golgi network, where maturation budding occurs. Little is known about the interaction except that it requires the leucine-isoleucine (LI) and acidic cluster motifs in UL11 and that no other viral proteins are involved. In particular, the important question of whether these two proteins bind to each other directly has not been addressed. Accordingly, UL11 and UL16 were expressed in bacteria, and the purified proteins were found to retain the ability to interact in a manner that was dependent upon the LI and acidic cluster. In an attempt to map the UL11-binding site contained in UL16, a large number of deletion mutants were constructed. The first 40 (nonconserved) amino acids were found to be dispensable, but all the other constructs failed to bind UL11 or had poor expression in transfected cells, suggesting that UL16 is very sensitive to alterations and probably lacks a multidomain structure. As an alternative strategy for identifying residues that are important for the interaction, the cysteines of UL16 were investigated, because many of these are highly conserved. Approximately half of the 20 cysteines in UL16 have been shown to be covalently modified by N-ethylmaleimide, and this treatment was found to block the interaction with UL11. Moreover, individual serine replacements of six of the most conserved cysteine residues were made, and four of these disrupted the interaction with UL11 without affecting protein stability. However, the UL11-UL16 interaction does not involve the formation of interspecies disulfide bonds, because binding occurred even when all the cysteines in UL11 were eliminated. Thus, UL16 directly interacts with UL11 and does so in a manner that requires free cysteines.
Herpesviruses have three morphological structures: the icosahedral capsid, which contains the viral DNA; the tegument, a proteinaceous compartment surrounding the capsid; and the lipid envelope embedded with virus-encoded glycoproteins. While the formation of the capsid has been studied in depth (22, 25, 26, 28), the assembly of tegument proteins and final envelopment remain poorly understood. It is thought that some tegument proteins are added to the capsid in the nucleus (7), whereas others are acquired after entering the cytoplasm or traveling to the site of final envelopment at the trans-Golgi network (TGN) (10, 20, 21). Several molecular interactions have been implicated in linking the virus capsid, tegument, and membrane during the envelopment process. Examples include VP22 (tegument)-gD (membrane), VP16 (tegument)-gH (membrane), and UL11 (membrane)-UL16 (tegument/capsid) (20, 21). The focus of this report is the interaction of the UL11 and UL16 tegument proteins of herpes simplex virus (HSV).
UL11, a 96-amino-acid protein, is conserved among all herpesviruses and thought to play a role in the virus budding process at the TGN during nucleocapsid envelopment (2, 12, 17, 29). UL11-null mutants are defective in virus replication and exhibit increased numbers of unenveloped capsids in the nucleus and cytoplasm (2, 6, 12, 17, 29). UL11 is a myristylated protein that accumulates on the cytoplasmic faces of nuclear and Golgi apparatus-derived membranes in infected cells and localizes primarily to the TGN when expressed alone (1, 13, 16, 27). In addition, UL11 has multiple sequences that are required for its proper membrane localization, including cysteines for palmitylation, a leucine-isoleucine (LI) motif, and an acidic cluster (AC) (3, 13). These sequences also regulate the amount of UL11 that is associated with detergent-resistant membranes (3).
UL16 is a 373-amino-acid protein that is also conserved among all herpesviruses (24, 32). It has been identified as a binding partner of UL11 by coimmunoprecipitation, glutathione S-transferase (GST) pull-down, and yeast two-hybrid assays, and the interaction requires the LI and AC motifs of UL11 (14, 31). ACs from other proteins are able to substitute for that of UL11 in trafficking assays, but they disrupt the interaction with UL16, suggesting a high specificity in the recognition (14, 15). More recently, we reported that UL16 dynamically interacts with the capsid (18). In particular, UL16 is stably associated with capsids in the cytoplasm but not those in extracellular virions. Free cysteines appear to play a critical role in this maturation event, because the addition of N-ethylmaleimide (NEM; a chemical that reacts with free cysteines) stabilizes UL16 on the capsids of extracellular virions (18).
We initially proposed that the interaction between UL11 (on the membrane) and UL16 (on the capsid) might provide a bridging function that contributes to the budding process at the TGN (14), but this hypothesis has never been proved. And, while it is clear that no other viral proteins are needed for binding, the crucial question of whether UL11 directly interacts with UL16 has never been addressed. It seemed possible that a cellular protein might be involved, because UL16 recognizes the same information (i.e., the LI and AC motifs) that is used by cellular machinery to traffic UL11 out of detergent-resistant membranes in the absence of other viral proteins (3, 13). That is, UL16 might be a viral homolog of a clathrin adaptor subunit, and if so, it might need to form a complex with particular cellular proteins to function (9, 11). The goals of the experiments described here were to address first whether the interaction is direct or indirect, and second, which part of UL16 is required for UL11 binding. The results prove that the interaction is indeed direct and suggest that the binding mechanism requires several (but not all) of the conserved cysteines within UL16.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Vero and human melanoma (A7) cell lines were maintained in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 5% fetal bovine serum (FBS), penicillin (65 μg/ml), and streptomycin (131 μg/ml). The KOS strain of HSV type 1 (HSV-1) (30) was used for these studies, along with recombinants that expressed UL11-green fluorescent protein (GFP) (3) or UL16-cyan fluorescent protein (CFP) (see below). For experiments with pseudorabies virus (PRV), the Becker strain was used. In all cases, infected cells were grown in Dulbecco's modified Eagle's medium supplemented with 2% FBS, 25 mM HEPES buffer, glutamine (0.3 μg/ml), penicillin, and streptomycin.
A His6-specific mouse monoclonal antibody was purchased from Novagen (product number 70796-3). The UL11-specific antibody was produced in rabbits and has been described previously (14). A GFP-specific antibody, produced by Cocalico Biologicals, Inc., was obtained from rabbits injected with purified His6-GFP (3). The anti-His6-GFP-specific serum (diluted 1:3,000) recognizes GFP, CFP, and also the His6 tag, which was fused to the N terminus of GFP and UL16 for purification from Escherichia coli (see Results).
Construction of KOS.UL16-CFP.
A derivative of HSV that expresses UL16-CFP was created by homologous recombination. Briefly, the gfp gene of plasmid pCMV.UL16-GFP (14) was replaced with the cfp gene by using flanking BamHI and BsrGI sites. Next, a fragment containing 300 bp downstream of UL16 was PCR amplified from the KOS genome with a forward primer containing a BsrGI site and a reverse primer containing a NotI site, and this fragment was inserted at the 3′ end of the UL16-cfp coding sequence. The resulting plasmid was linearized with EcoRI and transfected into A7 cells, along with purified KOS DNA. Plaques produced by the recombinant virus were identified by fluorescence microscopy, and these were picked for six rounds of purification. Confirmation that the desired virus was obtained was provided by PCR analyses using primers that flank the UL16-cfp coding sequence (yielding a larger product than untagged UL16) and the failure to express wild-type UL16 (as determined both by immunoblotting and radiolabeling-immunoprecipitation assays [data not shown]). Moreover, the recombinant was found to be identical to the wild type with regard to specific infectivity and plaque size, as well as subcellular localization and kinetics of UL16-CFP expression (data not shown).
Construction of BV.UL16-GFP.
To create a recombinant baculovirus that expresses UL16-GFP, the chimeric gene was first PCR amplified from pCMV.UL16-GFP (14) using a forward primer containing a BspHI site and a reverse primer containing a NotI site. This fragment was cloned into the pTriEx-1.1 vector (Novagen), which was used to produce the recombinant baculovirus by homologous recombination in insect cells via the BacVector-3000 transfection kit (Novagen). Spodoptera frugiperda cells (Sf21) were maintained in Grace's insect medium supplemented with Yeastolate (Mediatech), lactalbumin hydrolysate (Mediatech), penicillin, streptomycin, and 10% FBS in a humidified incubator at 28°C without CO2. Plaques produced by recombinant baculoviruses were identified by fluorescence microscopy, and these were picked for several rounds of purification. Virus stocks were amplified by infecting suspension cultures of Sf21 cells at a multiplicity of infection (MOI) of 0.2 PFU/cell. Virions were purified from culture medium at 5 to 7 days postinfection and concentrated, and titers were determined as described previously (5, 23).
Expression and purification of His6-tagged proteins.
To construct plasmids that encode His6-UL11 (wild-type and LI mutant versions) or His6-UL16, the alleles were cloned into pET-28 (Novagen). The resulting plasmids were transformed into E. coli BL21(DE3) cells (Novagen), and 0.1 mM isopropyl-beta-d-thiogalactopyranoside was added to the cultures to induce protein expression. Proteins were purified by using the His-Bind kit (Novagen). Briefly, approximately 1 g of bacteria was pelleted and resuspended in 10 ml of phosphate-buffered saline (PBS) containing protease inhibitors (P8340; Sigma). The bacteria were lysed by sonication and treatment with 1% Triton X-100 for 30 min at 4°C. After the removal of cell debris and insoluble material by centrifugation at 14,000 × g for 10 min, the lysates were incubated with nickel beads for 30 min. The beads were washed according to the His-Bind protocol, and proteins were eluted in 1 M imidazole and dialyzed overnight against 20 mM Tris-HCl (pH 7.9). Proteins were quantified in standard bicinchoninic acid assays or by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining. On average, the yields of His6-UL11 and His6-UL16 proteins from 1 g of bacteria were 0.2 mg and 0.1 mg, respectively.
GST-fusion constructs.
Plasmids encoding wild-type GST-UL11 and mutants lacking the LI, AC, or residues 51 to 96 were described previously (14). To construct a GST-UL11 mutant that lacks the three consecutive cysteines located near the N terminus, the UL11.CCC- allele was PCR amplified from pCMV.UL11-GFP.CCC- (13) and cloned into pGEX-4T-3 (GE Healthcare). The resulting plasmid was subsequently used to make GST-UL11.4C-, which does not contain any cysteines in UL11. This was accomplished by changing the codon for the remaining cysteine in UL11.CCC- (residue 83) to that for alanine (-GCA-) using QuikChange site-directed mutagenesis (Stratagene) according to the manufacturer's protocol. The wild-type UL16 gene was also cloned into pGEX-4T-3 to generate a plasmid that expresses GST-UL16. All GST-fusion proteins were purified from E. coli cells on glutathione beads using the standard methods described by the manufacturer (GE Healthcare).
UL16-GFP mutants.
For expression of UL16-GFP derivatives in mammalian cells, deletion mutations and alleles for single amino acid substitutions were created in pCMV.UL16-GFP (14) by either PCR technology or QuikChange site-directed mutagenesis. Codons for cysteines were individually replaced with ones for serine (-AGC-).
GST pull-down assay.
GST pull-down assays were done as described previously (14). To analyze the interaction of UL11 with UL16 mutants, pCMV.UL16-GFP derivatives were transfected into A7 cells by means of the calcium phosphate precipitation method. At 20 h posttransfection, the cells were harvested in NP-40 lysis buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0]), and glutathione bead-bound GST-UL11 was added. Proteins bound to the beads were separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by enhanced chemiluminescence-based immunoblotting (GE Healthcare). The same protocol was used to analyze the interaction of GST-UL11 with UL16 produced in Vero cells infected with HSV (wild-type KOS or KOS.UL16-CFP) or insect cells infected with BV.UL16-GFP (all produced in 60-mm plates at an MOI of 1). To detect proteins that were pulled down with GST-UL16 from HSV- or PRV-infected cells, the cells were first labeled with [35S]methionine, as described previously (14). To look for host proteins that might bind to GST-UL11 in complex with UL16-GFP produced in insect cells, SDS-PAGE gels were subjected to zinc staining following the manufacturer's instructions (Bio-Rad).
NEM treatment.
To covalently modify free cysteines in UL16, NEM was used at a final concentration of 10 mM (4). For in vitro binding assays, His6-UL16 proteins made in bacteria were purified on nickel beads, as described above. Before they were eluted from beads, His6-UL16 proteins were treated with NEM for 30 min at room temperature. The beads were washed thoroughly, and the proteins were eluted and dialyzed. For baculovirus- or HSV-infected cells, NEM was added to cells that had been resuspended in PBS, and after incubations of 30 min at room temperature, the cells were washed three times with PBS before NP-40 lysis. To examine the effect of NEM on the UL11-UL16 complex, GST pull-down assays were done as before. Then, glutathione bead-bound GST constructs and the associated proteins were treated with NEM for 30 min at room temperature before being analyzed on SDS-PAGE gels.
RESULTS
Search for host proteins that bind UL11 in complex with UL16.
It is quite clear that the interaction between UL11 and UL16 does not require any other viral proteins (14, 31); however, it seemed possible that host proteins might be involved in the interaction, because UL16 recognizes the membrane-trafficking signals of UL11 (i.e., UL16 might be a viral homolog of a clathrin adaptor subunit and function in complex with particular cellular proteins). To look for additional binding partners, a recombinant baculovirus was constructed that expressed UL16-GFP at high levels. To determine whether this source of UL16-GFP was still capable of binding specifically to GST-UL11, Sf21 cells were infected with BV.UL16-GFP, and NP-40 lysates were prepared 20 h postinfection. Equivalent amounts of glutathione bead-bound GST or GST-UL11 fusion proteins (Fig. 1A and B) were added to lysate samples, and bound proteins were analyzed by SDS-PAGE and immunoblotting for GFP. Like its mammalian-expressed counterpart (14), baculovirus-expressed UL16-GFP was pulled down by wild-type GST-UL11 and mutant Δ51-96 but not by the AC or LI mutants (Fig. 1C). These results show that no mammalian-specific host factor is necessary for the UL11-UL16 interaction. To look for host factors that might be in the complex, bound proteins were resolved in SDS-PAGE gels and subjected to zinc staining, but no distinct difference was observed between mock- and baculovirus-infected cells (data not shown). This negative result is consistent with the hypothesis that UL11 and UL16 may directly interact.
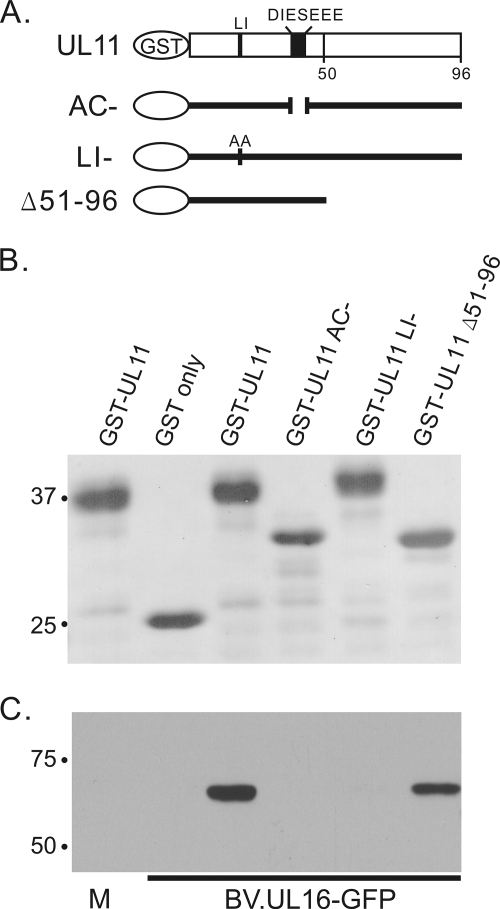
FIG. 1.
Baculovirus-expressed UL16-GFP interacts with UL11 via the AC and LI motifs. (A) Diagrams of GST-UL11 constructs. Approximately equal amounts of these constructs as estimated from a Ponceau S-stained gel (B) were mixed with lysates from uninfected (M, mock) or BV.UL16-GFP-infected Sf21 insect cells. The UL11 AC mutant has the approximately same mobility as the Δ51-96 mutant, which has been observed previously (14, 15). (C) Proteins bound to GST constructs were loaded onto SDS-PAGE gels and analyzed by immunoblotting for GFP. When analyzed using densitometry, approximately 40% of the amount of UL16-GFP protein in the lysates (input) was pulled down by GST-UL11.
Binding of purified UL11 and UL16.
To further address whether the UL11-UL16 interaction is direct or indirect, these two proteins were made and purified from bacteria to completely eliminate eukaryotic host factors. In the first attempt, His6-tagged UL11 was expressed in E. coli, purified on nickel beads, eluted, and dialyzed. A mutant that lacks the LI motif was also produced for use as a negative control for binding specificity. Various amounts of these His6-UL11 proteins were mixed with glutathione bead-bound GST-UL16 in NP-40 lysis buffer for 2 h at room temperature, conditions identical to those used for the baculovirus experiments. Although no His6-UL11 was pulled down by GST alone, the GST-UL16 construct pulled down only a small amount (less than 1%) of the input His6-UL11, as detected with a His6-specific monoclonal antibody, and very little difference was seen using the LI mutant (Fig. 2A). Because UL16 has many cysteines (18), it seemed likely that GST-UL16 from E. coli might be misfolded to some degree. If so, the misfolding must be specific for the UL11 interaction because GST-UL16 was capable of pulling down virus-specific proteins from HSV- and PRV-infected cell lysates (Fig. 2B), while no prominent proteins were pulled down by GST-UL16 from uninfected cell lysates (data not shown).
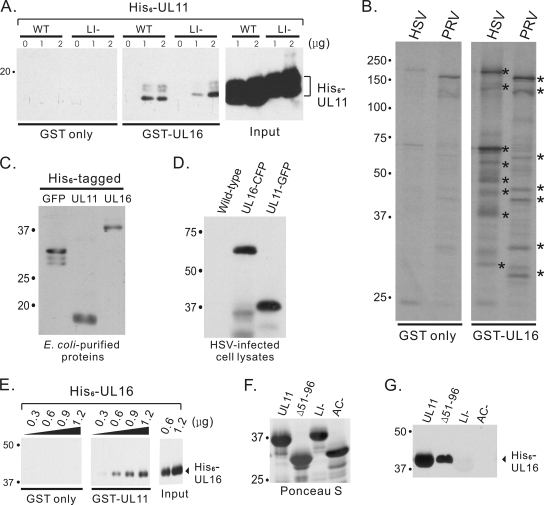
FIG. 2.
Direct interaction assays with UL11 and UL16. (A) The indicated amounts of His6-UL11 proteins were incubated with purified GST only or GST-UL16. Bound proteins were separated by SDS-PAGE and detected by immunoblotting using a monoclonal antibody specific for the His6 tag. WT, wild type. (B) To examine the abilities of purified GST or GST-UL16 to pull down virus-specific proteins, they were incubated with radiolabeled HSV- or PRV-infected cell lysates. Proteins bound were separated by SDS-PAGE, and radiolabeled proteins were detected by autoradiography. Examples of virus-specific proteins pulled down by GST-UL16 are indicated with asterisks. (C and D) Immunoblot analyses were used to show the specificity of rabbit anti-His6-GFP serum for various bacteria-expressed His6-tagged proteins (C) and viral proteins made by wild-type and recombinant HSV strains (D). (E) The indicated amounts of His6-UL16 proteins were incubated with purified GST only or GST-UL11. Bound proteins were separated by SDS-PAGE and detected by immunoblotting. (F and G) Aliquots (1.8 μg) of His6-UL16 protein were incubated with approximately equal amounts of GST-UL11 or mutants (Δ51-96 or LI or AC mutants) as estimated from a Ponceau S-stained gel (F), and bound proteins were analyzed by immunoblotting (G).
In a second attempt to look for direct interactions, the GST and His6 tags were switched. The hope was that His6-UL16 might be better able to fold if it was not linked to GST, which is well known to form a dimer. The switch of tags produced a minor problem in that the His6-specific monoclonal antibody (Novagen) (as used in Fig. 2A) unexpectedly cross-reacted with purified GST-UL11 (data not shown). Fortunately, this problem could be eliminated by using a polyclonal rabbit antibody against His6-GFP. This antibody reacts with His6-GFP, His6-UL11, and His6-UL16 (Fig. 2C), but it does not react with UL11 or UL16 (Fig. 2D, lane 1) unless they are tagged with CFP or GFP (Fig. 2D, lanes 2 and 3, respectively). Using this antiserum in the interaction assay, purified GST-UL11 was readily able to pull down purified His6-UL16 in a dose-dependent manner (Fig. 2E). Importantly, the Δ51-96 mutant could also bind to His6-UL16, but the LI and AC mutants could not (Fig. 2G), even though approximately equal amounts of each of the GST-fusion proteins were used (Fig. 2F). These data prove that UL11 directly interacts with UL16 and that both the acidic cluster and LI motifs are essential for the interaction.
Deletion analysis of UL16.
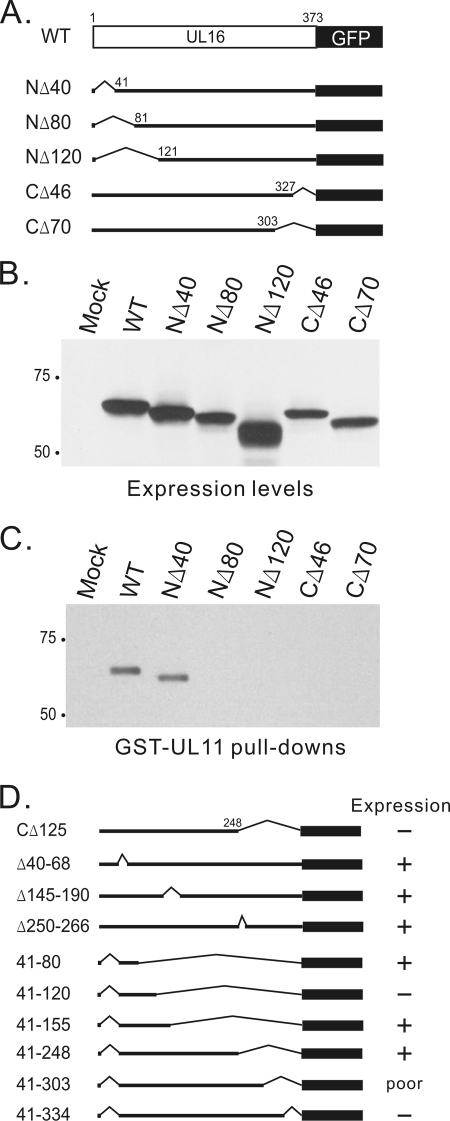
In an attempt to map the UL11-binding site contained in UL16, several N-terminal and C-terminal deletion mutants were constructed in the context of the eukaryotic expression vector, pCMV.UL16-GFP (Fig. 3A), and these were transfected into A7 cells. All mutants were expressed well (Fig. 3B); however, in the GST-UL11 pull-down experiment, only one mutant, NΔ40, was found to retain the ability to bind to UL11 (Fig. 3C). Because the N-terminal 40 amino acids reside in the least-conserved region among UL16 homologs (not shown), it is not surprising that they were found dispensable for UL11 binding. Numerous other truncation and internal deletion mutants were constructed (Fig. 3D), but many of these had no or poor expression (summarized in Fig. 3D), and all those that did express were unable to bind to UL11 (data not shown). These results suggest that UL16 is very sensitive to mutations, at least with regard to the requirements for UL11 binding.
FIG. 3.
The first 40 amino acids of UL16 are not required for UL11 binding. (A) Diagrams of UL16 wild-type (WT) and N- and C-terminal truncation mutants, which were all constructed as GFP-fusion proteins. (B) Expression levels of indicated constructs in transiently transfected A7 cells. At 20 h posttransfection, cells were harvested in sample buffer, loaded onto SDS-PAGE gels, and analyzed by immunoblotting for GFP. (C) Transfected cells were lysed and incubated with purified GST-UL11 proteins. Bound proteins were analyzed by SDS-PAGE and immunoblotting for GFP. (D) Summary of expression levels of other UL16 mutants.
Sensitivity of the interaction to NEM treatment.
As an alternative strategy for identifying residues that are important for the interaction, the cysteines of UL16 were investigated because many of these are highly conserved (32). NEM is a very small (125-Da) membrane-permeable molecule that has long been used to chemically modify free cysteines (8). Recent studies have shown that about half of the 20 cysteines in UL16 can be modified by NEM, and this treatment stabilizes the interaction of the protein with the extracellular capsid (18). To examine the possibility that free cysteines might also be important for UL11 binding, UL16 was treated with NEM and then tested for binding.
Vero cells were infected with a recombinant HSV that expresses UL16-CFP. At 20 h postinfection, infected cells were treated with NEM for 30 min at room temperature, washed, and then lysed in NP-40 buffer to test for binding in GST-UL11 pull-down assays (Fig. 4A and B). A blockage of the UL11-UL16 interaction was observed (Fig. 4B, lanes 1 and 3). Identical results were obtained using wild-type HSV (data not shown). The same blockage was also observed when baculovirus-expressed UL16-GFP was treated with NEM in the GST-UL11 pull-down assay (data not shown). In these assays, the cells were washed thoroughly to remove excess NEM before being lysed, but it was important to consider the possibility that residual NEM might disrupt the interaction. To address this, UL11 and UL16 proteins were allowed to bind prior to the addition of NEM, and the complex was found to be stable (Fig. 4B, lane 2). Hence, it is clear that modification of UL16 with NEM blocks UL11 binding.
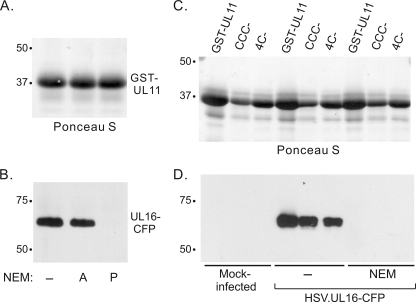
FIG. 4.
UL11 cannot interact with NEM-modified UL16 from infected cell lysates. (A and B) Vero cells were infected with HSV.UL16-CFP at an MOI of 1. At 20 h postinfection, cells were scraped into PBS. One set of cells were treated with NEM for 30 min before detergent lysis. Cell lysates were incubated with approximately equal amounts of glutathione bead-bound GST-UL11 as estimated from a Ponceau S-stained gel (A). Bound proteins were analyzed by SDS-PAGE and immunoblotting (B). For lanes 2 and 3, the addition of NEM was either after (A) or prior (P) to the 2-h incubation. (C and D) GST-UL11 fusion proteins as detected in a Ponceau S-stained gel (C) were mixed with either untreated or NEM-treated HSV.UL16-CFP- infected cell lysates, and bound proteins were analyzed by SDS-PAGE and immunoblotting.
The NEM treatment results raised the possibility that free cysteines in UL11 and UL16 might form irrelevant disulfide bonds when mixed together. To examine this, cysteine mutants of UL11 were constructed. Of the four cysteines in UL11, three are located near the N terminus (residues 11 to 13), and the fourth is located in the dispensable second half of the molecule (residue 83). UL11 mutants lacking three (CCC-) or all four (4C-) of the cysteines were expressed as GST-fusion proteins and purified from bacteria. Both mutants retained the ability to interact with UL16-CFP produced in infected cells (Fig. 4C and D). Identical results were obtained using untagged UL16 expressed by wild-type HSV (data not shown). Moreover, pretreatment of the infected cells with NEM blocked the interaction (Fig. 4C and D, lanes 7 to 9).
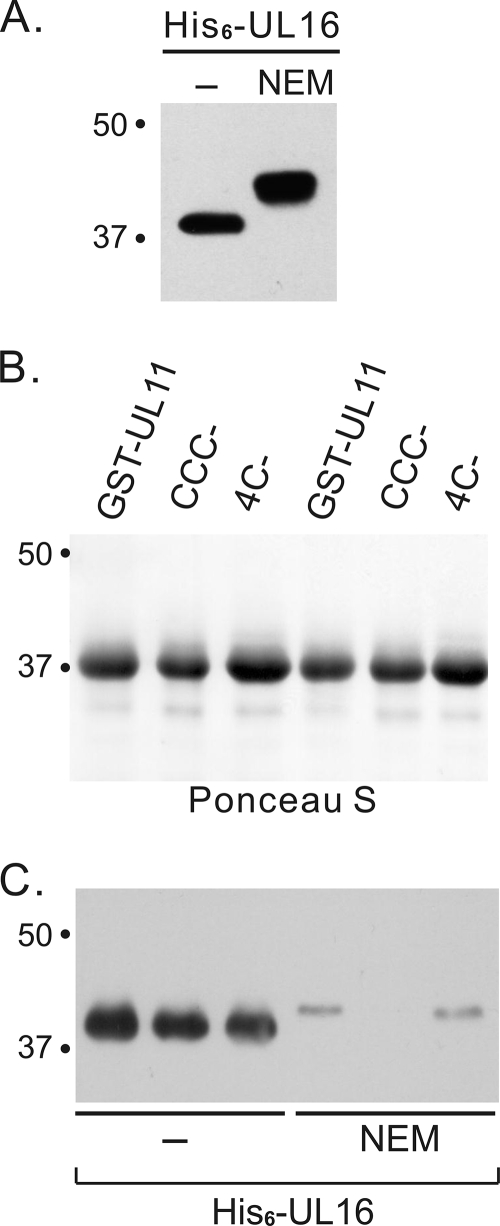
The UL11 mutants were also examined for their ability to interact with His6-UL16 purified from bacteria as described for Fig. 2, and as expected, direct binding was observed (Fig. 5B and C). When the His6-UL16 protein was treated with NEM prior to its elution from the nickel beads, a large increase in its mass was observed (Fig. 5A), indicating the presence of multiple free cysteines. As predicted, the modified protein lost its ability to bind UL11 (Fig. 5B and C). Collectively, these results demonstrate that interspecies disulfide bonds are not required for a stable interaction.
FIG. 5.
NEM modification of UL16 inhibited the in vitro UL11-UL16 interaction. (A) His6-UL16 proteins purified from bacteria were treated with or without NEM and analyzed by immunoblotting. (B and C) Aliquots (1.2 μg) of untreated or NEM-treated His6-UL16 proteins were incubated with approximately equal amounts of GST-UL11 fusion proteins as estimated from a Ponceau S-stained gel (B), and bound proteins were analyzed by immunoblotting (C).
Cysteine substitution mutants of UL16.
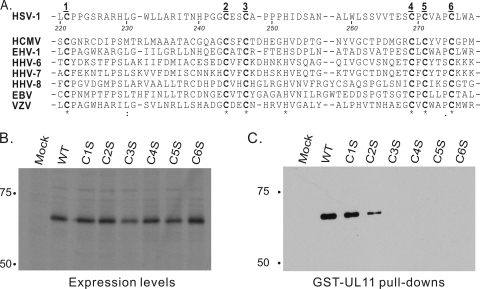
It would be a daunting task to analyze all 20 of the cysteines in UL16; however, a small subset of these are highly conserved (Fig. 6A). To determine whether any of these are important for the interaction with UL11, six were individually replaced with serine in the context of UL16-GFP. Plasmids encoding the resulting mutants, designated C1S to C6S, were transfected into A7 cells, and each was found to be expressed as well as wild-type UL16-GFP (Fig. 6B). Mutants C1S and C2S retained the ability to interact with GST-UL11, but the other four were completely inactive for binding (Fig. 6C). These results further support the hypothesis that free cysteines (perhaps conserved ones) in UL16 play a critical role in the interaction with UL11 (see below).
FIG. 6.
Conserved cysteines within UL16 are important for UL11 binding. (A) Alignment of regions in UL16 homologs containing six highly conserved cysteine residues. The residue numbers in this region of HSV-1 are indicated. HCMV, human cytomegalovirus; EHV, equine herpesvirus; HHV, human herpesvirus; EBV, Epstein-Barr virus; VZV, varicella-zoster virus. (B) Expression levels of HSV-1 UL16-GFP wild type (WT) and six cysteine-to-serine substitution mutants (C1S to C6S). Codons for the six cysteines indicated in panel A were individually replaced with codons for serine and constructed into the GFP expression vector. The alleles were transfected into A7 cells, and protein expression was examined by immunoblotting for GFP. (C) GST-UL11 pull-down assays. Transfected cells were lysed with detergent and incubated with glutathione bead-bound GST-UL11. Bound proteins were analyzed by SDS-PAGE and immunoblotting for GFP.
DISCUSSION
Although the UL11 and UL16 proteins of HSV are conserved among all the herpesviruses, very little is known about their structure and function. Based on the available information, we previously hypothesized that an interaction between these proteins (either directly or indirectly) might provide a bridging function that contributes to the budding process at the TGN (14). The goal of this study was to more precisely define the molecular interaction that was previously predicted to occur (14, 31). When purified from bacteria, the two proteins were found to be capable of interacting directly and in a manner that is dependent upon the acidic cluster and LI motifs of UL11, which were previously shown to have roles in the interaction with UL16 (14, 15). However, the molecular tags used for protein purification were found to be of crucial importance to the success of the experiments, a finding that provides important guidance for future studies. In this regard, it is also important to have learned that UL16 is highly sensitive to deletions throughout its length, and hence, it probably does not have a separate domain that is devoted to binding UL11. This means that elucidation of the residues in UL16 that actually contact UL11 will be difficult. Nevertheless, the mutational analyses described here provide evidence that free cysteines somehow play a role (see below).
Implications for virion maturation.
It was previously shown that the interaction of UL16 with capsids changes during egress (18). In particular, UL16 is stably associated with cytoplasmic capsids isolated from NP-40-treated cell lysates, but this interaction is very different in extracellular virions, where disruption with NP-40 under identical conditions releases UL16 from the capsid. More recently, we showed that the natural trigger for the release of UL16 from the capsid occurred upon the attachment of virus to the host cell (19). Free cysteines appear to play a critical role in this release mechanism because the addition of NEM stabilizes UL16 on the capsids of extracellular virions (i.e., it is no longer released from the capsid by NP-40 treatment). Stabilization is also observed when extracellular virions are exposed to low pH (5.0 to 5.5), conditions that protonate free cysteines, making them less reactive (18). However, there are likely to be many proteins in the virion that have free cysteines, and hence, the change in the UL16-capsid association properties seen with NEM treatments could be complex.
In the experiments described here, NEM modification of purified UL16 was found to directly block the interaction with purified UL11, a finding that was further supported by the inability of some (but not all) UL16 cysteine substitution mutants to bind UL11. Based on these observations, we can now hypothesize that the interaction of UL16 with the capsid may be destabilized as a result of binding to UL11 during budding. In other words, when the capsid traverses the cytoplasm and arrives at the TGN, UL16 may engage UL11 and undergo a conformational change, making it sensitive to release by NP-40. Although these ideas are highly speculative, our findings represent the initial glimpses of a molecular mechanism that is present in the tegument.
Possible roles for cysteines in UL16.
There are 20 cysteines in UL16, and sequence alignments revealed a cluster in the second half of the protein that is highly conserved (Fig. 6A) (32). The substitution mutants described here reveal that some, but not all, of these residues are important for UL11 binding. While it is possible that these cysteines directly contact UL11, there are several other (perhaps more likely) possibilities for what they do. Some cysteines may be needed merely for the proper folding of the protein, much like any other amino acid in UL16. Other cysteines might serve more complex structural roles, either as participants in disulfide bonds or as components of metal-binding domains (e.g., zinc fingers). It is also possible that UL16 is an enzyme that utilizes one or more free cysteines in its active site. For example, protein disulfide isomerases, which catalyze the formation and breakage of disulfide bonds, generally have a C-X-X-C motif in their active site, and this motif is also found among the conserved residues of UL16 (Fig. 6A). Further complicating the study of this protein is the possibility that all of these roles for cysteines will be found in UL16. Moreover, the role of any particular cysteine may change during virion budding and egress (e.g., particular disulfide bonds may be formed, broken, and rearranged along the way). Hence, much caution is needed when interpreting the results of any cysteine mutants, especially within the far more complicated context of recombinant viruses.
Acknowledgments
We extend special thanks to our colleagues Nicholas Baird, Jacob Marsh, and Carol Wilson for helpful discussions and encouragement. Additional thanks go to Harriot Isom for providing Sf21 cells and to Richard Heipertz, Jr., and Susan Kocher for their expertise in the construction and growth of the recombinant baculovirus.
This work was supported by NIH grants to J.W.W. (AI071286) and in part by a training grant from the National Cancer Institute (CA60395) for support of D.G.M.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 665168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baird, N. L., P. C. Yeh, R. J. Courtney, and J. W. Wills. 2008. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology 374315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolen, J. B., and M. A. Israel. 1983. Inhibition of polyoma virus middle T antigen-associated tyrosyl kinase activity by N-ethylmaleimide. J. Biol. Chem. 25815135-15140. [PubMed] [Google Scholar]
- 5.Boyce, F. M., and N. L. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 932348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britt, W. J., M. Jarvis, J. Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucks, M. A., K. J. O'Regan, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creighton, T. E. 1993. Proteins structures and molecular properties. W. H. Freeman Co., New York, NY.
- 9.Crump, C. M., Y. Xiang, L. Thomas, F. Gu, C. Austin, S. A. Tooze, and G. Thomas. 2001. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 202191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchhausen, T. 2002. Clathrin adaptors really adapt. Cell 109413-416. [DOI] [PubMed] [Google Scholar]
- 12.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 775339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 7512209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 7711417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2006. Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 8010534-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 703147-3157. [DOI] [PubMed] [Google Scholar]
- 17.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73539-547. [DOI] [PubMed] [Google Scholar]
- 18.Meckes, D. G., Jr., and J. W. Wills. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 8113028-13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meckes, D. G., Jr., and J. W. Wills. 2008. Structural rearrangement within an enveloped virus upon binding to the host cell. J. Virol. 8210429-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus. Res. 106167-180. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263432-446. [DOI] [PubMed] [Google Scholar]
- 23.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. Oxford University Press, New York, NY.
- 24.Oshima, S., T. Daikoku, S. Shibata, H. Yamada, F. Goshima, and Y. Nishiyama. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch. Virol. 143863-880. [DOI] [PubMed] [Google Scholar]
- 25.Remillard-Labrosse, G., G. Guay, and R. Lippe. 2006. Reconstitution of herpes simplex virus type 1 nuclear capsid egress in vitro. J. Virol. 809741-9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 735714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schimmer, C., and A. Neubauer. 2003. The equine herpesvirus 1 UL11 gene product localizes to the trans-Golgi network and is involved in cell-to-cell spread. Virology 30823-36. [DOI] [PubMed] [Google Scholar]
- 28.Sherman, G., and S. L. Bachenheimer. 1988. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology 163471-480. [DOI] [PubMed] [Google Scholar]
- 29.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 7710594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, K. O. 1964. Relationship between the envelope and infectivity of herpes simplex virus. Proc. Soc. Exp. Biol. Med. 115814-816. [DOI] [PubMed] [Google Scholar]
- 31.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 799566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wing, B. A., G. C. Lee, and E. S. Huang. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 703339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]